Introduction
Signal transducers and activators of transcription 5
(STAT5) can be activated by numerous cytokines and growth factors
in various cell lines and tissues. Furthermore, STAT5 has a variety
of regulatory roles, which control different cell functions,
including growth, survival, differentiation and invasion (1). However, the role of STAT5 in the
pathogenesis of glioblastoma multiforme (GBM) has not been examined
fully and the function of hepatocyte growth factor (HGF), the
physiological activator of STAT5, in stimulating STAT5 in GBM cells
remains to be elucidated. The present study, to the best of our
knowledge, demonstrated for the first time that HGF induces
phosphorylation of STAT5 at Tyr-694/699, nuclear translocation of
STAT5 and increases proliferation of the U87-MG cell line. To
directly assess the biological significance of STAT5 signaling in
GBM cells, using small interfering RNA (siRNA) to deplete STAT5 in
the human GBM cell line (U87-MG) and to inhibit the HGF-induced
STAT5 activation, changes in cell viability and cell cycle
progression were examined. Changes in the expression of several
genes, including Cyclin D1, p21 and p27, that are directly
associated with cell cycle regulation were also assessed. The aim
of the present study was to determine the role of STAT5 signaling
in GBM progression and to test the hypothesis that STAT5 signaling
may serve as a therapeutic target.
Materials and methods
Cell culture
The human U87-MG cell line was obtained from the
Cell Bank of Type Culture Collection of the Chinese Academy of
Sciences (CBTCCCAS; Shanghai, China). The human U87-MG cell line
was cultured in Dulbecco’s modified Eagle’s medium (DMEM;
Gibco-BRL, Carlsbad, CA, USA) supplemented with 2.0 g/l sodium
bicarbonate and 10% fetal bovine serum (FBS; Gibco-BRL) in a
humidified atmosphere containing 5% CO2 and 95% air at
37°C. For experimental purposes, confluent cultures of U87-MG cells
were serum-starved for 12 h prior to treatment with 40 ng/ml HGF
(R&D Systems, Minneapolis, MN, USA).
Cell stimulation with HGF
Confluent cultures of U87-MG cells were washed twice
with DMEM without serum, equilibrated in the same medium at 37°C
for at least 30 min and then collected at different time-points (0,
15, 30, 60, 120, 180 and 240 min) following HGF treatment. A total
of 2×106 cells was grown in 60 mm dishes containing 4 ml
of DMEM for each experimental condition.
Transient transfection of STAT5
siRNA
siRNA oligos for knockdown of endogenous STAT5
proteins were prepared by using the ON-TARGETplus SMARTpool
siRNA from Dharmacon, Inc. (Lafayette, CO, USA). Cells were
transfected with STAT5 siRNA (100 nM) by using the DharmaFECT siRNA
transfection reagent (Dharmacon, Inc.) according to the
manufacturer’s instructions. ON-TARGETplus non-targeting siRNA
(Dharmacon, Inc.) was used as a negative control (control siRNA)
and the selective silencing of STAT5 was confirmed by western blot
analysis.
Cellular protein preparation and western
blot analysis
U87-MG cells were treated as described. Nuclear cell
protein for studying p-STAT5 was extracted with the ProteoJET™
Cytoplasmic and Nuclear Protein Extraction kit (Fermentas, Vilnius,
Lithuania) according to the manufacturer’s instructions. Total cell
protein was extracted with the Total Cell Protein Extraction kit
(Millipore, Billerica, MA, USA). Protein concentrations were
determined using the Coomassie (Bradford) protein assay kit (Thermo
Fisher Scientific, Waltham, MA, USA). Equal amounts of protein were
separated by SDS-PAGE using 8% separating gels followed by transfer
to nitrocellulose membranes. Following transfer, membranes were
blocked using 5% non-fat dried milk in phosphate-buffered saline
(PBS; pH 7.2) and incubated overnight at 4°C with the primary
antibody (pAb), including STAT5, p-STAT5a/b, Cyclin D1, p21, p27
and β-actin (1:1,000 dilution; Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA). The membranes were washed three times with PBS,
0.1% Tween 20 and then incubated with secondary Abs (horseradish
peroxidase-conjugated, goat antibodies to rabbit and goat
antibodies to mouse; Santa Cruz Biotechnology, Inc., Santa Cruz,
CA, USA; dilution of 1:5,000) for 2 h at 24°C. Following washing
three times with PBS/0.1% Tween 20, the immunoreactive bands were
visualized using enhanced chemiluminescence detection reagents.
Autoradiograms were scanned and the labeled bands were quantified
using the Sigma-Gel software (Sigma, St. Louis, MO, USA).
RNA isolation and quantitative polymerase
chain reaction (qPCR)
Total RNAs from cells were extracted and cDNA
synthesis and amplification were performed as described previously
(2). Primers were designed as:
p21Cip1 forward, 5′-CGATGCCAACCTCCTCAACGA-3′ and
reverse, 5′-TCGCAGACCTCCAGCATCCA-3′; p27Kip1 forward,
5′-TGCAACCGACGAT TCTTCTACTCAA-3′ and reverse,
5′-CAAGCAGTGATGTATCTGATAAACAAGGA-3′; CyclinD1 forward,
5′-AACTACCTGGACCGCTTCCT-3′ and reverse, 5′-CCACTTGAGCTTGTTCACCA-3′;
GAPDH forward, 5′-GACTCATG ACCACAGTCCATGC-3′ and reverse,
3′-AGAGGCAGGGATGATGTTCTG-5′. Comparative qPCR was performed in
triplicate, including no template controls. Relative expression was
calculated using the comparative Ct method.
MTT assay for cell viability
U87-MG cells (5×103 per well) were
incubated in 96-well plates each containing 200 μl of medium and
cultured overnight in growth medium. Then the culture medium was
replaced with DMEM supplemented with 10% FBS and HGF (40 ng/ml). To
assay the effect of STAT5 knockdown on cell proliferation, cells
were transfected in 96-well plates. The rate of cellular
proliferation was measured every 24 h for 96 h. At the end of each
time-point, 20 μl of 5 mg/ml of MTT (Sigma) was added to each well
and incubated for 4 h at 37°C. Following removal of the culture
medium from each well, 150 μl of dimethylsulfoxide was added to the
MTT-treated wells and the absorption at 570 nm was determined using
an ELISA spectrophotometer (model 3550; Bio-Rad Laboratories,
Richmond, CA, USA). Each experimental condition was conducted in
triplicate.
Cell cycle analysis
Approximately 1×106 cells were harvested
at specified time-points, washed twice with PBS and fixed in cold
ethanol for 12 h at 4°C and then incubated with propidium iodide
for 30 min. Cells were treated with siRNA as described. Following
72 h, the cells were added to a conical tube and spun at 1,000 × g
for 3 min. The pellet was vortexed at a low speed and 0.5 ml of
cold PBS was added. Then, the pellet was vortexed again for 2–3 sec
and resuspended in 5 ml of cold PBS. The cells were centrifuged for
6 min at 1,000 × g and the PBS was then aspirated off. Cold PBS
(0.5 ml) was added and pipetted up and down to achieve a single
cell suspension. The tube was prepared with 4.5 ml ice-cold 100%
ethanol. Cells (0.5 ml) were permeablized by adding ice-cold 100%
methanol slowly to pre-chilled cells whilst gently vortexing.
Subsequently, cells were incubated on ice or at 4°C for 12 h and
then stained with propidium iodide. Thereafter, cells were analyzed
using a flow cytometer (BD FACSCanto II; BD Biosciences Franklin
Lakes, NJ, USA).
Reporter gene assay
The pGL3-CyclinD1 vector and the control vector were
prepared as described previously (3). Briefly, 0.4 μg of reporter gene
constructs was transfected into U87-MG cells using Lipofectamine
(Invitrogen Life Technologies, Carlsbad, CA, USA) reagent according
to the manufacturer’s instructions. This transfection was performed
concurrently with the transfection of STAT5 siRNA. Cells
co-transfected with pRL-TK served as controls. The results are
expressed as the percentage of relative luciferase activity of the
control group without HGF stimulation, which was set to 1.
Immunofluorescence microscopy
U87-MG cells were grown on coverslips, washed with
serum free-DMEM and treated with 40 ng/ml HGF for 1 h at 37°C. The
cells were fixed for 10 min with 4% paraformaldehyde in PBS,
permeabilized with 0.5% Triton X-100 in PBS and blocked for 1 h in
5% fetal bovine serum in PBS. Cells were incubated overnight at 4°C
with p-STAT5 Ab (1:100 dilution), as indicated, followed by
fluorescein isothiocyanate (FITC)-labelled anti-rabbit secondary Ab
for 2 h. Finally, Hoechst (1:1,000 dilution) was used for
nonspecific staining of the nucleus. Cells were viewed using a
confocal microscope (Leica, Mannheim, Germany).
Tissue samples and patients
Tumor specimens were obtained from patients admitted
for diagnosis and treatment at the Fourth Affiliated Hospital of
Harbin Medical University (Harbin, China). The diagnosis was made
according to World Health Organization criteria. The present study
was approved by the ethics committee of the Fourth Affiliated
Hospital of Harbin Medical University and was based on the criteria
of the Helsinki convention. Approval for use of the tissue in the
present study was obtained from the institutional review board.
Fresh surgical samples from glioma patients and non-neoplastic
brain tissues (temporal lobectomy from epilepsy surgery) were
immediately snap-frozen in liquid nitrogen upon surgical removal.
The formalin-fixed, paraffin-embedded archival tissue blocks were
retrieved and the matching HE-stained slides were screened for
representative tumor regions by a neuropathologist. The tissue
included 25 diffuse astrocytomas (grade II), 25 anaplastic
astrocytomas (grade III) and 50 glioblastomas (grade IV). In
addition, ten non-neoplastic brain tissues from epilepsy surgical
resections were also included.
Immunohistochemical (IHC) analysis
The perfused brains were cryoprotected in a solution
of 20% sucrose in 0.1 M of potassium phosphate buffer overnight.
The brain sections were cut on a freezing microtome (Leica SM2000
R) and mounted on gelatinized slides. The sections were dried at
40–50°C for 2 h and were maintained at −20°C until analysis. IHC
analysis followed as described briefly: The sections were incubated
at room temperature overnight with the primary antibody (p-STAT5,
1:100) diluted in PBS with Tween 20 (PBST). The negative controls
received only PBST. The slides were washed with PBST and incubated
with the secondary antibodies (1:1,000 in PBST) for 90 min. The
slides were washed again with PBST and incubated with
streptavidin-horseradish peroxidase (1:200 in PBST) for 60 min. The
reactions were developed with 0.04% 3,3′-diaminobenzidine
(DAB)+0.03% H2O2. The DAB reactions were
intensified with an OsO4 solution (0.04%) for 30 min.
The slides were counterstained with hematoxylin, dehydrated and
mounted with Permount. Results were visualized and images were
captured under a light microscope (Olympus BX-51; Olympus Optical,
Tokyo, Japan).
The degree of immunostaining of sections was viewed
and scored separately by two independent investigators, the scores
were determined by combining the proportion of positively stained
tumor cells and the intensity of staining. Scores from the two
investigators were averaged for further comparative evaluation of
the p-STAT5 expression. The proportion of positively stained tumor
cells was graded as described previously (4).
Statistical analysis
All the data are presented as the mean ± standard
deviation. All analyses were performed with one-way analysis of
variance using SPSS 13.0 software (SPSS, Inc., Chicago, IL, USA).
P<0.05 was considered to indicate a statistically significant
difference.
Results
HGF induces nuclear translocalization of
STAT5
Activated STATs are known to translocate from the
cytoplasm to the nucleus (5).
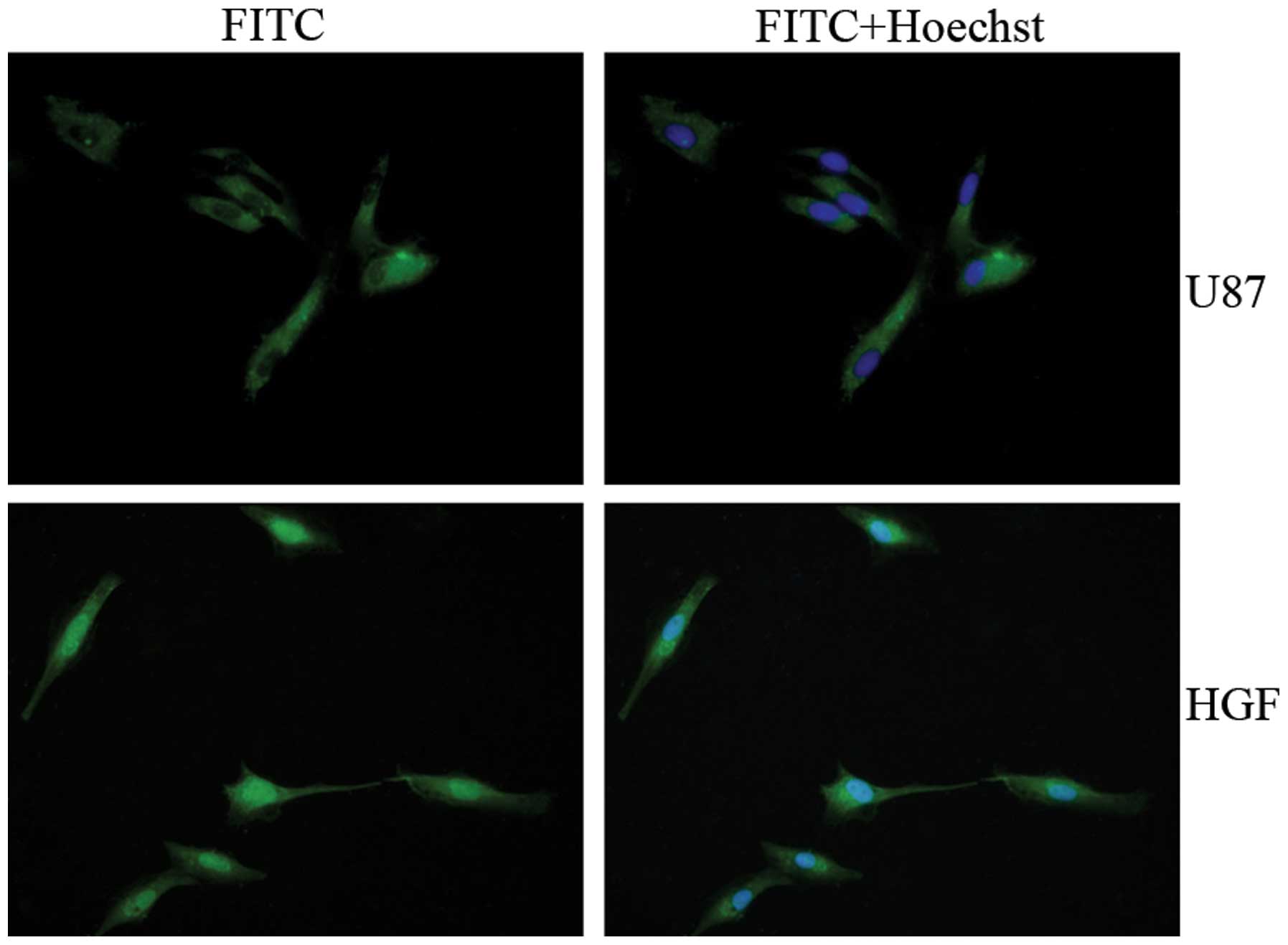
Immunofluorescent analysis of treated and untreated U87-MG cells
indicated that p-STAT5 accumulated in the nucleus with HGF
treatment (Fig. 1). HGF
preferentially activated STAT5, which subsequently translocated to
the nucleus. This observation prompted us to examine whether STAT5
was constitutively activated in glioma tissues.
STAT5 is constitutively activated in GBM
but inactivated in the U87-MG cell line
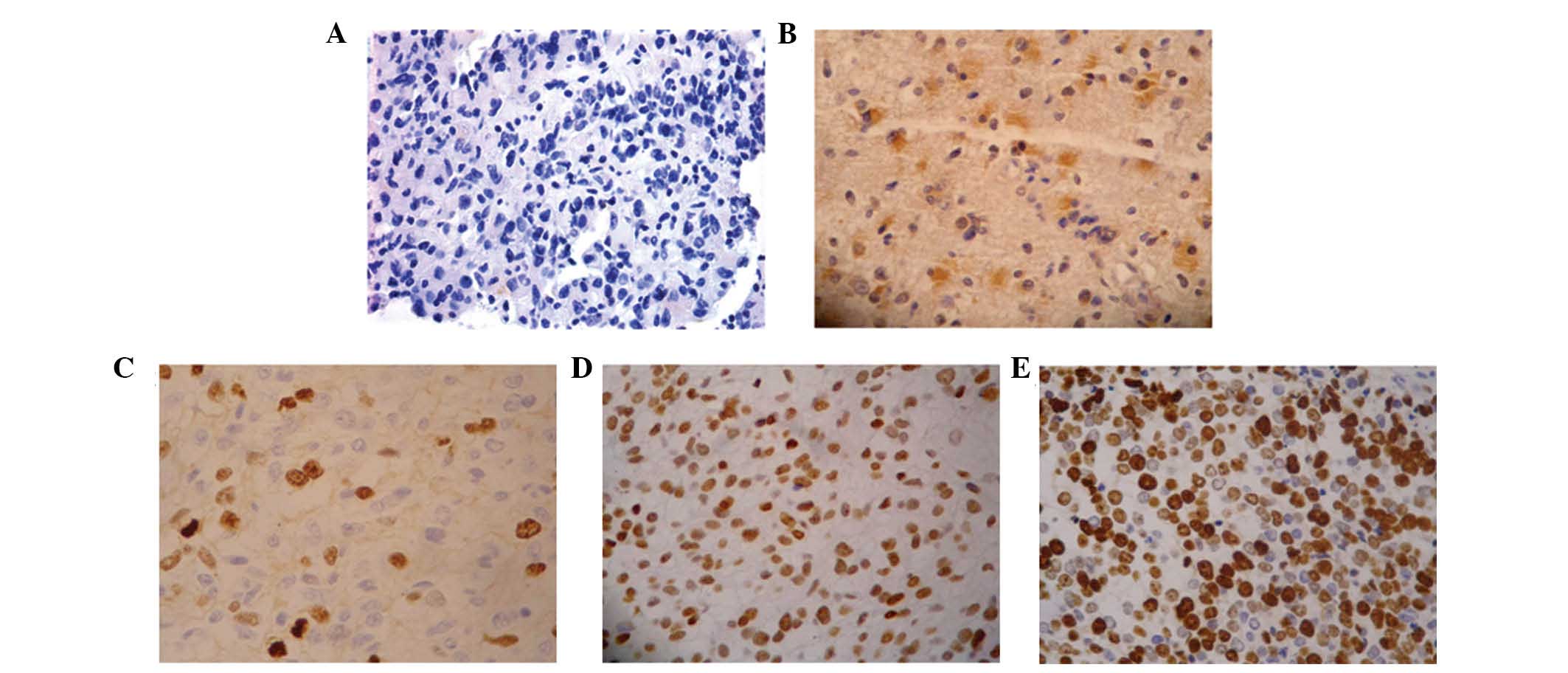
To determine whether STAT5 is constitutively
activated in glioma, the levels of p-STAT5 expression were assessed
by immunohistochemistry using anti-p-STAT5a/b (Tyr694/Tyr699)
antibody on glioma tissues. Among glioma samples 92% demonstrated
positive nuclear staining, whereas normal tissue did not stain. The
p-STAT5 staining was predominantly nuclear in vivo (Fig. 2). There was no significant
difference in constitutive activation frequency between low and
high grade gliomas. However, the expression levels of p-STAT5 were
significantly higher in high grade gliomas (grade III and grade IV)
compared with low grade gliomas (grade II) (P<0.05), which
supports the hypothesis that STAT5 activation is associated with
the progression of glioma.
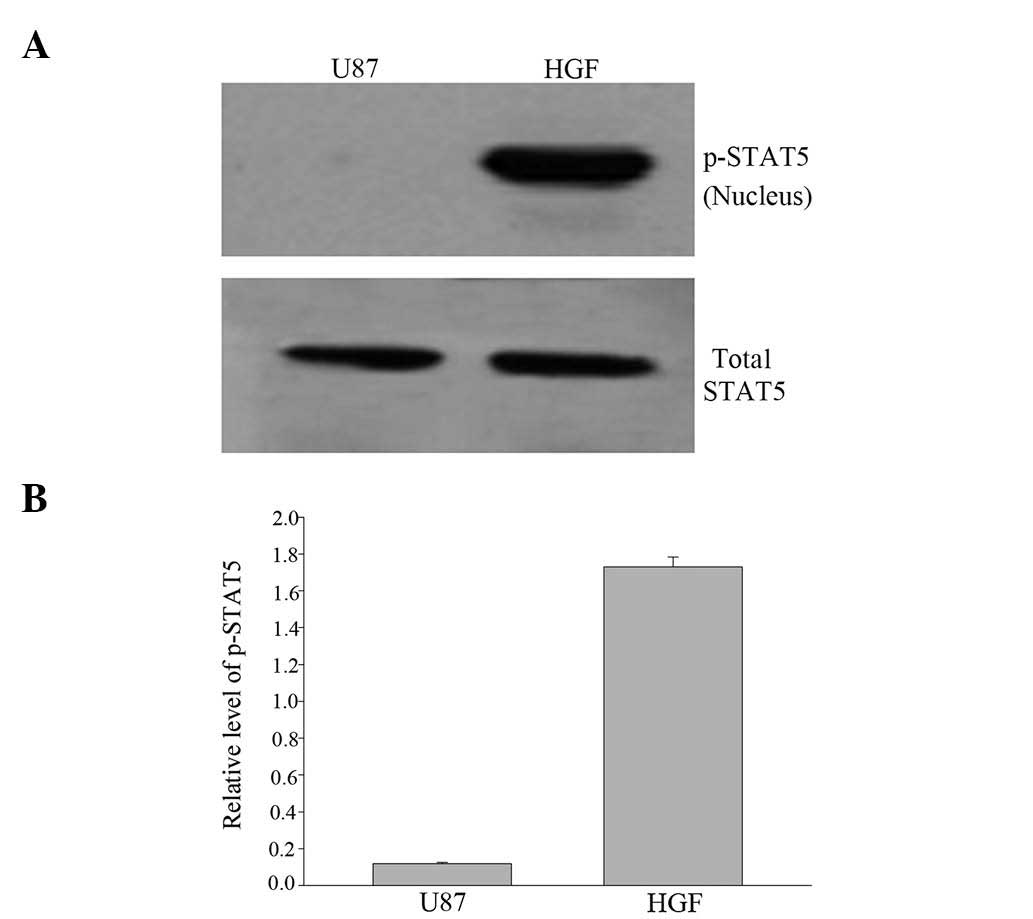
Western blot analysis with anti-STAT5 and
anti-p-STAT5 antibodies revealed that STAT5 was inactivated in the
U87-MG cell line but tyrosine-phosphorylated with HGF treatment
(Fig. 3).
HGF treatment induces STAT5 activation in
the U87-MG cell line
Western blot analysis of nuclear p-STAT5
demonstrated that p-STAT5 was not localized in the nuclei of
untreated cells. As shown in Fig.
4, treatment of cells with HGF induced an increase in STAT5
phosphorylation at Tyr-694/699, which reached a maximum within 15
min and declined toward base-line levels at 4 h of HGF treatment.
Western blot analysis with anti-STAT5 Ab confirmed that similar
amounts of STAT5 protein were present following treatment in the
absence or in the presence of HGF.
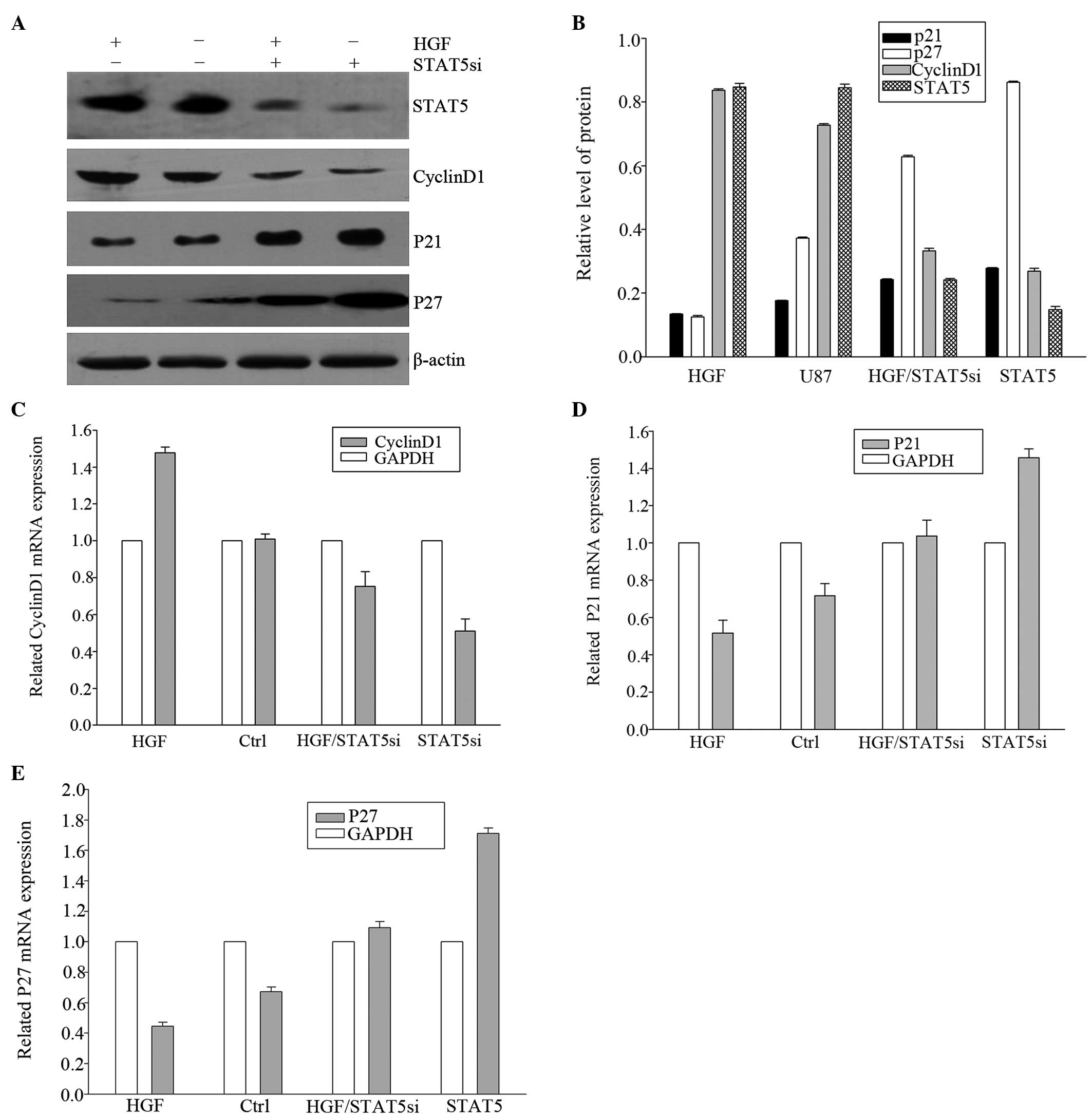
STAT5 siRNA inhibits the expression of
STAT5 and HGF-induced STAT5 phosphorylation in the U87-MG cell
line
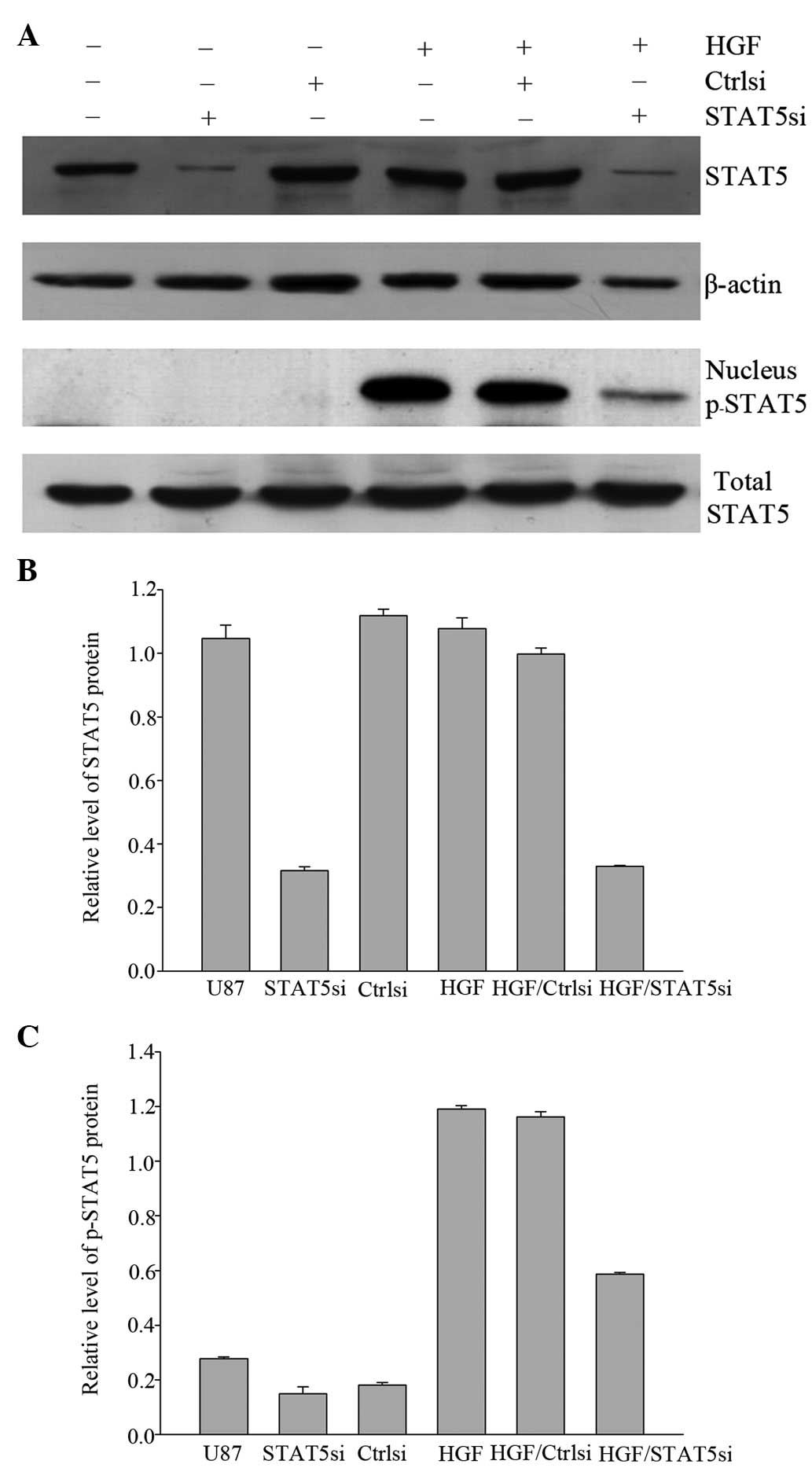
Fig. 5 presents
immunoblots revealing a decrease in STAT5 expression in cells
transiently transfected with STAT5 siRNA, whereas STAT5 expression
was not altered by control siRNA. As another control, the ability
of STAT5 siRNA to inhibit STAT5 activation induced by HGF was also
assessed. It was demonstrated that STAT5 siRNA effectively
inhibited HGF-induced STAT5 phosphorylation. These results
suggested that siRNA may be a useful tool in limiting STAT5
expression and activation.
HGF and STAT5 siRNA induce changes in
expression of downstream cell-cycle regulators at the
transcriptional level
To elucidate the effects of HGF and RNAi treatment
on STAT5 downstream genes, the expression of CyclinD1,
p21Cip1 and p27Kip1 were examined by qPCR and
western blot analyses. HGF was able to downregulate the expression
of p21Cip1 and p27Kip1 and upregulate
CyclinD1 at the transcriptional level. To confirm that the effect
of HGF on CyclinD1, p21Cip1 and p27Kip1
expression proceeds via STAT5 activation, these cell cycle
regulators were assessed in cells that were transiently transfected
with STAT5 siRNA. As displayed in Fig.
6, transfection of U87-MG cells with STAT5 siRNA resulted in
diminished CyclinD1 and upregulated p21Cip1 and
p27Kip1 RNA expression even in the presence of HGF,
suggesting that this effect is at least in part mediated through
STAT5.
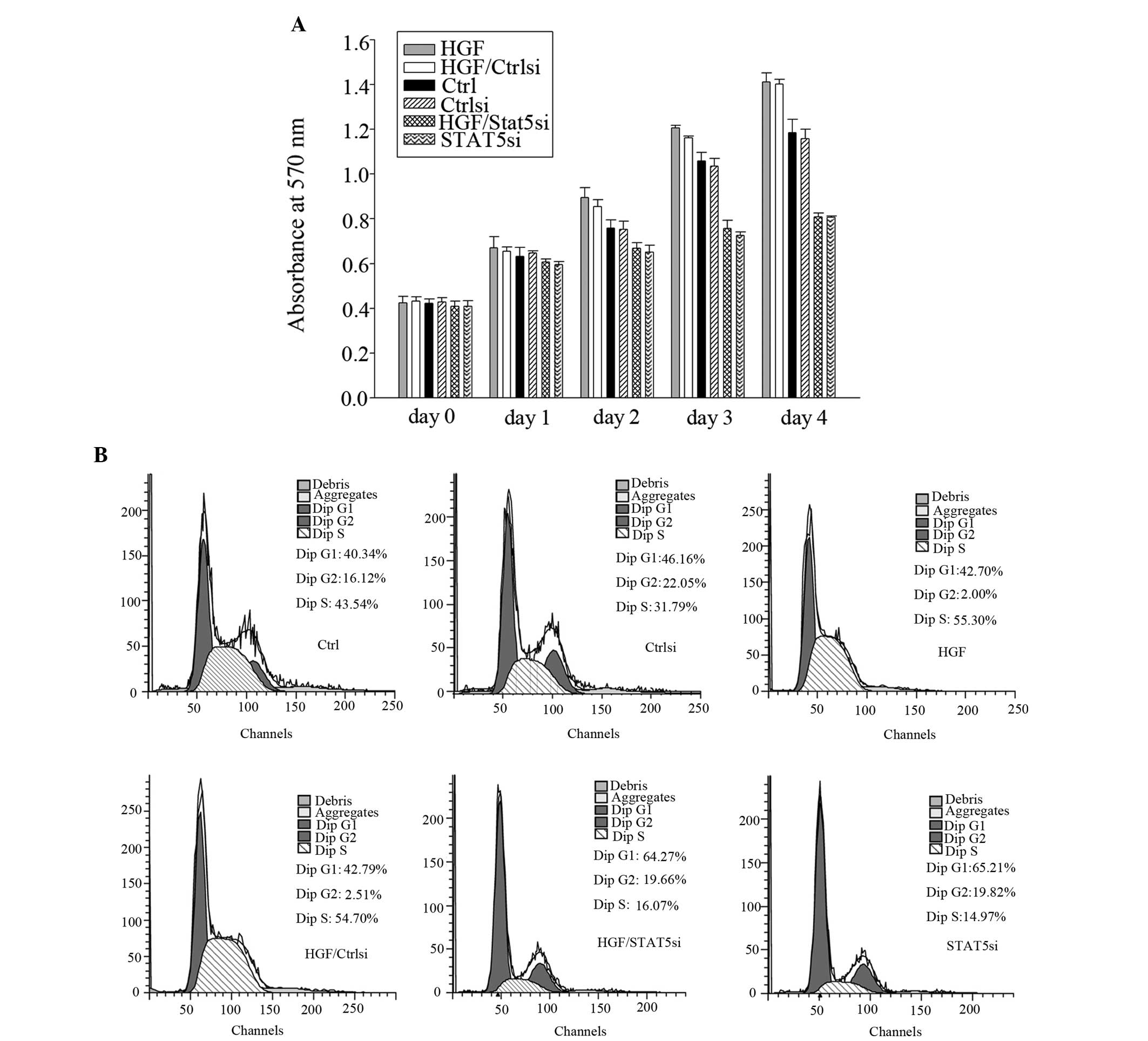
STAT5 is important in the proliferation
of U87-MG cells
The results obtained by the MTT assay suggested that
HGF is able to act as a growth factor in GBM cells (Fig. 7a). It was demonstrated that HGF
caused an increase in the cell number in U-87-MG cells during the
entire four-day incubation period. The increase in proliferation
for HGF was observed at a concentration of 40 ng/ml, which was
demonstrated to stimulate p-STAT5 protein expression. Transfecting
U87-MG cells with STAT5 siRNA resulted in a reduction in cell
number compared with the control. The viability of U87-MG cells was
significantly affected by STAT5 siRNA treatment. Additionally, the
viability of U87-MG cells following treatment with 40 ng/ml HGF was
significantly greater and this increase was suppressed by STAT5
siRNA, but not by control siRNA. The decrease in cell number caused
by STAT5 siRNA treatment of GBM cells implies that STAT5
participates in cell cycle progression, cell survival, or both.
Cell cycle analysis demonstrated that HGF treatment of U87-MG cells
increased the number of cells in S phase compared with the
untreated cells, and that this increase was suppressed by STAT5
siRNA, but not altered by control siRNA. Treating U87-MG cells with
STAT5 siRNA resulted in an increased proportion of cells in early
G1 phase. This may be due to a G1 phase cell cycle arrest (Fig. 7b).
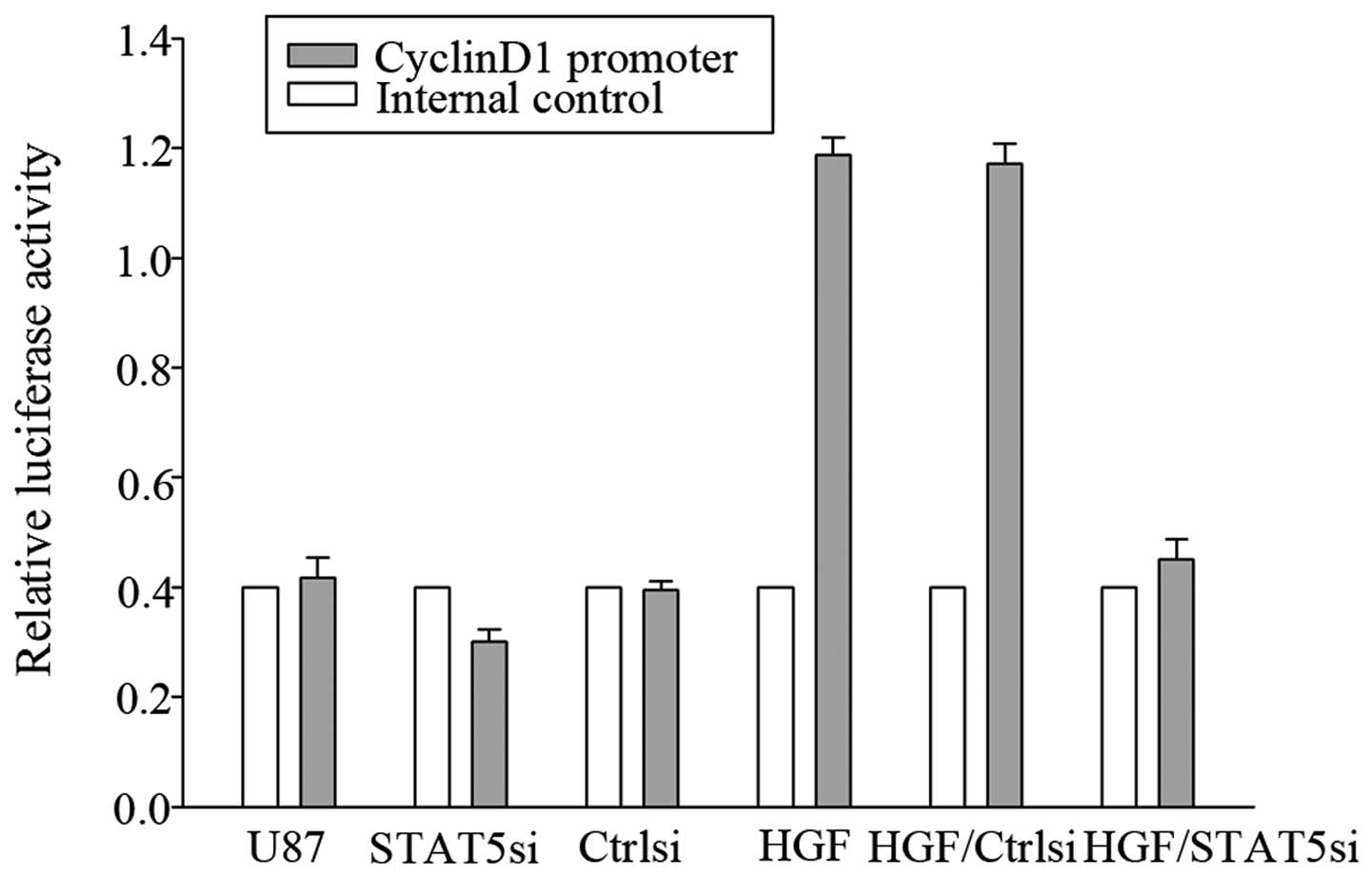
To evaluate whether CyclinD1 was a genuine target of
STAT5, a luciferase reporter assay was performed. As shown in
Fig. 8, co-transfection of STAT5
siRNA with the CyclinD1 reporter gene led to significantly
decreased CyclinD1 promoter activity, suggesting that STAT5 may
target CyclinD1.
Discussion
Persistent activation of STAT5 occurs in a number of
human cancer types, including multiple myelomas, breast, ovarian,
prostate carcinomas and head and neck tumors. Human tumors are
often characterized by an amplification of either growth factors or
cytokines, including interleukin (IL)-2 (6), IL-3 (7), IL-5 (7), IL-7 (8), granulocyte-macrophage
colony-stimulating factor (9),
insulin (10), erythropoietin
(11), thrombopoietin (11), growth hormone (11) and epidermal growth factor (2) that may lead to constitutive
activation of STAT5. The activation of STAT5 depends on
phosphorylation of a tyrosine residue (Tyr694/699) in the
c-terminal domain. Subsequent STAT5 translocation to the nucleus
results in transcriptional activation of a variety of genes,
including cell cycle regulators CyclinD1, CyclinD2,
p21Cip1 and p27Kip1 and antiapoptotic genes,
including B-cell lymphoma 2 and B-cell lymphoma 2 extra large
protein.
A number of studies indicated that STAT5 has a
pro-proliferative role in human hepatocellular liver carcinoma,
breast cancer, head and neck cancer, prostate cancer and lung
adenocarcinoma (12–17). Since STAT5 mediates cancer cell
proliferation, identification of factors that increase STAT5
activity is critical to identifying potential therapeutic targets.
The mechanisms of STAT5 tyrosine phosphorylation and
transcriptional activation may provide important insights into the
potential role of STAT5 in the process of tumorigenesis.
The present study demonstrated that STAT5 is
constitutively activated in GBM tumors. Furthermore, HGF was able
to induce tyrosine phosphorylation of Tyr-694/699 and
transcriptional activation of STAT5 in the U87-MG cell line. By
using immunofluorescence and western blot analysis, it was also
demonstrated that HGF induces nuclear translocation of STAT5 in the
U87-MG cell line. This is in agreement with a previous study, which
demonstrated that STAT is tyrosine phosphorylated in other cancer
cells stimulated by HGF (18).
Numerous studies demonstrated that the process of STAT5
phosphorylation is rapid and transient in vitro (19,20).
In accordance with these results, the present study demonstrated
that HGF promoted the nuclear translocation of STAT5, which was
able to be detected within 15 min, reached a maximum and declined
toward base-line levels following 4 h of treatment.
The present study revealed that HGF also exerted
proliferative effects in the majority of U87-MG cells. Flow
cytometric cell cycle analysis and cell counting revealed that HGF
increased the cell number and S-phase fraction. In parallel,
increased levels of CyclinD1 were observed, which may explain the
increase in cell proliferation. Since CyclinD1 is a downstream
target of STAT5, it was expected that increased activity of STAT5
may increase cell growth. Transfection of U87-MG cells with STAT5
siRNA resulted in a diminished effect of HGF on cell proliferation,
suggesting that STAT5 is required for HGF-induced cell
proliferation in GBM cells.
The characterization of HGF-induced tyrosine
phosphorylation and transcriptional activation of STAT5 led to the
following conclusions: HGF was able to induce the phosphorylation
of STAT5 on Tyr694/699 in the U87-MG cell line and the activated
STAT5 (p-STAT5) increased the proliferative ability in the U87-MG
cell line, which indicated that STAT5 was necessary for HGF-induced
proliferation. These findings suggested that STAT5 signaling may be
a new target for limiting GBM cell proliferation. qPCR analysis
determining which transcription units are altered by HGF in a
STAT5-dependent manner demonstrated that the expression of CyclinD1
was upregulated; however, p21Cip1 and p27Kip1
were diminished upon HGF exposure. These findings suggested a
molecular basis for the STAT5 dependence of HGF-mediated GBM
progression.
STAT5 pathways were silenced with siRNA in the
U87-MG cell line. Suppression of cell growth and a reduced cell
number were observed in U87-MG cells following silencing of STAT5.
These data support that the STAT5 pathway may serve as a
therapeutic target in GBM using siRNA to block the expression of
genes encoding STAT5. It has been demonstrated that interference of
the STAT5 signaling pathway led to the inhibition of cancer cell
proliferation. Silencing of STAT5 in U87-MG cells caused
corresponding changes in the cell cycle, which was blocked at G1
stage. These data supported that suppression of cell growth in GBM
cells is possibly due to the antagonizing effects of silencing
STAT5 on cell proliferation that is promoted by elevated STAT5
phosphorylation. Based on these findings, it is hypothesized that
GBM cell proliferation is the crucial factor for STAT5 involved in
GBM cell tumorigenesis. Certain studies agreed with this
hypothesis, which also observed an alteration in STAT5 expression
affecting cell proliferation in cancer cells (21,22).
In conclusion, the present study demonstrated for
the first time, to the best of our knowledge, that STAT5 was
associated with GBM proliferation. STAT5 activation was at least
partially mediated by HGF. The present study not only provided a
molecular basis for the role of STAT5 in GBM but also suggested a
novel therapeutic target for the treatment of GBM.
Acknowledgements
This study was supported by the Science Foundation
of Heilongjiang Health Department (no. 2011-157), the Science
Foundation of Heilongjiang Education Department (no. 12521268) and
the China Postdoctoral Science Foundation (no. 106941).
References
|
1
|
Buitenhuis M, Coffer PJ and Koenderman L:
Signal transducer and activator of transcription 5 (STAT5). Int J
Biochem Cell Biol. 36:2120–2124. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cao S, Wang C, Zheng Q, et al: STAT5
regulates glioma cell invasion by pathways dependent and
independent of STAT5 DNA binding. Neurosci Lett. 487:228–233. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang H, Li M, Han Y, et al:
Down-regulation of miR-27a might reverse multidrug resistance of
esophageal squamous cell carcinoma. Dig Dis Sci. 55:2545–2551.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang C, Cao S, Yan Y, et al: TLR9
expression in glioma tissues correlated to glioma progression and
the prognosis of GBM patients. BMC Cancer. 10:4152010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Heim MH: The Jak-STAT pathway: cytokine
signalling from the receptor to the nucleus. J Recept Signal
Transduct Res. 19:75–120. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hou J, Schindler U, Henzel WJ, Wong SC and
McKnight SL: Identification and purification of human Stat proteins
activated in response to interleukin-2. Immunity. 2:321–329. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mui AL, Wakao H, O’Farrell AM, Harada N
and Miyajima A: Interleukin-3, granulocyte-macrophage colony
stimulating factor and interleukin-5 transduce signals through two
STAT5 homologs. EMBO J. 14:1166–1175. 1995.
|
|
8
|
Foxwell BM, Beadling C, Guschin D, Kerr I
and Cantrell D: Interleukin-7 can induce the activation of Jak 1,
Jak 3 and STAT 5 proteins in murine T cells. Eur J Immunol.
25:3041–3046. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Barahmand-pour F, Meinke A, Eilers A,
Gouilleux F, Groner B and Decker T: Colony-stimulating factors and
interferon-gamma activate a protein related to MGF-Stat 5 to cause
formation of the differentiation-induced factor in myeloid cells.
FEBS Lett. 360:29–33. 1995. View Article : Google Scholar
|
|
10
|
Wartmann M, Cella N, Hofer P, et al:
Lactogenic hormone activation of Stat5 and transcription of the
beta-casein gene in mammary epithelial cells is independent of p42
ERK2 mitogen-activated protein kinase activity. J Biol Chem.
271:31863–31868. 1996. View Article : Google Scholar
|
|
11
|
Pallard C, Gouilleux F, Bénit L, et al:
Thrombopoietin activates a STAT5-like factor in hematopoietic
cells. EMBO J. 14:2847–2856. 1995.PubMed/NCBI
|
|
12
|
Joung YH, Lee MY, Lim EJ, et al: Hypoxia
activates the IGF-1 expression through STAT5b in human HepG2 cells.
Biochem Biophys Res Commun. 358:733–738. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sultan AS, Xie J, LeBaron MJ, Ealley EL,
Nevalainen MT and Rui H: Stat5 promotes homotypic adhesion and
inhibits invasive characteristics of human breast cancer cells.
Oncogene. 24:746–760. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Weaver AM and Silva CM: Modulation of
signal transducer and activator of transcription 5b activity in
breast cancer cells by mutation of tyrosines within the
transactivation domain. Mol Endocrinol. 20:2392–2405. 2006.
View Article : Google Scholar
|
|
15
|
Xi S, Zhang Q, Dyer KF, et al: Src kinases
mediate STAT growth pathways in squamous cell carcinoma of the head
and neck. J Biol Chem. 278:31574–31583. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xi S, Zhang Q, Gooding WE, Smithgall TE
and Grandis JR: Constitutive activation of Stat5b contributes to
carcinogenesis in vivo. Cancer Res. 63:6763–6771. 2003.PubMed/NCBI
|
|
17
|
Cao S, Yan Y, Zhang X, et al: EGF
stimulates cyclooxygenase-2 expression through the STAT5 signaling
pathway in human lung adenocarcinoma A549 cells. Int J Oncol.
39:383–391. 2011.PubMed/NCBI
|
|
18
|
Spiekermann K, Biethahn S, Wilde S,
Hiddemann W and Alves F: Constitutive activation of STAT
transcription factors in acute myelogenous leukemia. Eur J
Haematol. 67:63–71. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chin H, Nakamura N, Kamiyama R, Miyasaka
N, Ihle JN and Miura O: Physical and functional interactions
between Stat5 and the tyrosine-phosphorylated receptors for
erythropoietin and interleukin-3. Blood. 88:4415–4425.
1996.PubMed/NCBI
|
|
20
|
Pircher TJ, Petersen H, Gustafsson JA and
Haldosén LA: Extracellular signal-regulated kinase (ERK) interacts
with signal transducer and activator of transcription (STAT) 5a.
Mol Endocrinol. 13:555–565. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Koppikar P, Lui VW, Man D, et al:
Constitutive activation of signal transducer and activator of
transcription 5 contributes to tumor growth, epithelial-mesenchymal
transition, and resistance to epidermal growth factor receptor
targeting. Clin Cancer Res. 14:7682–7690. 2008. View Article : Google Scholar
|
|
22
|
Xiong H, Su WY, Liang QC, et al:
Inhibition of STAT5 induces G1 cell cycle arrest and reduces tumor
cell invasion in human colorectal cancer cells. Lab Invest.
89:717–725. 2009. View Article : Google Scholar : PubMed/NCBI
|