Introduction
Acute leukemia is a hematological malignant tumor
characterized by the rapid accumulation of myeloid blasts in the
bone marrow and peripheral blood. Acute leukemia is rapidly fatal
without treatment and has a poor prognosis. Over recent decades,
the treatment of acute leukemia has developed, with advances in
anthracycline-based chemotherapy and stem-cell transplantation
(1,2) improving the outlook for patients with
acute leukemia. However, although the prognosis of acute leukemia
has improved, the five- and 10-year relative survival estimates for
patients with leukemia are only 21.4 and 18.7%, respectively
(3). Drug resistance is the
primary cause of disease relapse or lack of remission in patients
with acute leukemia. Therefore, studies into the mechanisms
associated with chemotherapy resistance are required.
microRNAs (miRNAs), a novel class of small noncoding
RNAs, ranging between 19 and 25 nucleotides in size, regulate
specific target genes through translational repression or direct
mRNA degradation. miRNAs thereby regulate numerous cellular
functions, including cell proliferation, differentiation and
apoptosis (4–6). A previous study showed that the
deregulated expression of specific miRNAs that modulate the
expression of oncogenes and tumor suppressors is associated with
the development of malignancies, and that specific miRNA expression
signatures can be used to effectively classify human tumors
(7). miR-125b is the ortholog of
lin-4 in Caenorhabditis elegans and is highly conserved in
numerous species, from nematodes to humans. In humans, there are
two homologs: hsa-miR-125b-1 and hsa-miR-125b-2, transcribed from
two loci on chromosomes 11q23 and 21q21, respectively. miR-125b has
been found to be an oncomiR in hepatocellular carcinoma (8), breast cancer (9), lung cancer (10), melanoma (11) and gastrointestinal cancer (12). Furthermore, miR-125 is involved in
myelodysplastic syndrome, acute myeloid leukemia (AML), B-cell
acute lymphoid leukemia, megakaryoblastic leukemia and chronic
lymphocytic leukemia (CLL) (13–17).
In addition, miR-125 contributes to leukemogenesis (18) and increases drug resistance in
pediatric acute promyelocytic leukemia (18) and acute lymphoblastic leukemia
(ALL) cells treated with vincristine and daunorubicin (DNR)
(19).
The present study aimed to investigate the value of
miR-125b in determining acute leukemia prognosis and the role of
miR-125b in inducing DNR resistance in leukemia cell lines through
the inhibition of apoptosis.
Materials and methods
Patients and sample collection
A total of 46 patients with acute leukemia from
Shanghai Jiaotong University Affiliated Shanghai First People’s
Hospital (Shanghai, China) were enrolled in this study between May
2011 and November 2012. All of the patients were primary or
no-remission patients. Patient exclusion criteria were concomitant
or previous cancer and acute promyelocytic leukemia. The
characteristics of the patients included in this study are listed
in Table I. Bone marrow was
collected from patients by bone marrow puncture at diagnosis or at
follow-up subsequent to therapy. All patients were followed for at
least four months or until recurrence. Event-free survival (EFS)
and overall survival (OS) were the end-points used for the analysis
of the treatment results. EFS (20) was measured from the date of entry
into the present study to the date of induction failure, relapse
from complete remission (CR), CR with incomplete hematologic
recovery or death from any cause. OS was calculated from the date
of entry into the study to the date of death from any cause. Data
were censored if patient had no event at the last follow-up.
Healthy samples were collected from three healthy adults. Written
informed consent was obtained from the patients for the biological
studies. This study was approved by the Ethics Committee of
Shanghai Jiaotong University Affiliated Shanghai First People’s
Hospital.
 | Table ICharacteristics of patients with acute
leukemia. |
Table I
Characteristics of patients with acute
leukemia.
| Characteristics | Value |
|---|
| Age in years at
diagnosis, median (range) | 44 (15–67) |
| Gender, n (%) |
| Male | 26 (56.52) |
| Female | 20 (43.48) |
| WBC count, n (%) |
|
<4×109/l | 13 (28.26) |
|
≥4×109/l,
<10×109/l | 6 (12.77) |
|
≥10×109/l,
<50×109/l | 13 (28.26) |
|
≥50×109/l | 14 (30.43) |
| FAB +
immunophenotyping, n (%) |
| ALL | 11 (23.91) |
| AML |
| M2 | 8 (17.39) |
| M4 | 8 (17.39) |
| M5 | 12 (26.09) |
| Others | 3 (6.50) |
| Mixed lineage
acute leukemia, n (%) | 4 (8.70) |
| Primary, n (%) | 31 (67.39) |
| Recurrence/no
remission, n (%) | 15 (32.61) |
RNA extraction and quantitative
polymerase chain reaction (qPCR)
To evaluate miR-125b expression in bone marrow
mononuclear cells, qPCR for miRNA was performed. Mononuclear cells
were separated using a Ficoll-Hypaque centrifugation gradient from
2 ml bone marrow samples. Total RNA was isolated with
TRIzol® reagent (Invitrogen Life Technologies, Carlsbad,
CA, USA) according to the manufacturer’s instructions. To detect
miR-125b expression, 10 ng total RNA was reverse transcribed with
miRNA-specific primers using a TaqMan® MicroRNA Reverse
Transcription kit (Applied Biosystems, Foster City, CA, USA). qPCR
was then performed using TaqMan® MicroRNA Assays
(Applied Biosystems) according to the manufacturer’s instructions,
using an StepOnePlus™ Real-Time PCR System (Applied Biosystems).
The 20-μl PCR reaction contained the following: 1.3 μl reverse
transcription (RT) product, 10 μl TaqMan® Universal PCR
Master Mix and 1 μl primer and probe mix from the TaqMan miRNA
assays. The reactions were incubated in optical plates at 95°C for
10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 10
min. U6 small nuclear RNA was used as a control. miR-125b
expression relative to that in a healthy sample was calculated
using the 2−ΔΔCT method (21). All of the qPCR assays were
performed in triplicate.
Cell culture
The K562, THP-1, Jurkat and REH cell lines (The Cell
Bank of Type Culture Collection of Chinese Academy of Sciences,
Shanghai, China) were cultured in RPMI-1640 medium (HyClone, Logan,
UT, USA) supplemented with 10% fetal bovine serum (FBS; Gibco-BRL,
Carlsbad, CA, USA), 100 U/ml penicillin and 100 μg/ml streptomycin.
HEK293T cells (The Cell Bank of Type Culture Collection of Chinese
Academy of Sciences, Shanghai, China) were grown in Dulbecco’s
Modified Eagle Medium (HyClone) containing 10% FBS, 100 g/ml
L-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin.
Overexpression and knockdown of
miR-125b
In order to overexpress miR-125b in K562, THP-1 and
Jurkat cells, 10 μg expression vector or empty vector with 10 μg
packing plasmid (gag/pol and vesicular stomatitis virus G) was
incubated with Fugene 6® (Roche, Mannheim, Germany) for
15 min at room temperature and subsequently added to the HEK293T
cells. The viral supernatant was harvested after 48 h, and the
cells were treated by a spin infection with retroviral supernatant
(1 ml supernatant per 1×106 cells plus polybrene) in
six-well plates that were pre-coated with retronectin (Takara Bio,
Inc., Shiga, Japan). The percentage of green fluorescent
protein-positive cells was 70–90%.
In accordance with a previous study (22), miR-125 ‘sponge’ sequences (8–9
tandem repeats, each complementary to miR-125) were designed. An
imperfect base-pairing between the miRNA and the miR-125 ‘sponge’
was designed to impair endonucleolytic cleavage by Argonaute-2. The
miR-125 ‘sponge’ sequence was 5′-TCACAGGTTACTCAGGGA-3′ and was
cloned into the pRetroQ-mCherry-C1 retroviral vector (Clontech
Laboratories, Inc., Palo Alto, CA, USA). The miR-125 ‘sponge’
vector was then infected into REH cells using a retrovirus, in
order to generate miR-125b-knockdown REH cells. Subsequent to virus
infection, REH cells were screened using puromycin (Sigma-Aldrich,
St. Louis, MO, USA) at an final concentration of 2 μg/ml. The
percentage of red fluorescent protein-positive cells was ~100%.
Luciferase assay
G protein-coupled receptor kinase (GRK) 2 and
p53-upregulated modulator of apoptosis (PUMA) 3′ untranslated
region (3′UTR) luciferase reporters were individually generated by
inserting full-length human GRK2 and PUMA 3′UTR into the
XhoI and NotI sites in the psiCHECK-2 vector
(Promega, Madison, WI, USA) downstream from the Renilla luciferase
coding sequence. The GRK2 3′UTR was PCR amplified from human
genomic DNA (Roche) using the following primer sequences: Forward,
5′-TCGCTCGAGCCC GCCCACCCGCCTTTTA-3′ and reverse, 5′-TCGGCGGCC
GCAATCAGGCACCATTTT-3′. The PUMA 3′UTR was PCR amplified using the
following primer sequences: Forward, 5′-TCGCTCGAGGACTT
TCTCTGCACCAT-3′ and reverse, 5′-TAAGCGGCC GCGGCAAGCAGAAAGAGT-3′.
The sequences and cloning direction of the PCR products were
validated by DNA sequencing. The HEK293T cells were plated in
96-well plates at 5,000 cells/well the day prior to transfection.
Transfection was performed in triplicate with Fugene 6 (Roche) and
150 ng plasmid mixture (135 ng miR-125b expression vector and 15 ng
reporter vector). Luciferase assays for firefly and Renilla
luciferase were performed 48 h after transfection with a
Dual-Glo® Luciferase Assay kit (Promega). Luminescence
was quantified using a NOVOstar machine (BMG Labtech, Ortenberg,
Germany). The Renilla luciferase readings were normalized to the
firefly luciferase activity in the corresponding well.
Cell viability assay
Cells were seeded into 96-well plates in RPMI-1640
medium containing 10% FBS and were treated with serial dilutions of
DNR (Pfizer, Groton, CT, USA). Cell viability was determined 48 h
after DNR treatment using a luminescent cell viability assay
(Promega). Luminescence was quantified using a NOVOstar machine
(BMG Labtech).
Small interfering (si)RNA
transfection
K562 cells were transfected with GRK2 or PUMA siRNA
(Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) using Fugene
6 (Roche) according to the manufacturer’s instructions. The
transfection efficiency was ~80% and was calculated as the
percentage of fluorescein-labeled cells using fluorescence
microscopy. Forty-eight hours after transfection, the cells were
harvested for further analysis.
Western blot analysis
Cells were washed twice with phosphate-buffered
saline and then lysed in lysis buffer (Beyotime Institute of
Biotechnology, Shanghai, China) containing PhosSTOP (Roche). Cell
lysates (~40 μg protein) were loaded on a 10% sodium dodecyl
sulfate-polyacrylamide gel and subsequently transferred to a
polyvinylidene difluoride membrane (Millipore, Billerica, MA, USA).
Membranes were blocked in 5% non-fat dry milk in Tris-buffered
saline (TBS) for 2 h at room temperature and then incubated with
rabbit polyclonal anti-GRK2, anti-PUMA (Santa Cruz Biotechnology,
Inc.), anti-caspase-3 (Epitomics, Burlingame, CA, USA) and
anti-cleaved caspase-3 (Cell Signaling Technology, Inc., Danvers,
MA, USA) antibodies overnight at 4°C. Following three washes with
TBS supplemented with Tween 20 (TBST), the membranes were incubated
for 2 h with rabbit anti-human immunoglobulin G antibody (Cell
Signaling Technology, Inc.) conjugated to horseradish peroxidase.
Subsequent to washing with TBST, the membranes were exposed to
enhanced chemiluminescent reagent (Millipore) for 1 min and then to
Kodak X-ray film (Eastman Kodak, Rochester, NY, USA). Anti-β-actin
antibody (Cell Signaling Technology, Inc.) was used to detect
β-actin.
Statistical analysis
Data are presented as the mean ± standard error. EFS
was calculated using Kaplan-Meier analysis. The Student’s t-test
was performed for comparisons between two groups. Statistical
analysis was conducted using SPSS 13.0 software (SPSS, Inc.,
Chicago, IL, USA). A value of P<0.05 was considered to indicate
a statistically significant difference.
Results
miR-125b is highly expressed in acute
leukemia
To determine the differential expression pattern of
miR-125b, qPCR was performed in bone marrow samples from patients
with acute leukemia (n=46), including those with AML and ALL.
miR-125b expression was observed to be markedly higher in patients
with acute leukemia compared with that in the healthy controls,
demonstrating an average 4,375-fold increase (data not shown).
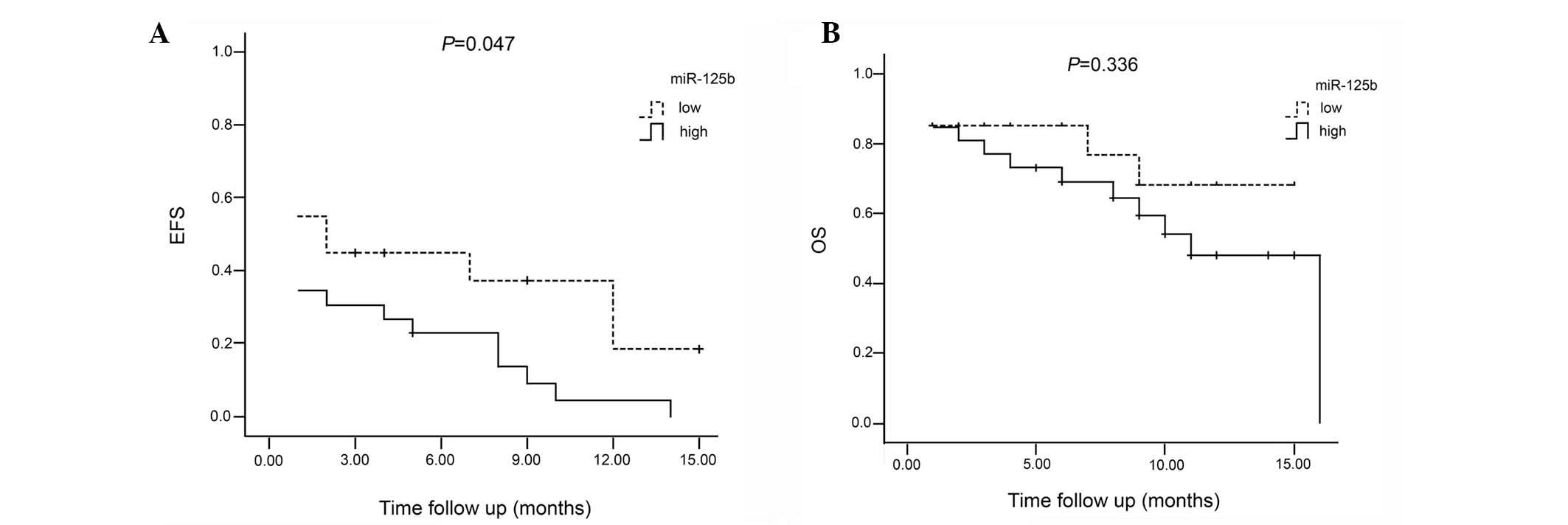
Patients were divided into two risk groups, low and high, using
median miR-125b expression. The median miR-125b expression value in
the patients with acute leukemia was 85.24-fold higher than that in
the samples from the healthy controls. Patients with an miR-125b
expression value below the median were assigned to the low group
and those with an expression value above the median were assigned
to the high group. Among the 20 patients in the low group, two
relapsed, two succumbed during therapy and nine did not achieve
remission. A total of 13 patients had an event, and the remaining
35% showed an EFS. The average EFS was 6.2 months [95% confidence
interval (CI), 3.5–8.9]. Among the 26 patients in the high group,
eight relapsed, three succumbed during therapy, one succumbed
during bone marrow transplantation subsequent to achieving complete
remission and 12 did not achieve remission. A total of 25 of the
patients had an event, and the remaining 3.8% showed an EFS. The
average EFS was 3.3 months (95% CI, 1.8–4.8). A significant
difference was observed in the EFS between patients in the high
group and those in the low group (P<0.05). However, the OS was
not observed to be significantly different between the two groups
(P=0.336) (Fig. 1).
miR-125b expression affects leukemia cell
sensitivity to DNR
To investigate the association between miR-125b and
DNR chemoresistance in leukemia cells, miR-125b was overexpressed
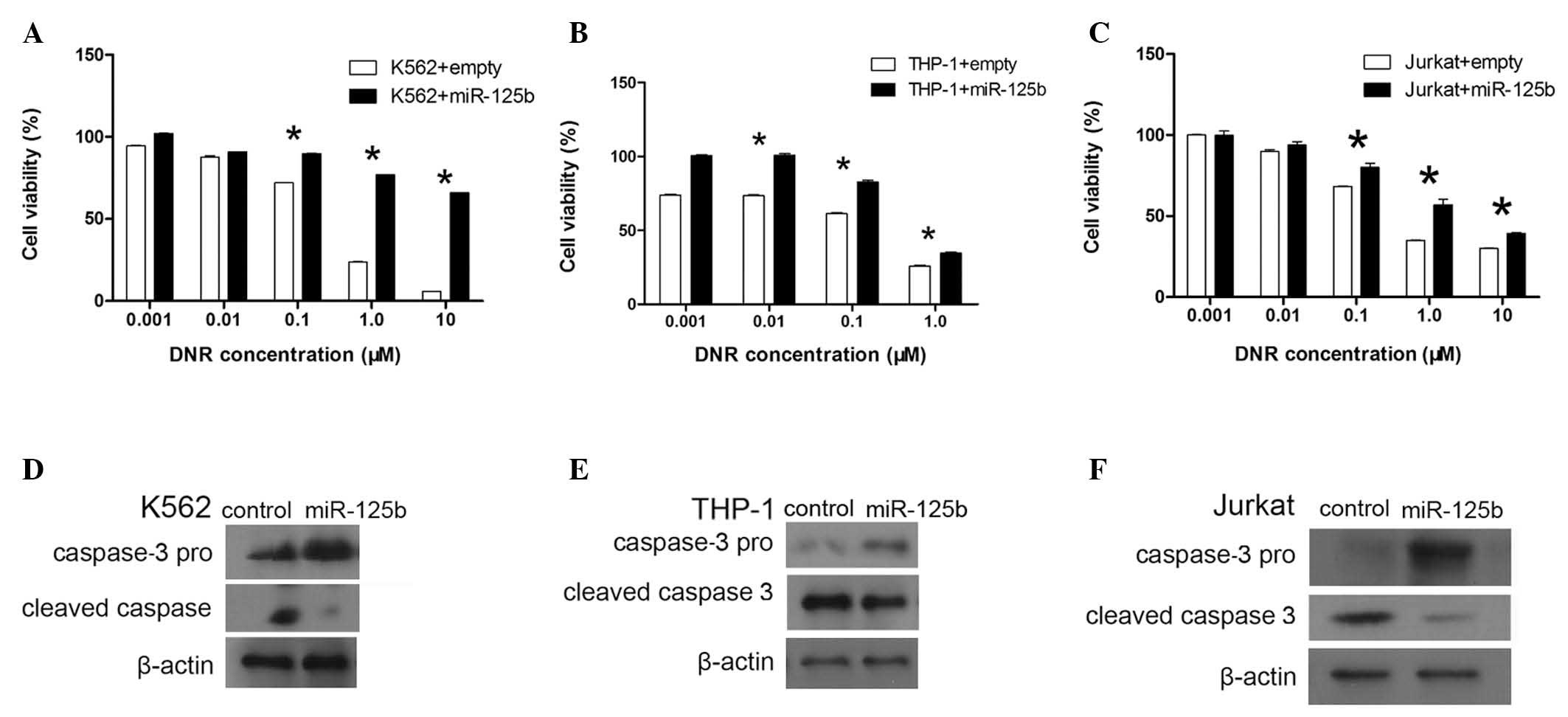
in K562, THP-1 and Jurkat cells and knocked down in REH cells.
K562, THP-1 and Jurkat cells were stably transduced with a murine
stem cell virus (MSCV)-miR-125b vector to overexpress miR-125b in
the cells (23). qPCR was used to
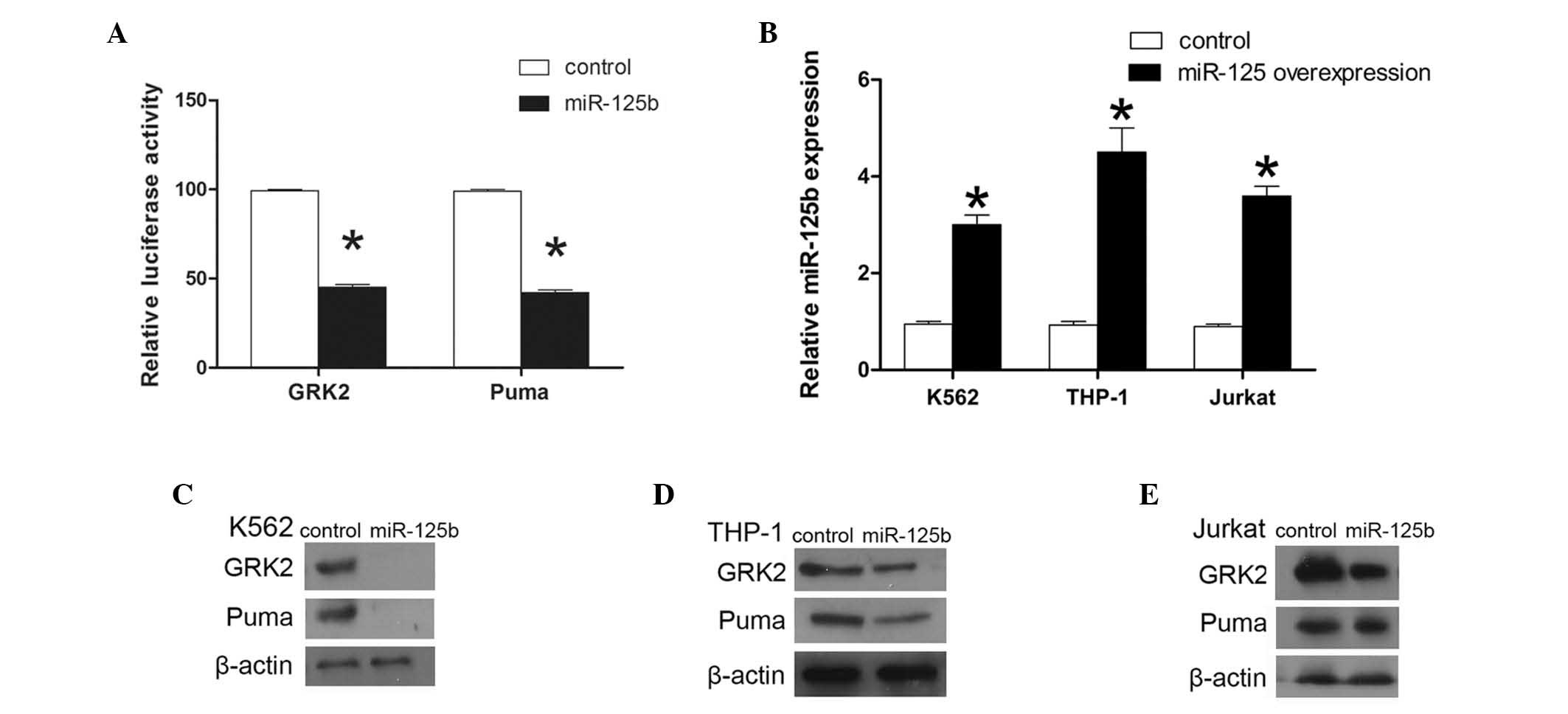
assess the expression of miR-125b. In K562 cells, miR-125b
expression was observed to be 3.2-fold higher in the cells
transduced with MSCV-miR-125b compared with the cells transduced
with an empty vector, and 4.9- and 4-fold higher in the THP-1 and
Jurkat cells, respectively. The K562, THP-1 and Jurkat cells
overexpressing miR-125b were treated with various doses of DNR.
Overexpression of miR-125b was associated with a significant
increase in the survival of K562, THP-1 and Jurkat cells (Fig. 2A–C). At DNR concentrations ≥0.1 μM,
the survival of K562 and Jurkat cells was significantly higher in
the miR-125b overexpression group than that in the control group
(P<0.05). At DNR concentrations ≥0.01 μM, the survival of THP-1
cells was significantly higher in the miR-125b overexpression group
than that in the control group (P<0.05).
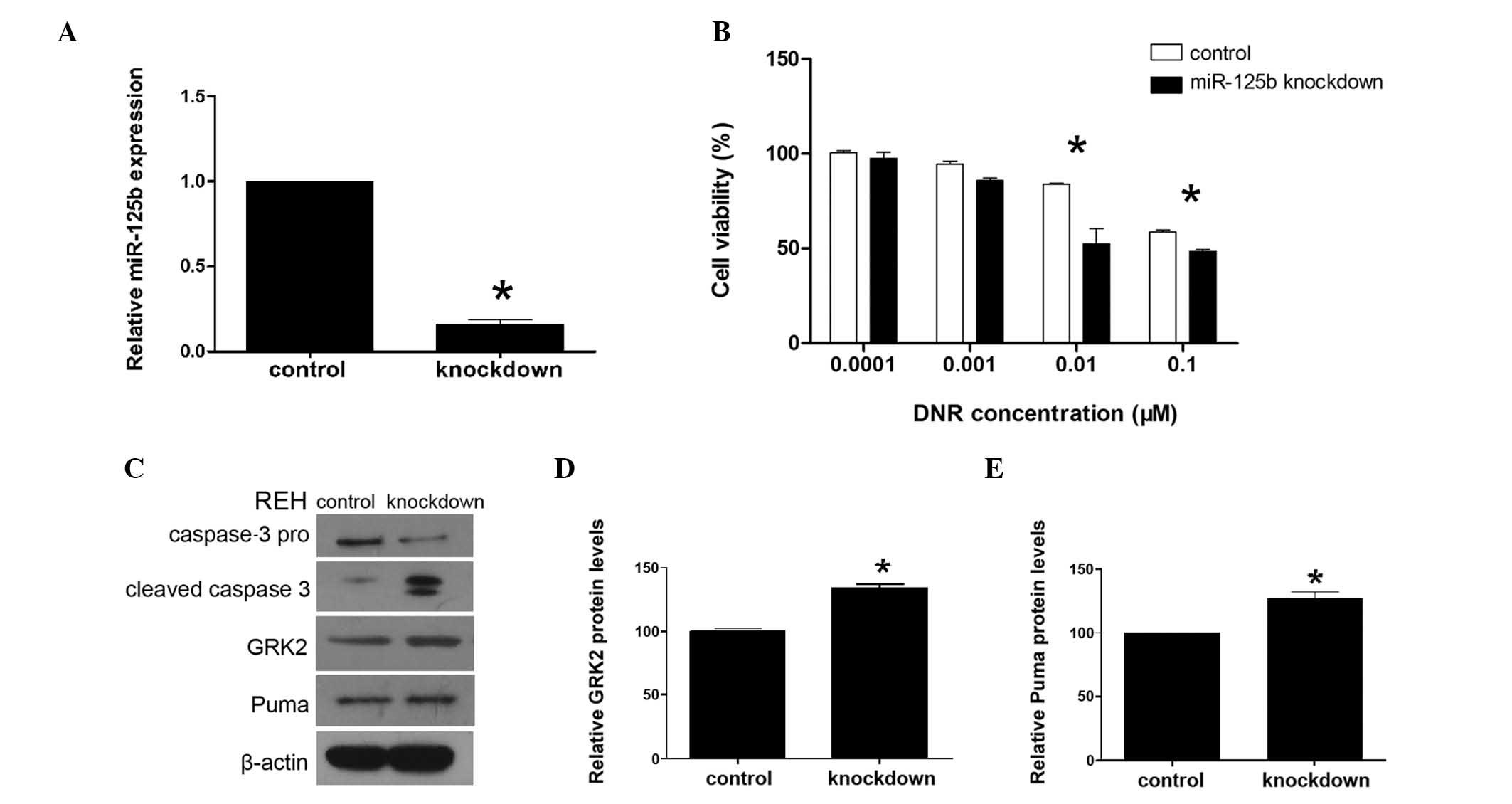
The effects of miR-125b-knockdown on REH cells were
investigated using transduction with an miR-125b ‘sponge’ vector.
The miR-125b ‘sponge’ vector effectively reduced the expression of
miR-125b in REH cells, which was verified using qPCR. The
expression of miR-125b in the miR-125b-knockdown REH cells was
reduced by 75%, compared with that in the control REH cells. The
expression of miR-125b was significantly lower in the knockdown
cells than that in the control cells (P<0.001) (Fig. 3A). Furthermore, knockdown of
miR-125b was associated with a significantly decreased survival
rate in the REH cells (Fig. 3B).
At DNR concentrations ≥0.01 μM, miR-125b-knockdown REH cells
exhibited a significantly lower survival rate than that of the
control cells (P<0.05). This suggests that overexpression of
miR-125b may contribute to DNR resistance in K562, THP-1 and Jurkat
cells and that knockdown of miR-125b may contribute to DNR
sensitivity in REH cells. Therefore, these findings suggest that a
correlation exists between miR-125b expression and sensitivity to
DNR in leukemia cells.
High expression of miR-125b inhibits
apoptosis
In order to determine whether overexpression of
miR-125b affects apoptosis, the release of activated caspase-3 was
assessed. Caspase-3 pro and cleaved caspase-3 protein levels were
analyzed using western blot analysis. Upon treatment with DNR,
K562, THP-1 and Jurkat cells overexpressing miR-125b exhibited
higher expression of caspase-3 pro than the control group, and
lower expression of cleaved caspase-3 than the control group
(Fig. 2D–F). Knockdown of miR-125b
was observed to reduce the expression of caspase-3 pro and increase
that of cleaved caspase-3 in REH cells compared with the expression
in the control cells (Fig. 3C).
These results show that overexpression of miR-125b induced DNR
resistance through the inhibition of apoptosis and that the
inhibition of miR-125b increased the sensitivity of REH cells to
DNR by increasing apoptosis. These findings suggest that miR-125b
increases the resistance of leukemia cells to DNR through
inhibiting apoptosis.
miR-125b downregulates GRK2 and PUMA
expression
To investigate the effect of miRNA on mRNA
expression, miR-125b target mRNAs were obtained from the miRBase
(http://www.mirbase.org) and the TargetScan
(http://www.targetscan.org) databases
(24). The correlation between
miR-125b expression and the expression of its predicted target
mRNAs, including the number of binding sites in the respective
mRNAs and the variation in the correlation coefficients, were
statistically analyzed. Among these potential gene targets, the
present study focused on GRK 2 and PUMA, which may have effects on
cell apoptosis (Fig. 3). It was
hypothesized that GRK2 and PUMA are direct targets of miR-125b.
To investigate this hypothesis, HEK293T cells were
separately cotransfected with miR-125b and either GRK2 or PUMA
3′UTR luciferase reporters. Transfections with control vector were
performed in parallel. Cotransfection resulted in a 55 and 56.6%
reduction in the reporter activity for GRK2 and PUMA, respectively
(P<0.01; Fig. 4A). These
results demonstrate that GRK2 and PUMA 3′UTR are targets of
miR-125b. The role of miR-125b in the regulation of GRK2 and PUMA
expression in K562, THP-1 and Jurkat cells was then assessed. In
the present study, it was hypothesized that overexpression of
miR-125b was likely to reduce the GRK2 and PUMA protein expression.
GRK2 and PUMA protein levels were observed to be lower in K562,
THP-1 and Jurkat cells overexpressing miR-125b, compared with those
in the corresponding control cells (Fig. 4). In the miR-125b-knockdown REH
cells, GRK2 and PUMA protein levels were found to be significantly
higher than those in the control cells (Fig. 3C–E). These findings show that GRK2
and PUMA are targets of miR-125b in K562, THP-1, Jurkat and REH
cells.
GRK2 and PUMA have a key role in DNR
resistance
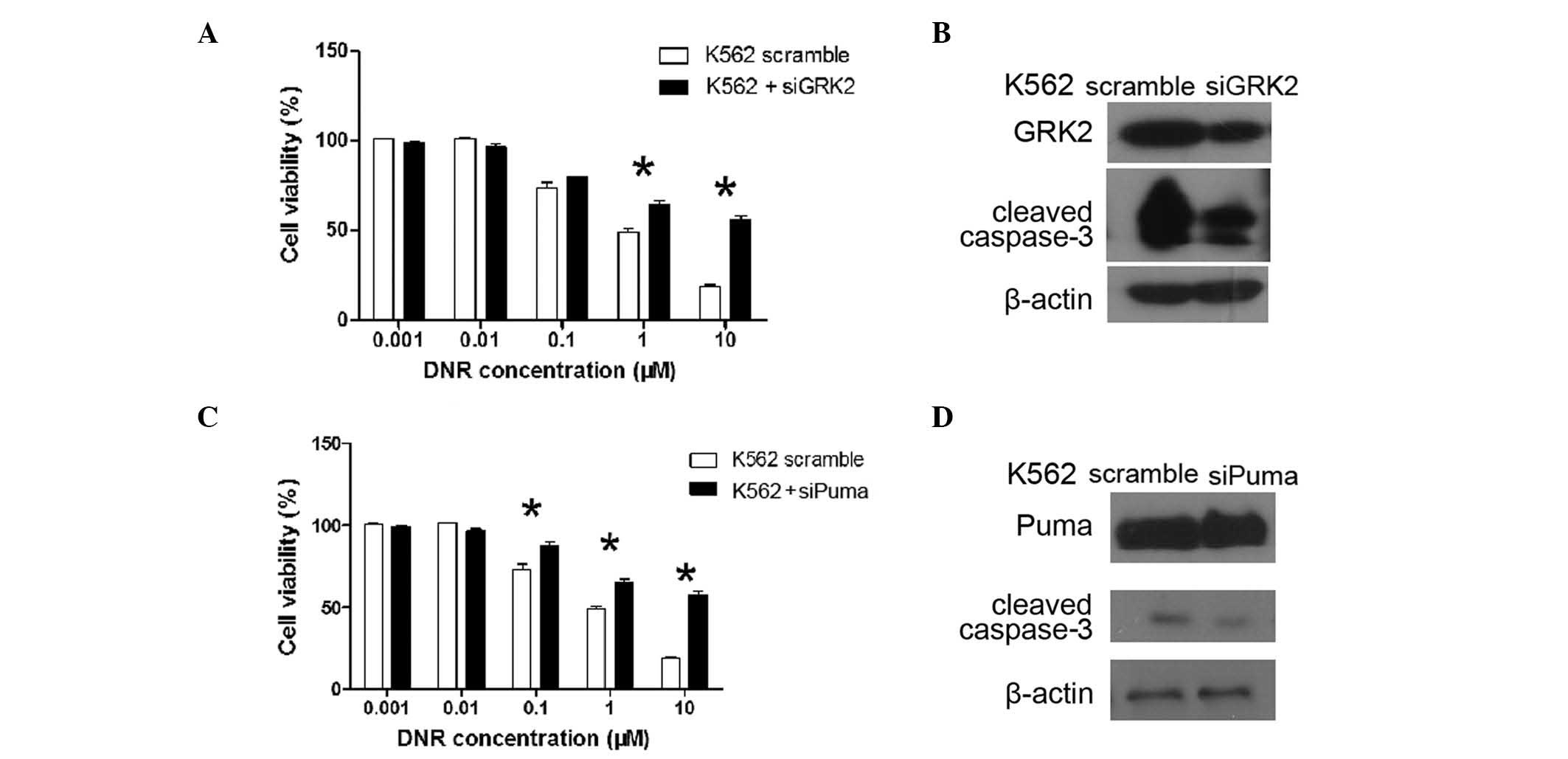
GRK2 and PUMA siRNA was used to silence the
expression of GRK2 and PUMA in K562 cells, respectively. GRK2
expression was observed to be significantly decreased upon
transfection with GRK2 siRNA. Of note, K562 cells transfected with
GRK2 siRNA exhibited a similar survival pattern to cells
overexpressing miR-125b. The level of cleaved caspase-3 protein was
also decreased upon DNR treatment in the cells transfected with
GRK2 siRNA. Similarly, the expression of PUMA was significantly
decreased following transfection of PUMA siRNA, and these cells
exhibited a similar survival pattern to cells overexpressing
miR-125b. The level of cleaved caspase-3 protein was also observed
to decrease upon DNR treatment in PUMA siRNA-transfected cells.
These findings show that GRK2 and PUMA have a key role in DNR
resistance (Fig. 5), and indicate
that miR-125b contributes to DNR resistance through reducing the
expression of GRK2 and PUMA in leukemia cells.
Discussion
To the best of our knowledge, this is the first
study to show that miR-125b expression is significantly increased
in patients who undergo an event (P<0.05) in acute leukemia,
with the exception of those classified as M3 (promyelocytic
leukemia). Patients in remission may have a longer EFS than
patients who undergo an event. DNR-induced drug resistance is
associated with the upregulation of miR-125b in K562, THP-1 and
Jurkat cell lines. Furthermore, miR-125b may regulate the survival
of leukemia cells by targeting GRK2 and PUMA proteins and inducing
changes in apoptosis in K562, THP-1, Jurkat and REH cells.
Overexpression of miR-125b occurs in patients with
myelodysplasia, megakaryoblastic leukemia (15) and APL (18), as well as those with AML who carry
the t(2;11)(p21;q23) translocation (13). High expression of miR-125b is
correlated with treatment response, as well as relapse in pediatric
APL (18). miR-125b may predict
poor prognosis in non-small-cell lung (10) and colorectal cancer (12). The present study employed primary
patients and relapse or no-remission patients; therefore, the
average expression level of miR-125b was extremely high. Based on
induction therapy, the improved survival time of patients with APL
and the short follow-up duration in the present study, patients
with APL were not included in this investigation. In the present
study, event-free patients were observed to exhibit longer EFS than
therapy-failure patients. Although there was no significant
difference in OS between patients in remission and event patients,
patients in remission were found to have an increased survival
compared with other patients. These findings suggest that miR-125b
may be an important prognostic marker for patients with acute
leukemia.
The association between miR-125b and drug resistance
has been investigated in numerous types of human cancer. However,
contradictory data exist regarding the expression of miR-125b in
different tumor types, and miR-125b has been reported to have
different roles in drug sensitivity and drug resistance. miR-125b
is downregulated in resistant Ehrlich ascites tumor cells (25) and has been shown to increase the
apoptotic response to cisplatin treatment in breast cells (26). Conversely, miR-125b has been found
to be upregulated in Taxol-resistant cancer (9), doxorubicin-resistant Ewing sarcoma
(27), cisplatin-resistant ovarian
cancer (28) and AML (18) cells. In the present study, miR-125b
upregulation was found to increase cell survival compared with that
in the control cells, whereas miR-125b downregulation was observed
to decrease cell survival. These findings indicate that high
expression of miR-125b induces resistance to DNR treatment.
The mechanism underlying patient resistance to
chemotherapeutic treatment is yet to be elucidated. Therefore,
identifying the cause of drug resistance and developing biomarkers
are critical. Anthracyclines have been shown to intercalate with
DNA and indirectly inhibit the activity of the enzyme topoisomerase
II, resulting in DNA strand breaks and induction of apoptosis
(29). In the present study, it
was hypothesized that miR-125b induced DNR resistance in leukemia
cells through regulating apoptosis. Previous studies have shown
that members of the B-cell lymphoma (Bcl)-2 family and other
factors involved in apoptosis are important targets of miR-125.
Numerous proteins have been shown to affect apoptosis, including
anti-apoptotic members of the Bcl-2 family, such as Bcl-w (30), Bcl-2 (31), myeloid cell leukemia sequence 1
(30,32) and Bcl-2-antagonist/killer (Bak)-1
(9,28,30,32),
acting as the Bcl-2 homologous antagonist, and pro-apoptotic
targets, including P53 (33),
tumor protein p53-inducible nuclear protein 1 (TP53INP1) (34), tumor necrosis factor α-induced
protein 3 (35) and p38α (36). High miR-125 expression has been
shown to downregulate Bak-1 and TP53INP1, and consequently protects
cells from apoptosis and promotes tumorigenesis (37). In the present study, GRK2 and PUMA
were identified to be direct targets of miR-125b in leukemia cells.
GRK2 and PUMA protein expression was found to be downregulated by
miR-125b in K562, THP-1 and Jurkat cells. Furthermore, miR-125b
upregulation inhibited DNR-induced apoptosis in K562, THP-1 and
Jurkat cells. GRK2 and PUMA protein expression was also upregulated
by miR-125b in REH cells. Downregulation of miR-125b increased cell
apoptosis following treatment with DNR. These results indicate that
miR-125b downregulated GRK2 and PUMA, which inhibited apoptosis and
induced leukemia cell resistance to DNR. However, the detailed
mechanism underlying this action is yet to be elucidated.
In conclusion, this study has shown that DNR
resistance is associated with miR-125b upregulation and the
consequent downregulation of the miR-125b target genes GRK2 and
PUMA. Therefore, upregulation of miR-125b may inactivate the
caspase pathway and inhibit apoptosis, and dysregulation of
miR-125b may lead to the acquisition of DNR resistance in AML. Of
note, miR-125b expression was found to be associated with disease
development and treatment response. These results suggest that
miR-125b may represent a potential biomarker for predicting the
effect of chemotherapy in patients with leukemia.
Acknowledgements
This study was supported by a grant from the
Shanghai Jiaotong University Affiliated Shanghai First People’s
Hospital (no. 061138). The authors would like to thank the staff of
the Department of Hematology of Shanghai Jiaotong University
Affiliated First People’s Hospital.
References
|
1
|
Kimby E, Nygren P and Glimelius B;
SBU-group. Swedish Council of Technology Assessment in Health Care.
A systematic overview of chemotherapy effects in acute myeloid
leukaemia. Acta Oncol. 40:231–252. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tallman MS, Gilliland DG and Rowe JM: Drug
therapy for acute myeloid leukemia. Blood. 106:1154–1163. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pulte D, Gondos A and Brenner H: Expected
long-term survival of patients diagnosed with acute myeloblastic
leukemia during 2006–2010. Ann Oncol. 21:335–341. 2010.
|
|
4
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lim LP, Lau NC, Garrett-Engele P, et al:
Microarray analysis shows that some microRNAs downregulate large
numbers of target mRNAs. Nature. 433:769–773. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lu J, Getz G, Miska EA, et al: MicroRNA
expression profiles classify human cancers. Nature. 435:834–838.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim JK, Noh JH, Jung KH, et al: Sirtuin7
oncogenic potential in human hepatocellular carcinoma and its
regulation by the tumor suppressors MiR-125a-5p and MiR-125b.
Hepatology. 57:1055–1067. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhou M, Liu Z, Zhao Y, et al:
MicroRNA-125b confers the resistance of breast cancer cells to
paclitaxel through suppression of pro-apoptotic Bcl-2 antagonist
killer 1 (Bak1) expression. J Biol Chem. 285:21496–21507. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yuxia M, Zhennan T and Wei Z: Circulating
miR-125b is a novel biomarker for screening non-small-cell lung
cancer and predicts poor prognosis. J Cancer Res Clin Oncol.
138:2045–2050. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kappelmann M, Kuphal S, Meister G,
Vardimon L and Bosserhoff AK: MicroRNA miR-125b controls melanoma
progression by direct regulation of c-Jun protein expression.
Oncogene. 32:2984–2991. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nishida N, Yokobori T, Mimori K, et al:
MicroRNA miR-125b is a prognostic marker in human colorectal
cancer. Int J Oncol. 38:1437–1443. 2011.PubMed/NCBI
|
|
13
|
Bousquet M, Quelen C, Rosati R, et al:
Myeloid cell differentiation arrest by miR-125b-1 in
myelodysplastic syndrome and acute myeloid leukemia with the
t(2;11)(p21;q23) translocation. J Exp Med. 205:2499–2506. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Enomoto Y, Kitaura J, Hatakeyama K, et al:
Eμ/miR-125b transgenic mice develop lethal B-cell malignancies.
Leukemia. 25:1849–1856. 2011.
|
|
15
|
Klusmann JH, Li Z, Böhmer K, et al:
miR-125b-2 is a potential oncomiR on human chromosome 21 in
megakaryoblastic leukemia. Genes Dev. 24:478–490. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gefen N, Binder V, Zaliova M, et al:
Hsa-mir-125b-2 is highly expressed in childhood ETV6/RUNX1
(TEL/AML1) leukemias and confers survival advantage to growth
inhibitory signals independent of p53. Leukemia. 24:89–96. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Calin GA, Liu CG, Sevignani C, et al:
MicroRNA profiling reveals distinct signatures in B cell chronic
lymphocytic leukemias. Proc Natl Acad Sci USA. 101:11755–11760.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang H, Luo XQ, Feng DD, et al:
Upregulation of microRNA-125b contributes to leukemogenesis and
increases drug resistance in pediatric acute promyelocytic
leukemia. Mol Cancer. 10:1082011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Schotte D, De Menezes RX, Akbari Moqadam
F, et al: MicroRNA characterize genetic diversity and drug
resistance in pediatric acute lymphoblastic leukemia.
Haematologica. 96:703–711. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Döhner H, Estey EH, Amadori S, et al;
European LeukemiaNet. Diagnosis and management of acute myeloid
leukemia in adults: recommendations from an international expert
panel, on behalf of the European LeukemiaNet. Blood. 115:453–474.
2010.PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Starczynowski DT, Kuchenbauer F,
Argiropoulos B, et al: Identification of miR-145 and miR-146a as
mediators of the 5q-syndrome phenotype. Nat Med. 16:49–58. 2010.
View Article : Google Scholar
|
|
23
|
Hughes MS, Yu YY, Dudley ME, et al:
Transfer of a TCR gene derived from a patient with a marked
antitumor response conveys highly active T-cell effector functions.
Hum Gene Ther. 16:457–472. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Stark A, Brennecke J, Russell RB and Cohen
SM: Identification of Drosophila MicroRNA targets. PLoS
Biol. 1:397–409. 2003.
|
|
25
|
Husted S, Søkilde R, Rask L, et al:
MicroRNA expression profiles associated with development of drug
resistance in Ehrlich ascites tumor cells. Mol Pharm. 8:2055–2062.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rajabi H, Jin C, Ahmad R, McClary C, Joshi
MD and Kufe D: Mucin 1 oncoprotein expression is suppressed by the
miR-125b oncomir. Genes Cancer. 1:62–68. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Iida K, Fukushi J, Matsumoto Y, et al:
miR-125b develops chemoresistance in Ewing sarcoma/primitive
neuroectodermal tumor. Cancer Cell Int. 13:212013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kong F, Sun C, Wang Z, et al: miR-125b
confers resistance of ovarian cancer cells to cisplatin by
targeting pro-apoptotic Bcl-2 antagonist killer 1. J Huazhong Univ
Sci Technolog Med Sci. 31:543–549. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Richardson DS and Johnson SA:
Anthracyclines in haematology: preclinical studies, toxicity and
delivery systems. Blood Rev. 11:201–223. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gong J, Zhang JP, Li B, et al:
MicroRNA-125b promotes apoptosis by regulating the expression of
Mcl-1, Bcl-w and IL-6R. Oncogene. 32:3071–3079. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shi L, Zhang S, Feng K, et al:
MicroRNA-125b-2 confers human glioblastoma stem cells resistance to
temozolomide through the mitochondrial pathway of apoptosis. Int J
Oncol. 40:119–129. 2012.PubMed/NCBI
|
|
32
|
Balakrishnan A, Stearns AT, Park PJ, et
al: Upregulation of proapoptotic microRNA mir-125a after massive
small bowel resection in rats. Ann Surg. 255:747–753. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zeng CW, Zhang XJ, Lin KY, et al:
Camptothecin induces apoptosis in cancer cells via
microRNA-125b-mediated mitochondrial pathways. Mol Pharmacol.
81:578–586. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jiang F, Liu T, He Y, et al: MiR-125b
promotes proliferation and migration of type II endometrial
carcinoma cells through targeting TP53INP1 tumor suppressor in
vitro and in vivo. BMC Cancer. 11:4252011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kim SW, Ramasamy K, Bouamar H, Lin AP,
Jiang D and Aguiar RC: MicroRNAs miR-125a and miR-125b
constitutively activate the NF-κB pathway by targeting the tumor
necrosis factor alpha-induced protein 3 (TNFAIP3, A20). Proc Natl
Acad Sci USA. 109:7865–7870. 2012.PubMed/NCBI
|
|
36
|
Tan G, Niu J, Shi Y, Ouyang H and Wu ZH:
NF-κB-dependent microRNA-125b up-regulation promotes cell survival
by targeting p38α upon ultraviolet radiation. J Biol Chem.
287:33036–33047. 2012.
|
|
37
|
Bousquet M, Nguyen D, Chen C, et al:
MicroRNA-125b transforms myeloid cell lines by repressing multiple
mRNA. Haematologica. 97:1713–1721. 2012. View Article : Google Scholar : PubMed/NCBI
|