Introduction
The association between calorie restriction (CR) and
longevity was initially identified in a study by McCay et al
(1), who observed that laboratory
rodents maintained on a CR diet exhibited an increase in lifespan.
CR has since become an area of extensive investigation. Numerous
studies have shown that CR is capable of significantly increasing
the lifespan in a range of organisms, from yeast to mammals
(2,3). Furthermore, marked physiological
changes have been observed in CR rodents, including decreases in
blood glucose, insulin levels and body weight (BW), and increases
in insulin sensitivity, which have been suggested to be beneficial
with regard to preventing the onset of cardiovascular diseases
(CVDs) (3,4). Futhermore, it has been suggested that
CR may directly protect cardiomyocytes by reducing the levels of
reactive oxygen species and thereby attenuating the development of
atherosclerosis (5).
An enhanced understanding of CR may provide novel
approaches for the treatment of CVD. A previous study suggested
that some of the changes induced by CR require silent information
regulator 2 (Sir2), and that when Sir2 is deleted, CR is not
capable of lifespan extension (6).
Furthermore, it has been demonstrated that in yeast, CR may
activate Sir2 and consequently increase longevity by increasing
respiration, since the deacetylase activity of Sir2 is regulated by
the cellular oxidized nicotinamide adenine dinucleotide/reduced
nicotinamide adenine dinucleotide (NAD+/NADH) ratio
(6). It has been suggested that CR
is capable of reducing the levels of NADH or the
nicotinamide-degrading enzyme pyrazinamidase/nicotinamidase 1
(PNC1), which consequently activates the Sir2 deacetylase and
increases the lifespan (7,8).
The Sir2 gene encodes a highly conserved
NAD-dependent histone deacetylase (9). In mammals, there are seven homologues
of Sir2 known as sirtuins (SIRT1–7). SIRTs have diverse cellular
localizations, targeting multiple substrates and affecting a wide
range of cellular functions, including the regulation of oxidative
stress, DNA damage and metabolism (10). An increase in SIRT1 expression has
been observed in numerous tissues of CR animals (11). However, the roles of SIRT2–7 during
CR are yet to be elucidated. The present study investigated the
cellular location of SIRTs in H9c2 cells and rat cardiac tissues,
and is the first investigation, to the best of our knowledge, to
demonstrate that short-term CR activates not only SIRT1 but also
SIRT2–4 and -7 in cardiomyocytes in vivo and in
vitro.
Materials and methods
Materials
A total of 20 three-month-old Sprague Dawley (SD)
rats of both genders, weighing an average of 229.1 g each, were
purchased from the Experimental Animal Centre of Shantou University
Medical College (Shantou, China). Embryonic rat heart-derived H9c2
cells were obtained from the American Type Culture Collection
(Rockville, MD, USA). Primary antibodies against SIRT1–5 and -7 and
β-actin were purchased from Santa Cruz Biotechnology, Inc. (SIRT1,
sc-15404; SIRT2, sc-20966; SIRT3, sc-49743; SIRT4, sc-66296; SIRT5,
sc-66272; SIRT7, sc-66281, β-actin, sc-47778; Santa Cruz, CA, USA).
The anti-SIRT6 antibody was purchased from Abcam (SIRT6, ab62739;
Cambridge, MA, USA). An enhanced chemiluminescence (ECL) western
blot detection system was obtained from GE Healthcare (Amersham,
UK).
Animals and groups
SD rats were randomly divided into either a control
group (n=10) or a CR group (n=10), with five males and five females
in each group. Rats in the control group were fed ad libitum
(AL), while rats in the CR group were fed at 60% of AL. Rats were
housed individually. Sufficient vitamins and minerals were present
in each of the diets, and the rats had unlimited access to water
for three weeks. The components of the modified AL and CR diets are
further described in Table I. Rats
were monitored and weighed daily. All animal procedures were
approved by the Animal Care and Use Committee of Shantou University
Medical College.
 | Table INutrient composition of AL and CR
diets. |
Table I
Nutrient composition of AL and CR
diets.
| Nutrient
composition | AL diet (per 1 g
diet) | CR diet (per 0.62 g
diet) |
|---|
| Protein (g) | 0.19 | 0.19 |
| Carbohydrate
(g) | 0.67 | 0.29 |
| Fat (g) | 0.04 | 0.04 |
| Vitamin mix
(mg) | 9.48 | 9.48 |
| Mineral mix
(mg) | 9.48 | 9.48 |
| Cholesterol
(mg) | 0.20 | 0.20 |
| Cellulose (mg) | 47.38 | 47.38 |
| Dicalcium phosphate
(mg) | 12.32 | 12.32 |
| Calcium carbonate
(mg) | 5.21 | 5.21 |
| Potassium citrate
(mg) | 15.64 | 15.64 |
| Choline bitartrate
(mg) | 1.90 | 1.90 |
| Total calories | 3.80 | 2.28 |
Tissue processing
The rats (n=20) were sacrificed after three weeks by
cervical dislocation following anesthetisia. Hearts were harvested
and flushed with 0.9% normal saline solution, prior to being
sectioned into three parts. The central sections of each rat heart,
which included dual atriums and dual ventricles, were used for
immunohistochemical analysis. The two remaining sections were used
for western blot analysis and were immediately stored at −80°C.
Immunohistochemical staining
The central heart sections were immersed in 4%
parafomaldehyde for 24 h. Serial transverse heart sections were
deparaffinated in xylene and rehydrated in a graded ethanol series.
Sections were pre-incubated with 0.3% hydrogen peroxide in
phosphate-buffered saline (PBS) for 5 min, in order to inactivate
any endogenous peroxidase activity, prior to being blocked with 2%
bovine serum albumin (BSA) for 30 min. Specimens were then
incubated with primary antibodies against SIRT1–7 at a dilution of
1:25 (v/v), overnight at 4°C. Sections were subsequently incubated
with biotin-conjugated anti-rabbit or anti-goat immunoglobulin G
(IgG) (Kirkegaard & Perry Laboratories, Gaithersburg, MD, USA)
at a dilution of 1:100 (v/v) in PBS at room temperature for 1 h,
prior to the application of preformed avidin-biotin complex
conjugated to peroxidase (Vector laboratories, Inc., Burlingame,
CA. USA) for 30 min. The bound complexes were visualized using the
administration of a 0.05% solution of 3-3′-diaminobenzidine (DAB)
and counterstained with Harris’ hematoxylin. PBS was used instead
of the primary antibody for negative control reactions.
Western blot analysis
Equal quantities of protein were extracted from the
rat heart tissues according to standard protocols, prior to being
separated using 12% SDS-PAGE and then transferred to polyvinylidene
fluoride membranes according to standard procedures. Following
three washes with PBS, the membranes were soaked in 5% nonfat dry
milk for 2 h at room temperature and incubated with primary
antibodies against β-actin or SIRT1–7 overnight at 4°C. Membranes
were then incubated with horseradish peroxidase-conjugated
secondary antibodies for 1 h at room temperature. Immune complexes
were subsequently visualized using ECL and the band intensities
were measured and quantified using Quantity One®
software (Bio-Rad Laboratories Inc., Berkley, CA, USA).
Cell culture
H9c2 cells were cultured in high-glucose Dulbecco’s
modified Eagle medium (DMEM) supplemented with 10% fetal bovine
serum (FBS), 100 U/ml penicillin and 100 mg/ml streptomycin, in 5%
CO2 at 37°C for 48 h. Cells in the control group (Con)
were retained in DMEM containing 4.5 g/l glucose, while the cells
in the CR group were subsequently cultured in DMEM containing 1 g/l
glucose. Both groups were incubated in 5% CO2 at 37°C
for 24 h.
Immunocytochemical staining
Semiquantitative analysis of H9c2 cells was
performed following cell seeding onto coverslips. Following
washing, H9c2 cells were fixed using 4% paraformaldehyde in PBS for
15 min and incubated with 5% Triton X-100 in PBS at room
temperature for 20 min. Cells were then blocked using 2% BSA for 30
min, prior to incubation with primary antibodies against SIRT1–7 at
a dilution of 1:50 (v/v) overnight at 4°C. Following multiple
washes with PBS, slides were incubated with biotin-conjugated
anti-rabbit or anti-goat immunoglobulin G at dilutions of 1:100
(v/v) for 1 h at room temperature. The slides were then incubated
with the preformed avidin-biotin complex conjugated to peroxidase
for 30 min. The bound complexes were visualized using the
application of a 0.05% solution of DAB, and counterstained with
Harris’ hematoxylin. PBS was used instead of the primary antibody
for negative control reactions.
Quantitative polymerase chain reaction
(qPCR)
Total RNA was extracted from the H9c2 cells using
the RNAsimple Total RNA Kit (Tiangen Biotech Co., Beijing, China)
according to the manufacturer’s instructions. First-strand cDNAs
were synthesized by reverse transcription using oligos (dT) from
RNA samples. The primers (Genecore Biotechnologies Co., Shanghai,
China) used for PCR amplification are described in Table II. The PCR conditions used for all
primers were as follows: DNA denaturation at 94°C for 30 sec, 30
cycles of 94°C for 30 sec, 56°C for 30 sec and 72°C for 60 sec,
followed by a final extension step at 72°C for 6 min. PCR products
were electrophoresed in 2% agarose gels and visualized using
ethidium bromide. Relative mRNA expression was quantified
densitometrically using the Gel Image System version 3.74 (Tianon,
Shanghai, China).
 | Table IIPrimer sequences used in quantitative
polymerase chain reaction. |
Table II
Primer sequences used in quantitative
polymerase chain reaction.
| Gene | Forward sequences
(5′-3′) | Reverse sequences
(5′-3′) |
|---|
| SIRT1 |
CCAGATCCTCAAGCCATGT |
TTGGATTCCTGCAACCTG |
| SIRT2 |
TACCCAGAGGCCATCTTTGA |
TGATGTGTGAAGGTGCCGT |
| SIRT3 |
TACTTCCTTCGGCTGCTTCA | AAGGCG
AAATCAGCCACA |
| SIRT4 |
ACTGGGAGAAACTTGGGAAG |
CTGGTGCACAAAGTCAACCT |
| SIRT5 |
AGCAAGATCTGCCTCACCAT |
GGATTTCCAGCAGGTTCTTG |
| SIRT6 |
TTGTCAACCTGCAACCCA |
GCTTGGGCTTATAGGAACCA |
| SIRT7 |
TCTCTGAGCTCCATGGGAAT |
CATGAGGAGCCGCATTACAT |
| 18S | rRNA
ATTCCGATAACGAACGAGAC |
GGCATCACAGACCTGTTATTG |
Statistical analysis
All data are presented as the mean ± standard
deviation of at least three independent experiments. Statistical
analyzes were performed using one-way analysis of variance and
t-tests with a correction for multiple comparisons where
appropriate. A P-value of <0.05 was considered to indicate a
statistically significant difference.
Results
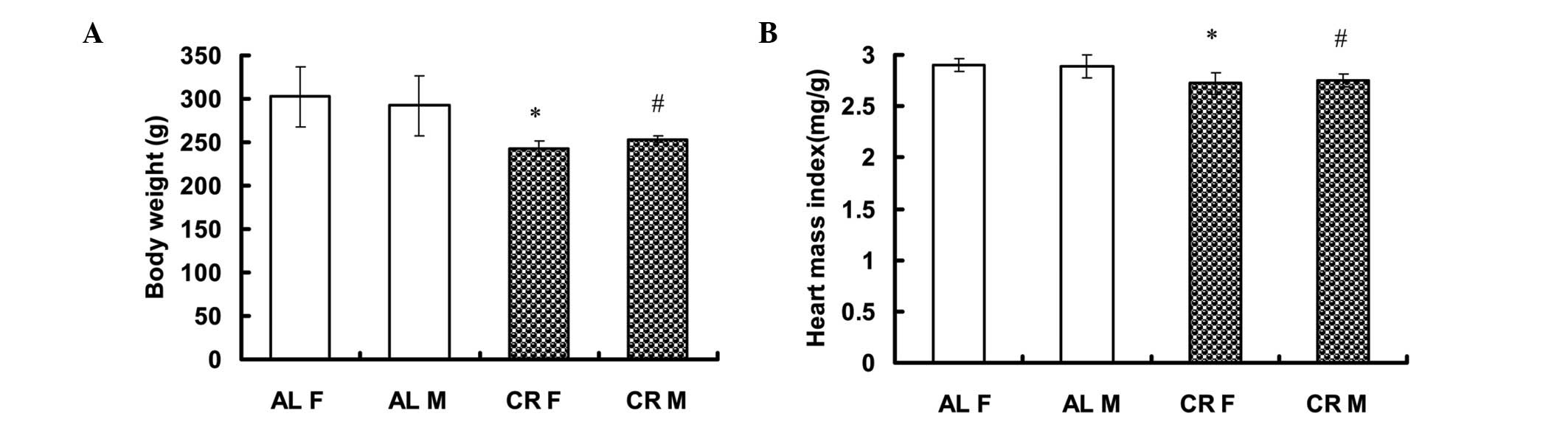
CR rats exhibit significantly lower BW
and heart weight
No significant differences were identified in BW of
male (M) or female (F) rats between the AL and CR groups prior to
the treatment conditions of the present study [AL F: 230.20±16.08 g
(n=5) vs. CR F: 219.20±5.63 g (n=5); AL M: 236.00±22.19 g (n=5) vs.
CR M: 231.00±10.89 g (n=5)]. Following three weeks on either the CR
or AL diets, BW was significantly higher in the AL group than in
the CR group [AL F: 302.60±34.60 g (n=5) vs. CR F: 243.00±8.60 g
(n=5; P<0.05); AL M: 292.20±34.85 g (n=5) vs. CR M: 253.40±3.78
g (n=5, P<0.05)] (Fig. 1A). The
heart mass index (HMI=Heart weight/BW) was also significantly
higher in the AL group [AL F: 2.90±0.06 mg/g (n=5) vs. CR F:
2.72±0.11 mg/g (n=5, P<0.05) and AL M: 2.89±0.11 mg/g (n=5) vs.
CR M: 2.75±0.06 mg/g (n=5, P<0.05)] (Fig. 1B).
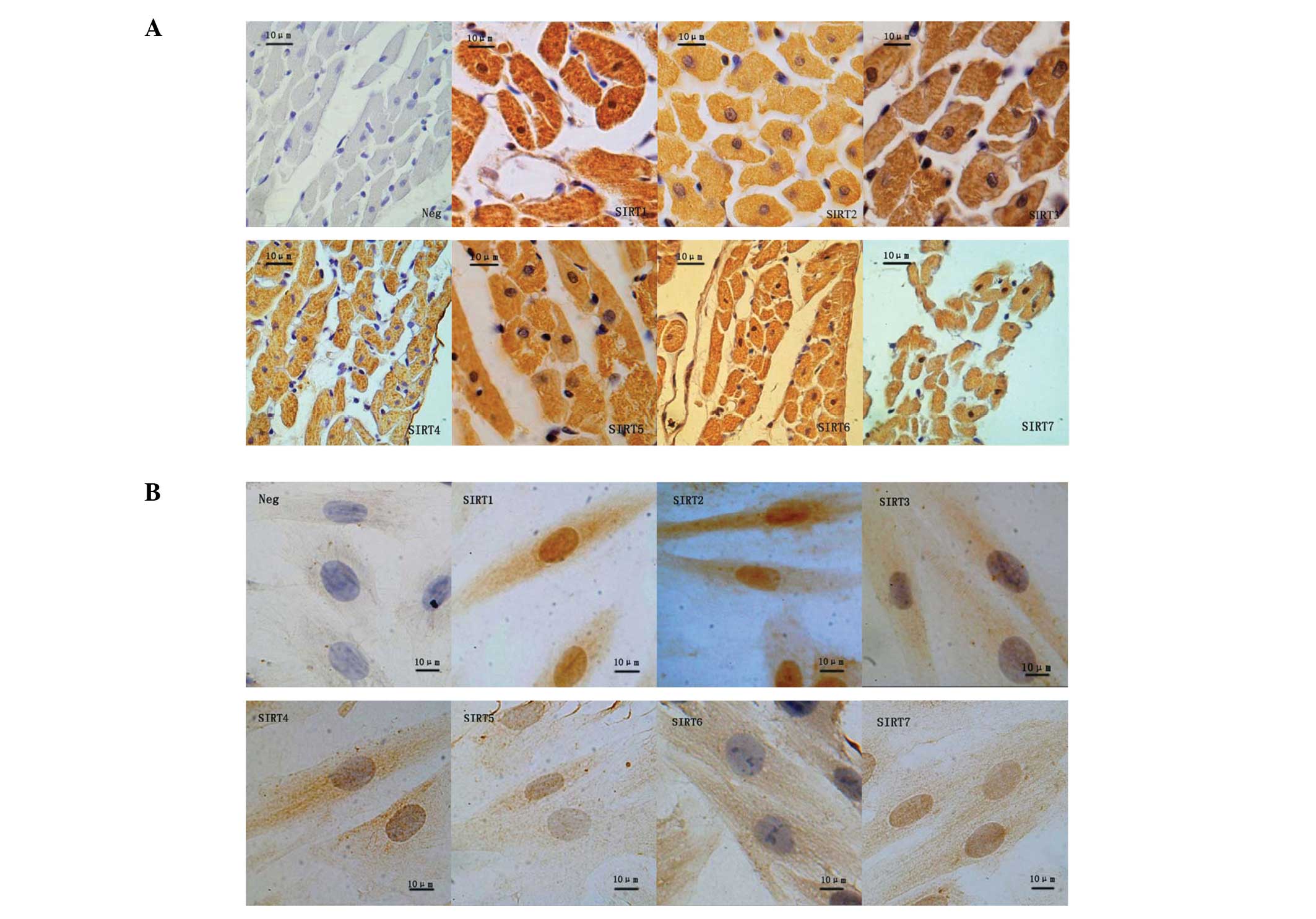
Cellular localization of SIRT1–7 in rat
cardiac tissues
SIRTs demonstrate diverse cellular locations and
various cellular functions. Using immunohistochemistry, seven SIRT
proteins were detected in the cardiac tissues of SD rats in the AL
and CR groups (Fig. 2A). SIRT1 was
observed to be distributed throughout the nucleus and the
cytoplasm, whereas SIRT2–5 were detected primarily in the
cytoplasm, and SIRT6 and -7 predominantly in the nucleus.
Cellular localization of SIRT1–7 in H9c2
cells
Immunocytochemistryrevealed that SIRT1, 3–5 and -7
exhibited a similar pattern of distribution in H9c2 cells as in rat
heart tissues (Fig. 2B). SIRT1 and
2 were identified in the nucleus and cytoplasm; however, SIRT3–6
were detected predominantly in the cytoplasm and SIRT7 in the
nucleus.
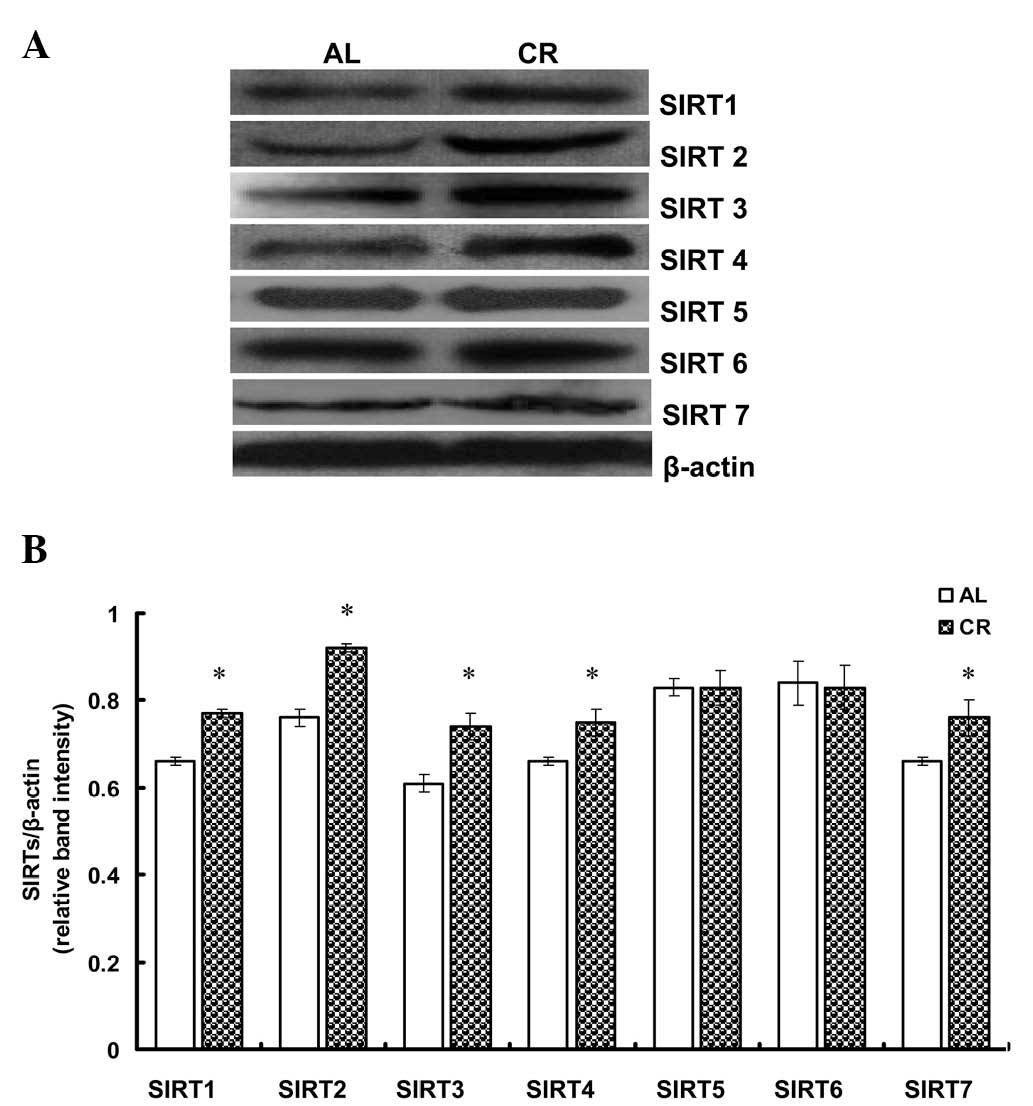
CR upregulates the protein expression of
SIRT1–4 and -7 in rat cardiac tissues
To investigate the effect of CR on SIRT expression
in rat hearts, 20 three-month-old SD rats were fed either AL or a
CR diet for three weeks. The changes in SIRT1–7 protein expression
were subsequently analyzed. Western blot analysis revealed that
SIRT1–4 and -7 expression were significantly upregulated in the CR
group compared with the AL group (P<0.05); however, no
significant difference was observed in the expression of SIRT5 and
6 (Fig. 3).
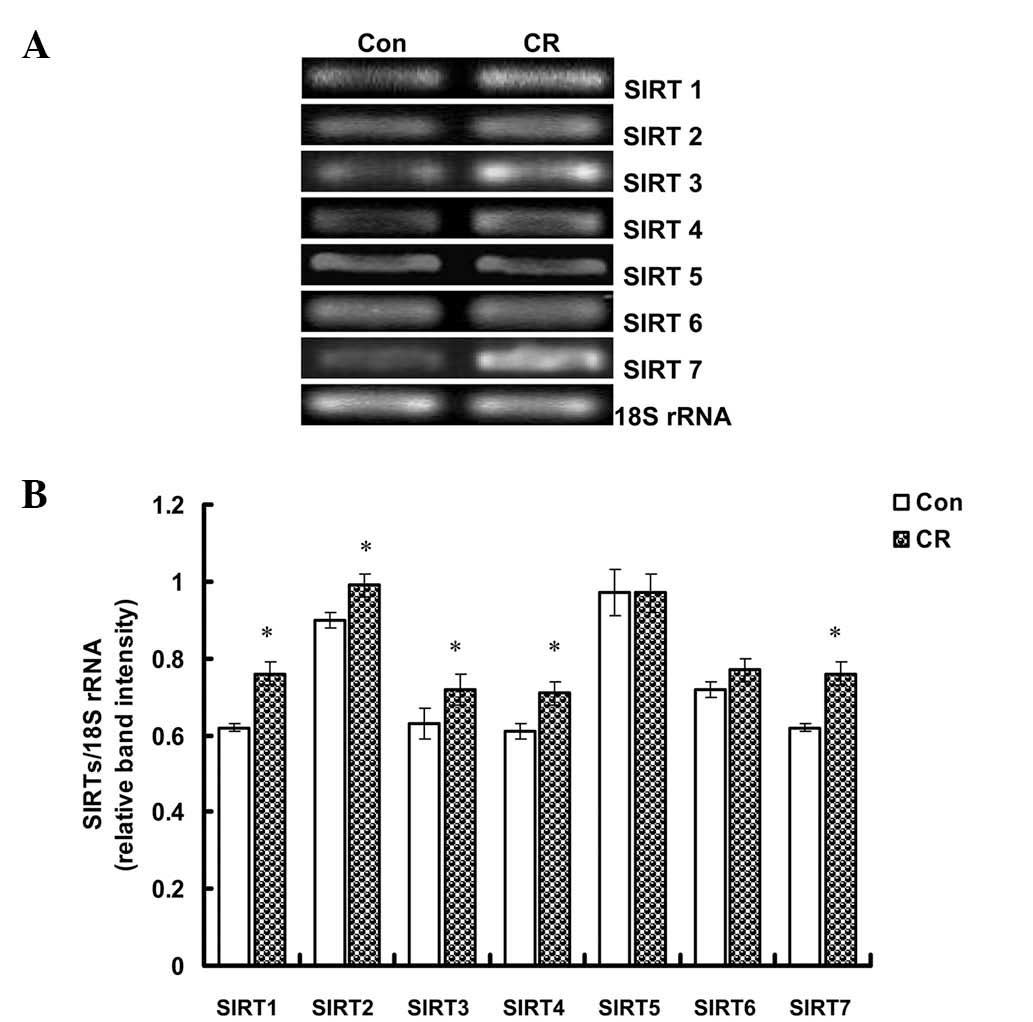
CR upregulates the mRNA expression of
SIRT1–4 and -7 in H9c2 cells
To support the results mentioned previously, H9c2
cells were cultured in either normal (4.5 g/l) or low (1 g/l)
concentrations of glucose for 24 h, representing control and CR
groups, respectively. qPCR analysis revealed that the mRNA
expression of SIRT1–4 and 7 was significantly higher in the CR
group compared with the control group (P<0.05); however, SIRT5
and 6 mRNA expression did not differ significantly (Fig. 4).
Discussion
CR has been observed to markedly extend the lifespan
in a wide range of organisms and attenuate numerous age-associated
diseases, i.e. CVD, partially through metabolic mechanisms,
including decreasing BW, body fat, blood glucose, insulin,
triglyceride and cholesterol, and increasing insulin sensitivity
and glucose tolerance (12).
Furthermore, CR has been demonstrated to directly affect the
cardiovascular system, improving the function of endothelial and
smooth muscle cells (13).
Moreover, long-term CR has been found to attenuate age-associated
diastolic dysfunction and ischemic damage in an experimental model
of myocardial infarction in rats (14,15).
It was observed in the present study that short-term CR reduced
heart weight and BW in rats, consistent with the metabolic effects
of CR.
Evidence suggests that Sir2 is a significant
regulator of responses to CR (2).
Sir2 is a highly conserved NAD-dependent histone deacetylase that
has been shown to modulate lifespan in numerous species (9,16).
Seven members of the Sir2 family, known as SIRTs (SIRT1–7), exist
in mammals, among which SIRT1 is the closest homologue of the yeast
Sir2 protein (17,18). In addition to histone
deacetylation, SIRT1 is capable of deacetylating other proteins,
including Forkhead transcription factors (FoxOs), myogenic
differentiation antigen (MyoD), peroxisome proliferator-activated
receptor-γ coactivator (PGC)-1α and the tumor suppressor p53
(19–24). SIRT1 is therefore capable of
regulating cellular metabolism and exerting corresponding effects
on gene expression. SIRT1 is a key regulator of cellular defense
mechanisms and survival under stress (11,21,22,25,26).
Furthermore, SIRT1 has been observed to improve vasodilatory and
regenerative functions in endothelial and smooth muscle cells of
the vascular wall, through regulation of the activity of
endothelial nitric oxide synthase, FoxO1, p53 and angiotensin II
type 1 receptor (AT1R), and is therefore suggested to have a
cardioprotective role (27).
SIRTs demonstrate diverse cellular localizations and
numerous cellular functions. SIRT1 is located in the nucleus and
cytoplasm in cardiomyocytes; however, its FoxO deacetylation
activty is restricted to the nucleus, where it is capable of
forming a protein complex with FoxOs (28,29).
The cellular localizations of SIRT2–7 in cardiomyocytes are
unknown. The present study examined the cellular localization of
SIRT1–7 in H9c2 cells and rat cardiac tissue. Consistent with a
previous study (30), SIRT1 was
identified in the nucleus and cytoplasm, while SIRT7 was
predominantly observed in the nucleus. SIRT3–5 were detected
primarily in the cytoplasm, in accordance with previous findings
(30,31). Significantly, the present study
identified that in the cardiac tissues of SD rats, SIRT6 was
predominantly identified in the nucleus, in accordance with a
previous report (32); however, a
high concentration of SIRT6 was detected in the cytoplasm in H9c2
cells. The embryonic nature of H9c2 cells is hypothesized to be
responsible for this discrepancy. Contrary results were obtained
for SIRT2.
It is well established that SIRTs are upregulated by
CR. Cohen et al (11)
suggested that in response to CR, SIRT1 is upregulated in numerous
tissues, including the brain, visceral fat pads, kidney and liver.
Furthermore, the expression of SIRT2 has been observed to increase
in the white adipose tissue and kidneys of mice in response to CR
(33). Moreover, Shi et al
(34) demonstrated that following
three months of CR, SIRT3 mRNA levels were elevated in both white
and brown adipose tissue in male mice. However, CR treatment has
been observed to decrease the expression of SIRT4 in pancreatic
β-cells (35,36). The roles of SIRT1–7 in CR responses
in cardiomyocytes are yet to be elucidated; however, the present
study revealed that CR significantly upregulated SIRT1–4 and 7 mRNA
and protein levels, suggesting that endogenous SIRT1–4 and -7 have
a significant role in CR responses in cardiomyocytes. The results
obtained for SIRT4 were inconsistent with the findings of
Mahlknecht and Voelter-Mahlknecht (35) and Chen et al (36), which may be a consequence of the
different cell lines employed.
In conclusion, the present study has identified the
presence of seven SIRTs in SD rat cardiac tissues and in H9c2
cells, and has demonstrated that endogenous SIRT1–4 and -7 may
participate, or have an essential role, in mediating CR responses
in cardiomyocytes in vivo and in vitro. These
findings may be of significance for attenuating age-associated
diseases, particularly CVD. Modulating SIRT1–4 and -7 activities is
likely to achieve similar biological effects to CR diet treatment.
These findings may aid the development of novel therapies for the
treatment of CVD, including drugs or biological methods that
activate SIRTs. Such treatments are anticipated to deliver broad
benefits for CVD.
Acknowledgements
The present study was supported by the Natural
Science Foundation of China (no. 81270382) and the Natural Science
Foundation of Guangdong Province, P.R. China (no.
10151503102000039).
Abbreviations:
|
AL
|
ad libitum
|
|
BW
|
body weight
|
|
CR
|
calorie restriction
|
|
CVDs
|
cardiovascular diseases
|
|
DAB
|
3-3′-diaminobenzidine
|
|
DMEM
|
Dulbecco’s modified Eagle medium
|
|
ECL
|
enhanced chemoluminescence
|
|
FBS
|
fetal bovine serum
|
|
FoxOs
|
Forkhead transcription factors
|
|
NAD
|
nicotinamide adenine dinucleotide
|
|
qPCR
|
quantitative polymerase chain
reaction
|
|
SD
|
Sprague Dawley
|
|
Sir2
|
silent information regulator 2
|
References
|
1
|
McCay CM, Crowell MF and Maynard LA: The
effect of retarded growth upon the length of life span and upon the
ultimate body size. 1935. Nutrition. 5:155–172. 1989.PubMed/NCBI
|
|
2
|
Lin SJ, Defossez PA and Guarente L:
Requirement of NAD and SIR2 for life-span extension by calorie
restriction in Saccharomyces cerevisiae. Science.
289:2126–2128. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ingram DK, Zhu M, Mamczarz J, Zou S, Lane
MA, Roth GS and deCabo R: Calorie restriction mimetics: an emerging
research field. Aging Cell. 5:97–108. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bordone L and Guarente L: Calorie
restriction, SIRT1 and metabolism: understanding longevity. Nat Rev
Mol Cell Biol. 6:298–305. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ungvari Z, Parrado-Fernandez C, Csiszar A
and de Cabo R: Mechanisms underlying caloric restriction and
lifespan regulation: implications for vascular aging. Circ Res.
102:519–528. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lin SJ, Kaeberlein M, Andalis AA, Sturtz
LA, Defossez PA, Culotta VC, Fink GR and Guarente L: Calorie
restriction extends Saccharomyces cerevisiae lifespan by
increasing respiration. Nature. 418:344–348. 2002.PubMed/NCBI
|
|
7
|
Anderson RM, Bitterman KJ, Wood JG,
Medvedik O and Sinclair DA: Nicotinamide and PNC1 govern lifespan
extension by calorie restriction in Saccharomyces
cerevisiae. Nature. 423:181–185. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lin SJ, Ford E, Haigis M, Liszt G and
Guarente L: Calorie restriction extends yeast life span by lowering
the level of NADH. Genes Dev. 18:12–16. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Imai S, Armstrong CM, Kaeberlein M and
Guarente L: Transcriptional silencing and longevity protein Sir2 is
an NAD-dependent histone deacetylase. Nature. 403:795–800. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Michan S and Sinclair D: Sirtuins in
mammals: insights into their biological function. Biochem J.
404:1–13. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cohen HY, Miller C, Bitterman KJ, Wall NR,
Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R and Sinclair
DA: Calorie restriction promotes mammalian cell survival by
inducing the SIRT1 deacetylase. Science. 305:390–392. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Koubova J and Guarente L: How does calorie
restriction work? Genes Dev. 17:313–321. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mattagajasingh I, Kim CS, Naqvi A,
Yamamori T, Hoffman TA, Jung SB, DeRicco J, Kasuno K and Irani K:
SIRT1 promotes endothelium-dependent vascular relaxation by
activating endothelial nitric oxide synthase. Proc Natl Acad Sci
USA. 104:14855–14860. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shinmura K, Tamaki K, Sano M, Murata M,
Yamakawa H, Ishida H and Fukuda K: Impact of long-term caloric
restriction on cardiac senescence: caloric restriction ameliorates
cardiac diastolic dysfunction associated with aging. J Mol Cell
Cardiol. 50:117–127. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ahmet I, Tae HJ, de Cabo R, Lakatta EG and
Talan MI: Effects of calorie restriction on cardioprotection and
cardiovascular health. J Mol Cell Cardiol. 51:263–271. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Smith JS, Brachmann CB, Celic I, Kenna MA,
Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer
C, Wolberger C and Boeke JD: A phylogenetically conserved
NAD+-dependent protein deacetylase activity in the Sir2
protein family. Proc Natl Acad Sci USA. 97:6658–6663. 2000.
|
|
17
|
Frye RA: Characterization of five human
cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins
(sirtuins) metabolize NAD and may have protein
ADP-ribosyltransferase activity. Biochem Biophys Res Commun.
260:273–279. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Frye RA: Phylogenetic classification of
prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res
Commun. 273:793–798. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Luo J, Nikolaev AY, Imai S, Chen D, Su F,
Shiloh A, Guarente L and Gu W: Negative control of p53 by Sir2alpha
promotes cell survival under stress. Cell. 107:137–148. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fulco M, Schiltz RL, Iezzi S, King MT,
Zhao P, Kashiwaya Y, Hoffman E, Veech RL and Sartorelli V: Sir2
regulates skeletal muscle differentiation as a potential sensor of
the redox state. Mol Cell. 12:51–62. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Brunet A, Sweeney LB, Sturgill JF, et al:
Stress-dependent regulation of FOXO transcription factors by the
SIRT1 deacetylase. Science. 303:2011–2015. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Motta MC, Divecha N, Lemieux M, Kamel C,
Chen D, Gu W, Bultsma Y, McBurney M and Guarente L: Mammalian SIRT1
represses forkhead transcription factors. Cell. 116:551–563. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rodgers JT, Lerin C, Haas W, Gygi SP,
Spiegelman BM and Puigserver P: Nutrient control of glucose
homeostasis through a complex of PGC-1alpha and SIRT1. Nature.
434:113–118. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cao C, Lu S, Kivlin R, Wallin B, et al:
SIRT1 confers protection against UVB- and
H2O2-induced cell death via modulation of p53
and JNK in cultured skin keratinocytes. J Cell Mol Med.
13:3632–3643. 2009.PubMed/NCBI
|
|
25
|
Vaziri H, Dessain SK, Ng Eaton E, Imai SI,
Frye RA, Pandita TK, Guarente L and Weinberg RA: hSIR2(SIRT1)
functions as an NAD-dependent p53 deacetylase. Cell. 107:149–159.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kobayashi Y, Furukawa-Hibi Y, Chen C,
Horio Y, Isobe K, Ikeda K and Motoyama N: SIRT1 is critical
regulator of FOXO-mediated transcription in response to oxidative
stress. Int J Mol Med. 16:237–243. 2005.PubMed/NCBI
|
|
27
|
Borradaile NM and Pickering JG: NAD(+),
sirtuins, and cardiovascular disease. Curr Pharm Des. 15:110–117.
2009.
|
|
28
|
Wang Y and Tissenbaum HA: Overlapping and
distinct functions for a Caenorhabditis elegans SIR2 and
DAF-16/FOXO. Mech Ageing Dev. 127:48–56. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen CJ, Yu W, Fu YC, Wang X, Li JL and
Wang W: Resveratrol protects cardiomyocytes from hypoxia-induced
apoptosis through the SIRT1-FoxO1 pathway. Biochem Biophys Res
Commun. 378:389–393. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Michishita E, Park JY, Burneskis JM,
Barrett JC and Horikawa I: Evolutionarily conserved and
nonconserved cellular localizations and functions of human SIRT
proteins. Mol Biol Cell. 16:4623–4635. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
North BJ, Marshall BL, Borra MT, Denu JM
and Verdin E: The human Sir2 ortholog, SIRT2, is an
NAD+-dependent tubulin deacetylase. Mol Cell.
11:437–444. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liszt G, Ford E, Kurtev M and Guarente L:
Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J
Biol Chem. 280:21313–21320. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang F, Nguyen M, Qin FX and Tong Q: SIRT2
deacetylates FOXO3a in response to oxidative stress and caloric
restriction. Aging Cell. 6:505–514. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shi T, Wang F, Stieren E and Tong Q:
SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial
function and thermogenesis in brown adipocytes. J Biol Chem.
280:13560–13567. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mahlknecht U and Voelter-Mahlknecht S:
Fluorescence in situ hybridization and chromosomal organization of
the sirtuin 4 gene (Sirt4) in the mouse. Biochem Biophys Res
Commun. 382:685–690. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen YR, Fang SR, Fu YC, Zhou XH, Xu MY
and Xu WC: Calorie restriction on insulin resistance and expression
of SIRT1 and SIRT4 in rats. Biochem Cell Biol. 88:715–722.
2010.PubMed/NCBI
|