Introduction
Hearing impairment is one of the most common types
of genetic diseases, with an estimated prevalence of 1–3/1,000
newborns, at least half of which is attributed to genetic factors
(1). ~One out of every 10 persons
with a hearing impairment are carriers of mutant genes associated
with deafness (1). Although not
life-threatening, loss of hearing is a debilitating disorder which
affects the life of patients in significant and occasionally
devastating ways. Mutations in a large variety of genes have been
identified as the cause of hereditary hearing loss. In 1998, Vahava
et al (2) determined the
molecular basis for an autosomal dominant progressive nonsyndromic
hearing loss, DFNA15 (MIM 602459), in an Israeli Jewish family. A
mutation in the Pou4f3 gene (encoding for the POU domain
transcription factor Brn3c/Brn3.1) was described, in which an
eight-base pair (bp) deletion in exon 2 results in the formation of
a premature stop codon in the first helix of this transcription
factor. The truncation of the Pou4f3 proteins affect their
ability to bind and activate downstream targets (3). The mechanism by which the
Pou4f3 mutation leads to DFNA15 hearing loss remains to be
elucidated. In Pou4f3−/− mice, the overall
architecture of the cochlea is unaffected and hair cell markers are
detected during early differentiation (4–6).
However, hair cells in Pou4f3−/− mice undergo
progressive loss compared with those of wild-type mice, with a
disorderly arrangement, and are misplaced among supporting cells
(6,7). Furthermore, the apical surface of
hair cells in Pou4f3−/− mice is devoid of
stereocilia, or with fused, disorganized, and occasionally giant
stereocilia (4–8).
Stereocilia are a type of organ-pipe-arrayed giant
stiff microvilli on the free surface of hair cells, containing a
parallel bundle of actin filaments at the core. Deletion or
mutation of the genes Espn, Myo15, Myo6, and
Myo7a accounts for the malformation of stereocilia (9–14).
Myosin VI, an anchoring protein encoded by Myo6, may aid to
hold down the membrane against surface tension forces and to anchor
the apical hair cell membrane to the actin cytoskeleton of the
cuticular plate, thus permitting the stereocilia to remain as
separate entities. Disturbance of myosin VI may be implicated in
the formation of fused and giant stereocilia, since inadequate
anchoring of the apical membrane between stereocilia may result in
the elevation of the membrane between adjacent stereocilia due to
surface tension (11,15,16).
These data suggest that Pou4f3 and
Myo6 are essential for the development and maintenance of
stereocilia. Mutation of either gene may lead to abnormal
stereocilial structures and hearing impairment in humans and mice.
To elucidate the pathological mechanisms that contribute to the
development of DFNA15, a form of hereditary deafness in humans, the
Myo6 promoter-driven expression in wild-type and mutant
cells, carrying an eight-bp deletion in exon 2 of Pou4f3,
were investigated in the present study using an in vitro
mouse cell-based model.
Materials and methods
Plasmids, strains and cell lines
The pGL3-Basic, renilla luciferase-thymidine kinase
(pRL-TK) control reporter, pIRES2-enhanced green fluorescent
protein (EGFP)-Pou4f3 (wild-type), and
pIRES2-EGFP-Pou4f3 (8-bp deletion in exon 2) vectors, C57/B6
mouse DNA, E. Coli DH5α competent cells, and the HEK293T
human embryonic kidney (293T) cells used in the present study were
kindly provided by Min-Sheng Zhu, Model Animal Research Center of
Nanjing University (Nanjing, China).
Primer design and polymerase chain
reaction (PCR) amplification
The promoter sequence of mouse Myo6 was
inferred from that of humans and consisted of a total of 2625 bp.
The 1899 bp upstream and 727 bp intron 1 sequences of the mouse
Myo6 gene were analyzed using Vector NTI 10 software
(Invitrogen Life Technologies, Carlsbad, CA, USA), which was also
used to design primers for amplification of the Myo6
promoter. The sequences of the forward and reverse primers were
5′-(ACGCGT)TTTAAAAACTAAAGTTCC CTTTCAG-3′ and
5′-(AAGCTT)CAGTATCTCCACATT GGGAT-3′, respectively, where ACGCGT and
AAGCTT are restriction sites for MluI and HindIII,
respectively. Primers were synthesized by GenScript Corporation
(Nanjing, China). The Myo6 promoter region was amplified
from the C57/B6 mouse tail DNA with high-fidelity polymerase ExTaq
using the above mentioned primers. The PCR conditions were as
follows: An initial denaturation step of 98°C for 5 min; followed
by 30 cycles at 98°C for 10 sec, 60°C for 15 sec and 72°C for 3.5
min, followed by a final elongation step at 72°C for 10 min and
maintenance at 16°C until the reactions were completed. Amplified
products were mixed with 0.5 μl Taq enzyme and incubated at 72°C
for 15 min followed by elongation at 4°C to add an A-tail directly
to the 3′-ends of the blunt-ended DNA fragment. Products were
analyzed by electrophoresis on 0.8% agarose gels. Target bands were
excised from the gel and the DNA was recovered using a DNA Gel
Extraction Kit (Corning Life Sciences-Axygen Inc., Union City, CA,
USA).
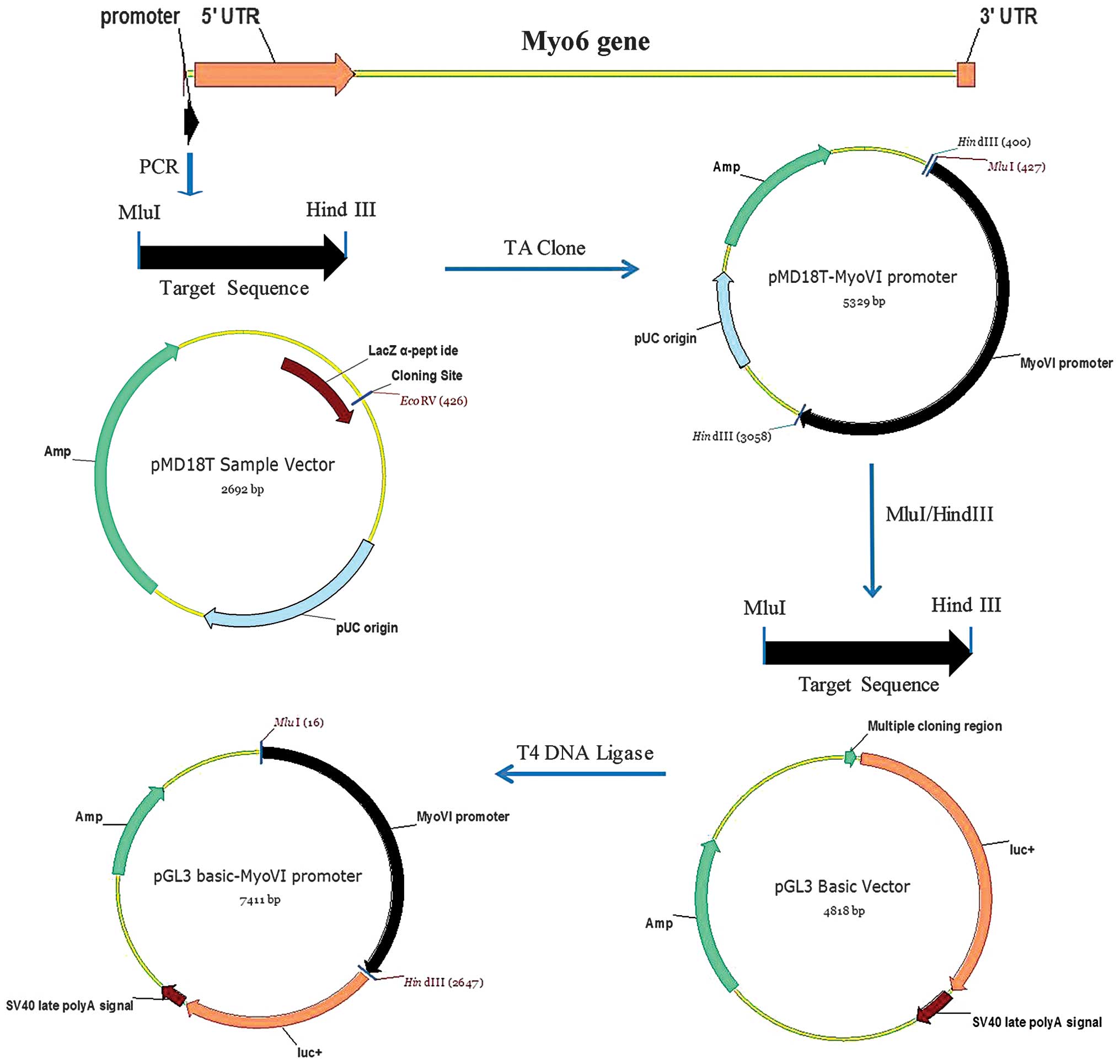
Plasmid constructs
The PCR products and pGL3-Basic vector were excised
using MluI and HindIII (Takara Bio, Inc., Shiga,
Japan) restriction enzymes. PCR products were ligated into the
pGL3-Basic vectors containing the luciferase reporter gene using T4
DNA ligase (Takara Bio, Inc.). Competent E. coli DH5α cells
were transformed with pGL3-Basic-Myo6 promoter vectors,
plated on Luria-Bertani (LB) medium containing ampicillin, and the
plates were incubated at 37°C for 12–20 h. Individual colonies were
selected and grown in the liquid culture medium containing
ampiccilin overnight. Plasmids were extracted using the AxyPrep
Plasmid Miniprep Kit (Corning Life Sciences-Axygen, Inc.) and
digested with MluI and HindIII, followed by
purification of the inserted sequences for further sequencing by
GenScript Corporation.
Cell culture
293T cells were grown and maintained in Dulbecco’s
modified Eagle medium (Gibco-BRL, Carlsbad, CA, USA) containing 10%
fetal bovine serum (PAA Laboratories, Cölbe, Germany) supplemented
with 100 IU/ml penicillin and 100 mg/l streptomycin at 37°C in an
atmosphere of 5% CO2.
Plasmid transfection and luciferase
reporter assays
The day prior to transfection, 293T cells were
suspended in fresh medium and plated in 96-well plates at a density
of 2×104 cells/well. When the cells reached 70%
confluency, they were transfected with 50 ng pGL3-Basic-Myo6
promoter and 0.5 ng pRL-TK (internal control), along with either
pIRES2-EGFP, pIRES2-EGFP-Pou4f3 (wild-type), or
pIRES2-EGFP-Pou4f3 (8-bp deletion in exon 2), using the
LipofectamineTM 2000 Transfection Reagent (Invitrogen
Life Technologies) according to the manufacturer’s instructions.
The total amount of plasmid DNA was maintained at 200 ng in each
well. The cells were harvested and lyzed 24 h post-transfection.
Firefly and Renilla luciferase activities were determined
with the Dual-Luciferase Reporter Assay System in a GloMax96
luminescence reader (both from Promega, Madison, WI, USA) according
to the manufacturer’s instructions. Relative luciferase activity
was expressed as the ratio of firefly luciferase activity to the
Renilla luciferase activity in each sample. All values were
obtained from at least three independent repetitions of the
transfection, with six wells for each transfection mixture/sample
in every experiment.
Statistical analysis
Data analysis was performed using the SPSS 16.0
statistical software (SPSS Inc., Chicago, IL, USA). The Student’s
t-test was used for comparing means between groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
Myo6 promoter amplification
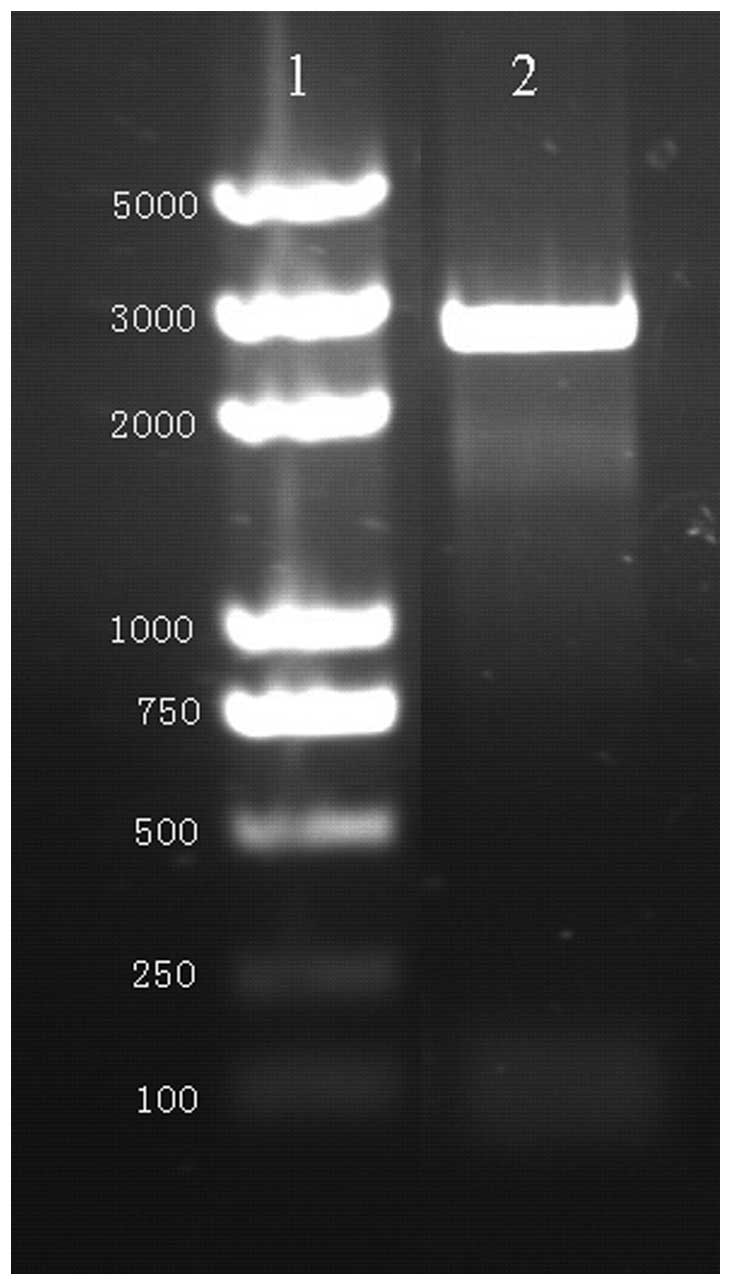
PCR was performed using primers with restriction
sites. Gel electrophoresis analysis revealed the presence of a 2625
bp fragment in the PCR product, as expected (Fig. 1). Non-specific amplification was
not detected (Fig. 1).
Furthermore, the sequencing analysis confirmed that the sequence of
the amplified Myo6 promoter was consistent with that of the
template.
Construction and confirmation of
pGL3-Basic-Myo6 promoter vectors
The Myo6 promoter-driven luciferase reporter
construct, pGL3-Basic-Myo6 promoter, was constructed as
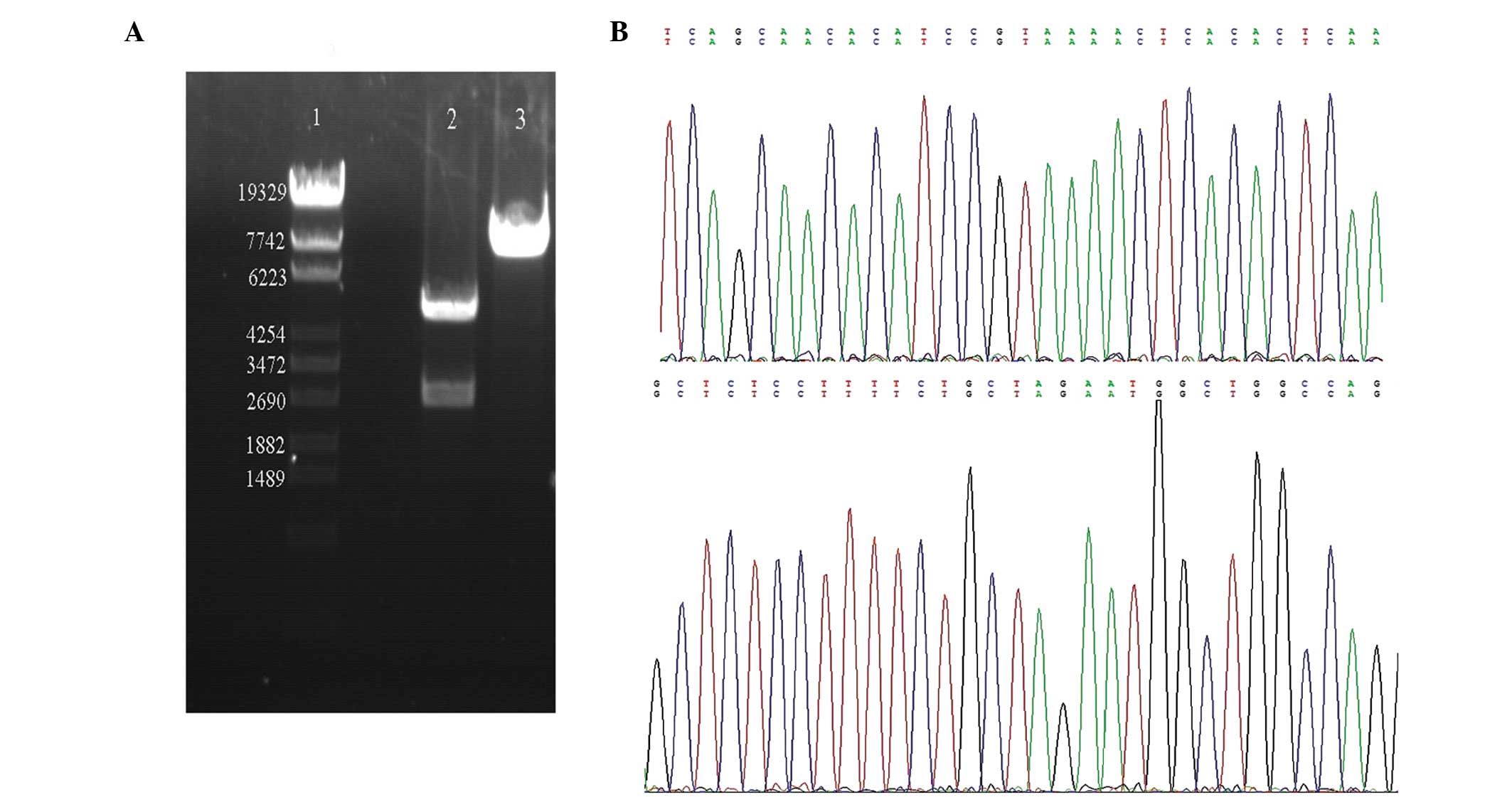
described in the materials and methods section (Fig. 2). To verify the sequences
introduced into the pGL3-Basic-Myo6 promoter vector, the
plasmid DNAs were digested with MluI and HindIII and
the DNA fragments were re-introduced into the same vectors, i.e.,
pGL3-Basic vector, followed by sequencing analysis. The sequences
were confirmed to be correct and devoid of any mutations,
demonstrating that the Myo6 promoter was successfully
inserted into pGL3-Basic vectors (Fig.
3).
Dual luciferase reporter assay
The activity of the Myo6 promoter in 293T
cells was assayed using the pGL3-Basic Myo6 promoter
reporter, in the presence of wild-type Pou4f3 or a truncated
mutant of Pou4f3, with the dual luciferase assay system. The
relative luciferase activity was expressed as the ratio of firefly
luciferase activity to Renilla luciferase activity to
evaluate the effect of Pou4f3 on the promoter activity of
Myo6. The relative luciferase activity of cells expressing
wild-type Pou4f3 was comparable to that of the control,
wherein the empty vector was co-transfected with the luciferase
reporters. Co-transfection of the construct expressing mutated
Pou4f3 downregulated the expression of luciferase from the
Myo6 promoter to almost half of that of the control
(P<0.001; Fig. 4), indicating
that mutated Pou4f3 has a negative role in the expression of
Myo6. These data indicate that the inhibition of expression
of Myo6 in the presence of the mutated Pou4f3 may be
an underlying mechanism of DFNA15 hearing loss.
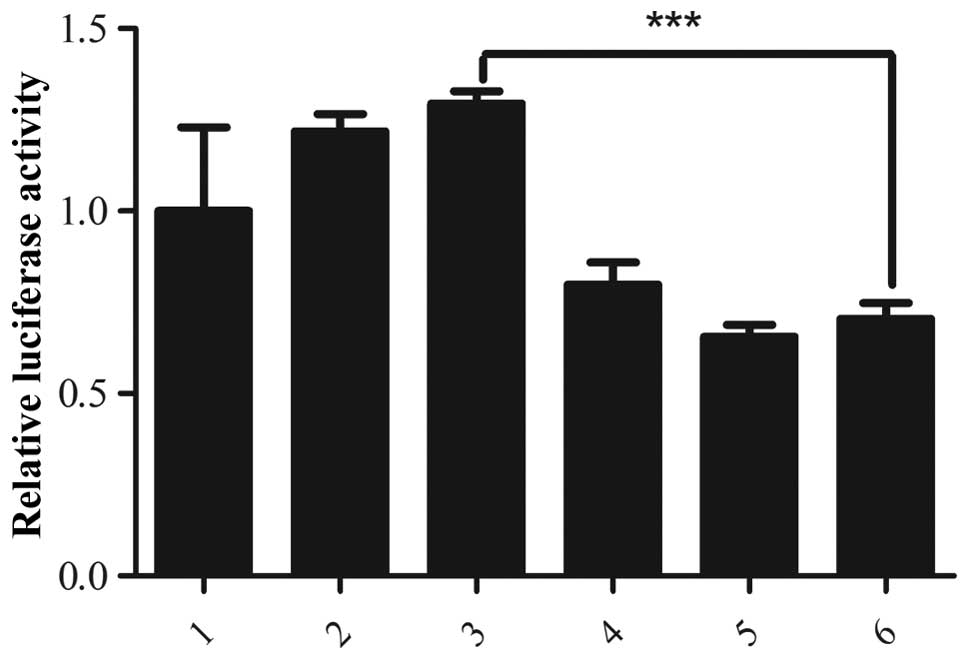 | Figure 4Regulation of the Myo6
promoter by Pouf4f3 as assessed using a luciferase reporter
assay. 293T cells were transfected with the indicated combinations
of vectors and the relative luciferase activity was measured to
evaluate the Myo6 promoter activity. 1, pIRES2-EGFP empty
vector, the ratio of firefly/Renilla luciferase activities
for this empty vector, used as the control, was normalized to 1.0
and the relative luciferase activity in all other samples is
calculated following normalization to the control value; 2–3, 100
and 150 ng of pIRES2-EGFP-Pouf4f3 (wild-type), respectively;
4–6, 50, 100 and 150 ng of pIRES2-EGFP-Pouf4f3 (8-bp
deletion in exon 2), respectively. No difference between in
relative luciferase activity was observed in cells transfected with
the wild-type Pouf4f3-expressing plasmid and the control. In
contrast, co-transfection with the mutant Pouf4f3-expressing
plasmid significantly inhibited the expression of luciferase from
the Myo6 promoter, reducing the relative luciferase activity
to almost half of that of the control (P<0.001).
***P<0.001, vs. control. Myo6, myosin IV
promoter; EGSP, enhanced green fluorescent protein. |
Discussion
Stereocilia of hair cells perform the process of
auditory transduction. They receive mechanical inputs derived from
airborne sound waves and transduce these signals into electrical
responses that are then relayed to the brain for interpretation.
Only stereocilia, arranged in rows of increasing height, are
capable of converting mechanical activity to electrical activity
(17–19). Damage to, or deterioration of the
staircase-like arrangement of stereocilia accounts for hearing loss
or balance disorders (15,20,21).
With the help of transgenic and knock-out mouse models of human
hearing loss (22), a variety of
genes responsible for inner ear development and deafness, including
genes encoding transcription factors, actins, ion channels,
membrane proteins, and structural proteins, have been identified
(2,21,23,24).
Specifically, transcription factors including Math1 (25–28)
and POU4f3 have been shown to be crucial for hair cell
development, differentiation, maturation and maintenance.
Structural proteins, including Myo6 and Myo7a are important for the
stability of the structure and function of stereocilia.
The POU-domain family of transcription factors is
widely expressed in the nervous system. For example, Pou4f1,
2 and 3 are essential for the development of sensory
ganglion, retinal ganglion and hair cells, respectively (29–33).
Pou4f3 (also known as Brn3.1 and Brn3c), an important
transcription factor of the POU-domain family, specifically binds
to a 9-bp recognition element, ATAATTAAT to activate downstream
gene expression and thus cell development (34,35).
Brn3c−/− mice deficient in Pou4f3 are
hearing impaired and exhibit vestibular dysfunction. Furthermore,
the hair cells fail to mature and stereociliary bundles are formed
or only malformed and disorganized stereocilia exist (6–8,36).
Mutation of unconventional myosins, including
myosins VIIa, VI, Ic, and XV, have been shown to be associated with
deafness. Normally present on cilia and actin-rich microvilli,
including the stereocilia on mammalian hair cells, unconventional
myosins are required for the cohesion and structural integrity of
the bundle (11,37–39).
Myosin VI, encoded by Myo6, is expressed in the inner and
outer hair cells of mouse cochlea, concentrating in the actin-rich
cuticular plate associated with stereociliary rootlets,
pericuticular necklaces, and cytoplasm (11,15).
As an anchoring protein, myosin VI may help anchor the membrane
against surface tension forces and to anchor the apical hair cell
membrane to the actin cytoskeleton of the cuticular plate, thus
permitting the stereocilia to remain as separate entities. Myosin
VI is crucial for the motility and shape change of hair cells
during the development of stereocilia, and mutation of myosin VI
results in the formation of giant stereociliary bundles and
degenerated hair cells (11).
The luciferase reporter assay system is widely used
to investigate the interaction between cis-acting regulatory
elements, including promoters, enhancers, silencers and
trans-acting regulatory proteins. This reporter assay has become
the choice of the majority of investigations due to the advantages
of rapidity, reliability and sensitivity. In the present study, the
Myo6 promoter was successfully inserted into an expression
vector and the Myo6 promoter-driven luciferase reporter
construct was transiently transfected into human embryonic kidney
293T cells, along with expression vectors carrying wild-type and
mutated Pou4f3 using liposomes. The present study showed no
differences between the Myo6 promoter activity in cells expressing
the wild-type Pou4f3 and the control cells. However,
co-transfection with the mutated Pou4f3-expressing construct
significantly inhibited the Myo6 promoter activity to almost
half of that of the control, indicating that mutated Pou4f3
has a negative role in the regulation of Myo6 expression.
The data suggest that the inhibition of the Myo6 promoter
and consequently the expression of myosin VI due to mutation of
Pou4f3 may be an underlying mechanism of DFNA15. These data
provide insights into the molecular basis of hearing impairment in
DFNA15 hearing loss and suggest that loss of expression from the
Myo6 promoter may explain, at least in part, the hearing
loss phenotype in the presence of the Pou4f3 truncation
mutation.
Acknowledgements
The authors would like to thank Professor Minsheng
Zhu, Drs Yajing Peng, Chen Chen, Chenghai Zhang, Yanning Qiao,
Caiping Chen and Pei Wang from the Model Animal Research Center of
Nanjing University in China. The present study was supported by
grants from the National Natural Science Foundation of China (nos
81371090 and 30973302), and the Key Project supported by Medical
Science and Technology Development Foundation, Nanjing Department
of Health (no. 201108019).
References
|
1
|
Piatto VB, Nascimento EC, Alexandrino F,
Oliveira CA, Lopes AC, Sartorato EL and Maniglia JV: Molecular
genetics of non-syndromic deafness. Braz J Otorhinolaryngol.
71:216–223. 2005.PubMed/NCBI
|
|
2
|
Vahava O, Morell R, Lynch ED, Weiss S,
Kagan ME, Ahituv N, Morrow JE, Lee MK, Skvorak AB, Morton CC,
Blumenfeld A, Frydman M, Friedman TB, King MC and Avraham KB:
Mutation in transcription factor POU4F3 associated with inherited
progressive hearing loss in humans. Science. 279:1950–1954. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Weiss S, Gottfried I, Mayrose I, Khare SL,
Xiang M, Dawson SJ and Avraham KB: The DFNA15 deafness mutation
affects POU4F3 protein stability, localization, and transcriptional
activity. Mol Cell Biol. 23:7957–7964. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gerrero MR, McEvilly RJ, Turner E, Lin CR,
O’Connell S, Jenne KJ, Hobbs MV and Rosenfeld MG: Brn-3.0: a
POU-domain protein expressed in the sensory, immune, and endocrine
systems that functions on elements distinct from known octamer
motifs. Proc Natl Acad Sci USA. 90:10841–10845. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ninkina NN, Stevens GE, Wood JN and
Richardson WD: A novel Brn3-like POU transcription factor expressed
in subsets of rat sensory and spinal cord neurons. Nucleic Acids
Res. 21:3175–3182. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xiang M, Gan L, Zhou L, Klein WH and
Nathans J: Targeted deletion of the mouse POU domain gene Brn-3a
causes selective loss of neurons in the brainstem and trigeminal
ganglion, uncoordinated limb movement, and impaired suckling. Proc
Natl Acad Sci USA. 93:11950–11955. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xiang M, Gan L, Li D, Chen ZY, Zhou L,
O’Malley BW Jr, Klein W and Nathans J: Essential role of POU-domain
factor Brn-3c in auditory and vestibular hair cell development.
Proc Natl Acad Sci USA. 94:9445–9450. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xiang M, Gan L, Li D, Zhou L, Chen ZY,
Wagner D, O’Malley BW Jr, Klein W and Nathans J: Role of the Brn-3
family of POU-domain genes in the development of the
auditory/vestibular, somatosensory, and visual systems. Cold Spring
Harb Symp Quant Biol. 62:325–336. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zheng L, Sekerková G, Vranich K, Tilney
LG, Mugnaini E and Bartles JR: The deaf jerker mouse has a mutation
in the gene encoding the espin actin-bundling proteins of hair cell
stereocilia and lacks espins. Cell. 102:377–385. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Karolyi IJ, Probst FJ, Beyer L, Odeh H,
Dootz G, Cha KB, Martin DM, Avraham KB, Kohrman D, Dolan DF,
Raphael Y and Camper SA: Myo15 function is distinct from Myo6,
Myo7a and pirouette genes in development of cochlear stereocilia.
Hum Mol Genet. 12:2797–2805. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Self T, Sobe T, Copeland NG, Jenkins NA,
Avraham KB and Steel KP: Role of myosin VI in the differentiation
of cochlear hair cells. Dev Biol. 214:331–341. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Holme RH and Steel KP: Stereocilia defects
in waltzer (Cdh23), shaker1 (Myo7a) and double waltzer/shaker1
mutant mice. Hear Res. 169:13–23. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Boëda B, El-Amraoui A, Bahloul A, Goodyear
R, Daviet L, Blanchard S, Perfettini I, Fath KR, Shorte S, Reiners
J, Houdusse A, Legrain P, Wolfrum U, Richardson G and Petit C:
Myosin VIIa, harmonin and cadherin 23, three Usher I gene products
that cooperate to shape the sensory hair cell bundle. EMBO J.
21:6689–6699. 2002.PubMed/NCBI
|
|
14
|
Belyantseva IA, Boger ET, Naz S, Frolenkov
GI, Sellers JR, Ahmed ZM, Griffith AJ and Friedman TB: Myosin-XVa
is required for tip localization of whirlin and differential
elongation of hair-cell stereocilia. Nat Cell Biol. 7:148–156.
2005. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hertzano R, Shalit E, Rzadzinska AK, Dror
AA, Song L, Ron U, Tan JT, Shitrit AS, Fuchs H, Hasson T, Ben-Tal
N, Sweeney HL, de Angelis MH, Steel KP and Avraham KB: A Myo6
mutation destroys coordination between the myosin heads, revealing
new functions of myosin VI in the stereocilia of mammalian inner
ear hair cells. PLoS Genet. 4:e10002072008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Avraham KB, Hasson T, Sobe T, Balsara B,
Testa JR, Skvorak AB, Morton CC, Copeland NG and Jenkins NA:
Characterization of unconventional Myo6, the human homologue of the
gene responsible for deafness in Snell’s waltzer mice. Hum Mol
Genet. 6:1225–1231. 1997.
|
|
17
|
Dallos P: Peripheral mechanisms of
hearing. Comprehensive Physiology. John Wiley & Sons, Inc;
2011, View Article : Google Scholar
|
|
18
|
Roberts WM, Howard J and Hudspeth AJ: Hair
cells: transduction, tuning, and transmission in the inner ear.
Annu Rev Cell Biol. 4:63–92. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fettiplace R and Fuchs PA: Mechanisms of
hair cell tuning. Annu Rev Physiol. 61:809–834. 1999. View Article : Google Scholar
|
|
20
|
Frolenkov GI, Belyantseva IA, Friedman TB
and Griffith AJ: Genetic insights into the morphogenesis of inner
ear hair cells. Nat Rev Genet. 5:489–498. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhu GJ, Wang F, Chen C, Xu L, Zhang WC,
Fan C, Peng YJ, Chen J, He WQ, Guo SY, Zuo J, Gao X and Zhu MS:
Myosin light-chain kinase is necessary for membrane homeostasis in
cochlear inner hair cells. PLoS One. 7:e348942012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Avraham KB: Mouse models for deafness:
lessons for the human inner ear and hearing loss. Ear Hear.
24:332–341. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Peters LM, Anderson DW, Griffith AJ,
Grundfast KM, San Agustin TB, Madeo AC, Friedman TB and Morell RJ:
Mutation of a transcription factor, TFCP2L3, causes progressive
autosomal dominant hearing loss, DFNA28. Hum Mol Genet.
11:2877–2885. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu XZ, Ouyang XM, Xia XJ, Zheng J, Pandya
A, Li F, Du LL, Welch KO, Petit C, Smith RJ, Webb BT, Yan D, Arnos
KS, Corey D, Dallos P, Nance WE and Chen ZY: Prestin, a cochlear
motor protein, is defective in non-syndromic hearing loss. Hum Mol
Genet. 12:1155–1162. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen P, Johnson JE, Zoghbi HY and Segil N:
The role of Math1 in inner ear development: Uncoupling the
establishment of the sensory primordium from hair cell fate
determination. Development. 129:2495–2505. 2002.PubMed/NCBI
|
|
26
|
Bermingham NA, Hassan BA, Price SD,
Vollrath MA, Ben-Arie N, Eatock RA, Bellen HJ, Lysakowski A and
Zoghbi HY: Math1: an essential gene for the generation of inner ear
hair cells. Science. 284:1837–1841. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zine A, Aubert A, Qiu J, Therianos S,
Guillemot F, Kageyama R and de Ribaupierre F: Hes1 and Hes5
activities are required for the normal development of the hair
cells in the mammalian inner ear. J Neurosci. 21:4712–4720.
2001.PubMed/NCBI
|
|
28
|
Kawamoto K, Ishimoto S, Minoda R, Brough
DE and Raphael Y: Math1 gene transfer generates new cochlear hair
cells in mature guinea pigs in vivo. J Neurosci. 23:4395–4400.
2003.PubMed/NCBI
|
|
29
|
Eng SR, Lanier J, Fedtsova N and Turner
EE: Coordinated regulation of gene expression by Brn3a in
developing sensory ganglia. Development. 131:3859–3870. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang EJ, Liu W, Fritzsch B, Bianchi LM,
Reichardt LF and Xiang M: Brn3a is a transcriptional regulator of
soma size, target field innervation and axon pathfinding of inner
ear sensory neurons. Development. 128:2421–2432. 2001.PubMed/NCBI
|
|
31
|
Badea TC, Cahill H, Ecker J, Hattar S and
Nathans J: Distinct roles of transcription factors brn3a and brn3b
in controlling the development, morphology, and function of retinal
ganglion cells. Neuron. 61:852–864. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu W, Khare SL, Liang X, Peters MA, Liu
X, Cepko CL and Xiang M: All Brn3 genes can promote retinal
ganglion cell differentiation in the chick. Development.
127:3237–3247. 2000.PubMed/NCBI
|
|
33
|
Sage C, Huang M, Vollrath MA, Brown MC,
Hinds PW, Corey DP, Vetter DE and Chen ZY: Essential role of
retinoblastoma protein in mammalian hair cell development and
hearing. Proc Natl Acad Sci USA. 103:7345–7350. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xiang M, Zhou L, Macke JP, Yoshioka T,
Hendry SH, Eddy RL, Shows TB and Nathans J: The Brn-3 family of
POU-domain factors: primary structure, binding specificity, and
expression in subsets of retinal ganglion cells and somatosensory
neurons. J Neurosci. 15:4762–4785. 1995.PubMed/NCBI
|
|
35
|
Xiang M, Gao WQ, Hasson T and Shin JJ:
Requirement for Brn-3c in maturation and survival, but not in fate
determination of inner ear hair cells. Development. 125:3935–3946.
1998.PubMed/NCBI
|
|
36
|
Keithley EM, Erkman L, Bennett T, Lou L
and Ryan AF: Effects of a hair cell transcription factor, Brn-3.1,
gene deletion on homozygous and heterozygous mouse cochleas in
adulthood and aging. Hear Res. 134:71–76. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Anderson DW, Probst FJ, Belyantseva IA,
Fridell RA, Beyer L, Martin DM, Wu D, Kachar B, Friedman TB,
Raphael Y and Camper SA: The motor and tail regions of myosin XV
are critical for normal structure and function of auditory and
vestibular hair cells. Hum Mol Genet. 9:1729–1738. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Holt JR, Gillespie SK, Provance DW, Shah
K, Shokat KM, Corey DP, Mercer JA and Gillespie PG: A
chemical-genetic strategy implicates myosin-1c in adaptation by
hair cells. Cell. 108:371–381. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kros CJ, Marcotti W, van Netten SM, Self
TJ, Libby RT, Brown SD, Richardson GP and Steel KP: Reduced
climbing and increased slipping adaptation in cochlear hair cells
of mice with Myo7a mutations. Nat Neurosci. 5:41–47. 2002.
View Article : Google Scholar : PubMed/NCBI
|