Introduction
Percutaneous coronary intervention and coronary
artery bypass grafting are the most important therapeutic
modalities for coronary heart disease. However, restenosis
occurring after coronary surgery or angioplasty seriously affects
the short- and long-term effects of the two treatments. Therefore,
restenosis has become an issue that should be addressed. In recent
years, a number of different therapies have been developed to treat
restenosis, but patients showed low response rates to all of these
(1). At present, effective
therapies for restenosis are still lacking. Previous studies
(2) have shown that the
proliferative and migrating abilities of vascular smooth muscle
cells (VSMCs) play a very important role in restenosis and are
regulated by an intricate signaling pathway network. Ras protein is
a critical component of numerous intertwined signal transduction
pathways and is a potentially important therapeutic target
(3). Ras protein can activate a
variety of signaling pathway proteins, such as mitogen-activated
protein kinases (MAPKs) and PI3K/Akt, to induce the proliferation
and migration of VSMCs. Therefore, Ras protein may play a crucial
role in restenosis. An intervening agent that can effectively
inhibit Ras protein activity would be thus highly suitable for the
development of preventive and therapeutic treatment for restenosis
occurring after coronary surgery or angioplasty. Aptamers have been
shown to effectively inhibit the activity of a variety of targets
in vitro and in vivo (4). They present a number of advantages
over antibodies and antibiotics. Being smaller than antibodies,
aptamers show improved cell penetration and blood clearance.
Aptamers have been used for pure recognition, inhibition,
diagnostic and therapeutic applications (5). To develop a high-affinity, specific
inhibitor of the Ras protein, we selected ssDNA aptamers using
systematic evolution of ligands by exponential enrichment (SELEX)
and analyzed their binding affinities and structures. The
high-affinity ssDNA aptamers identified in this study may be used
in the future to treat restenosis.
Materials and methods
Nucleic acids
The oligonucleotide library was synthesized as a
single-stranded (ssDNA), 78-bp long DNA fragment, with the
following sequence: 5′-GGGAGCTCAGAATAAACG
CTCAA-N35-TTCGACATGAGGCCCGGATC-3′, where N35
is random oligonucleotides based on the equal incorporation of A,
G, C and T at each position. The initial double-stranded DNA
(dsDNA) random library was generated by amplification using
polymerase chain reaction (PCR) and the primers: forward,
5′-GGGAGCTCAGAATAAACGCTCAA-3′ and reverse,
5′-GATCCGGGCCTCATGTCGAA-3′. The oligonucleotide library and the
primers were synthesized in vitro by Shanghai Sangon
Biological Engineering Co., Ltd. (Shanghai, China).
Ras protein
Recombinant, full-length human wild-type Ras protein
(H-Ras) was purchased from Abcam Ltd. (Hong Kong, China).
Optimization of PCR conditions
Symmetric PCR reactions were performed as follows:
PCR mixtures contained 1 pmol ssDNA library, 10 pmol forward
primer, 10 pmol reverse primer, 10 μl of 2X PCR Master mix (0.05
U/μl Taq DNA polymerase in reaction buffer, 4 mM MgCl2,
0.4 mM dATP, 0.4 mM dCTP, 0.4 mM dGTP and 0.4 mM dTTP) and
dsH2O, in a total volume of 20 μl. The mixtures were
thermally cycled 5, 8, 12, 16, 20, 24 and 28 times through 94°C for
3 min, 94°C for 40 sec and X°C for 30 sec, followed by a 7-min
extension at 72°C. X was the annealing temperature, which was 53.3,
53.6, 54.4, 55.7, 57.1, 58.6, 60.0, 61.4, 62.9, 64.2, 65.0 and
65.3°C, respectively. dsDNA PCR products were electrophoresed on a
3% agarose gel and the appropriate number of cycles and annealing
temperature for symmetric PCR were selected based on their
electrophoretic profiles. The asymmetric PCR conditions were then
optimized. The asymmetric PCR mixtures contained 1 μl dsDNA
template, 100, 80, 50 and 30 pmol of forward primer, 1 pmol reverse
primer, 10 μl 2X PCR Master mix and dsH2O, in a total
volume of 20 μl. The annealing temperature was 60°C (defined from
optimization of the symmetric PCR reactions) and the number of
heating cycles was 30, 35 and 40. The remaining conditions were
identical to the symmetric PCR reactions. ssDNA PCR products were
electrophoresed on a 3% agarose gel and electrophoretic profiles
were examined in order to choose the most appropriate concentration
of the forward primer and the appropriate number of heating cycles
for the asymmetric PCR.
In vitro selection
To initiate in vitro selection, 100 μl of Ras
protein and 100 μl sterile dsH2O (to remove
non-specifically bound DNA) were coated in Nunc™ 96-well plates at
4°C for 18 h using 100 μl coating buffer (0.05 mol/l
NaHCO3, pH 9.6), and were respectively labelled A and B.
After washing, the plates were blocked with 200 μl 3% bovine serum
albumin (BSA) at 37°C for 45 min. After washing, 100 μl of the
random ssDNA library were first added to B and incubated with 100
μl binding buffer (20 mM HEPES pH 7.35, 120 mM NaCl, 5 mM KCl, 1 mM
MgCl2 and 1 mM CaCl2) at 37°C for 45 min. The
mixture was then transferred to A and incubated at 37°C for 45 min.
A was washed six times with 200 μl washing buffer (binding buffer +
0.05% Tween-20). A total of 200 μl eluting buffer (20 mM Tris-HCl,
4 mol/l guanidine thiocyanate and 1 mM DTT, pH 8.3) was added to A
and incubated for 8 min in a water bath with temperature fixed at
80°C. The eluent was collected and DNA was extracted using the
UNIQ-10 DNA extraction kit (Shanghai Sangon Biological Engineering
Co., Ltd.). The amount of bound ssDNA was quantified on an
ultraviolet spectrophotometer. The bound ssDNA was amplified by
asymmetric PCR and PCR products were electrophoresed on a 3%
agarose gel. The corresponding bands were cut out and DNA fragments
from these were extracted using the EZ-10 Spin Column DNA Gel
extraction kit (Bio Basic Inc., Toronto, Canada). The concentration
of the extract was determined on an ultraviolet spectrophotometer
(UV762; Shanghai Jingke Scientific Instrument Co., Ltd., Shanghai,
China). This extract constituted the ssDNA pool for the following
round of selection, and was stored at −20°C. The selection and
amplification steps were reiterated until the binding rates and
affinities of the ssDNA pool no longer significantly increased.
Table I shows the doses of the
aptamer pools and Ras protein in each round of selection.
 | Table IDoses of aptamer pools and Ras
proteins in each round of selection. |
Table I
Doses of aptamer pools and Ras
proteins in each round of selection.
| Round | ssDNA (pmol) | Ras protein
(pmol) | ssDNA/Ras |
|---|
| 1 | 1,000 | 100 | 10 |
| 2 | 500 | 50 | 10 |
| 3 | 200 | 20 | 10 |
| 4 | 100 | 10 | 10 |
| 5 | 100 | 7.5 | 13 |
| 6 | 50 | 3.75 | 13 |
| 7 | 50 | 2.5 | 20 |
| 8 | 30 | 1.5 | 20 |
| 9 | 30 | 1.2 | 25 |
| 10 | 20 | 0.8 | 25 |
| 11 | 20 | 0.6 | 33 |
Binding rates and affinities between
aptamer pools and Ras protein
The rates of aptamer pool binding to Ras were
measured using the same method as in the in vitro selection,
with the following differences in the used quantities: 3 pmol (50
μl) of Ras protein or 50 μl sterile dsH2O were coated,
using 50 μl of coating buffer; 100 μl 3% BSA was used for blocking;
100 pmol (50 μl) random ssDNA was added to B and incubated with 50
μl binding buffer; A was washed with 100 μl washing buffer and 100
μl of eluting buffer was added to A. The binding rate was defined
as the proportion of bound ssDNA relative to 100 pmol of total
ssDNA.
The affinities of aptamer pool binding to Ras were
measured by ELISA. Each pool of aptamers was initially amplified by
asymmetric PCR using biotin-labeled primers to obtain
biotin-labeled ssDNA. The same procedure as for that used to
estimate the binding rates was followed, using the same quantities
of each reagent until the step of washing the A solution. Following
this step, a total of 100 μl streptavidin-peroxidase (1:10,000) was
added to A and incubated at 37°C for 60 min. After washing three
times using 100 μl washing buffer, A was incubated with the
3,3′,5,5′-tetramethylbenzidine (TMB) substrate provided in the
ELISA kit (Baosheng, Beijing, China). The optical density (OD) of
each sample was then measured at 450 nm on an ELISA reader (Victor
3 1420 Multilabel Counter; PerkinElmer, Waltham, MA, USA). The
equation Y = Bmax × X/(Kd + X) was used to calculate the
target-antigen binding equilibrium dissociation constant (Kd),
where Bmax is the maximum OD, Y is the mean OD and X is
the molar concentration of ssDNA.
Cloning and sequencing of selected
aptamers
The 11th pool of aptamers was cloned. Plasmid DNA
was isolated from individual clones, purified and analyzed by
sequencing. This procedure provided a total of 50 individual
aptamers.
Aptamer sequence analysis
The primary and secondary structures of individual
aptamers were analyzed based on their sequences using the DNAMAN
5.29 software (Lynnon Corp., Quebec, QC, Canada).
Binding affinity between individual
aptamers and Ras protein
Biotin-labeled individual aptamers were synthesized
based on their sequences. The binding affinities between individual
aptamers and Ras were measured by ELISA, followed by calculation of
Kd based on OD values. The same steps described above were
followed.
Results
Optimization of PCR conditions
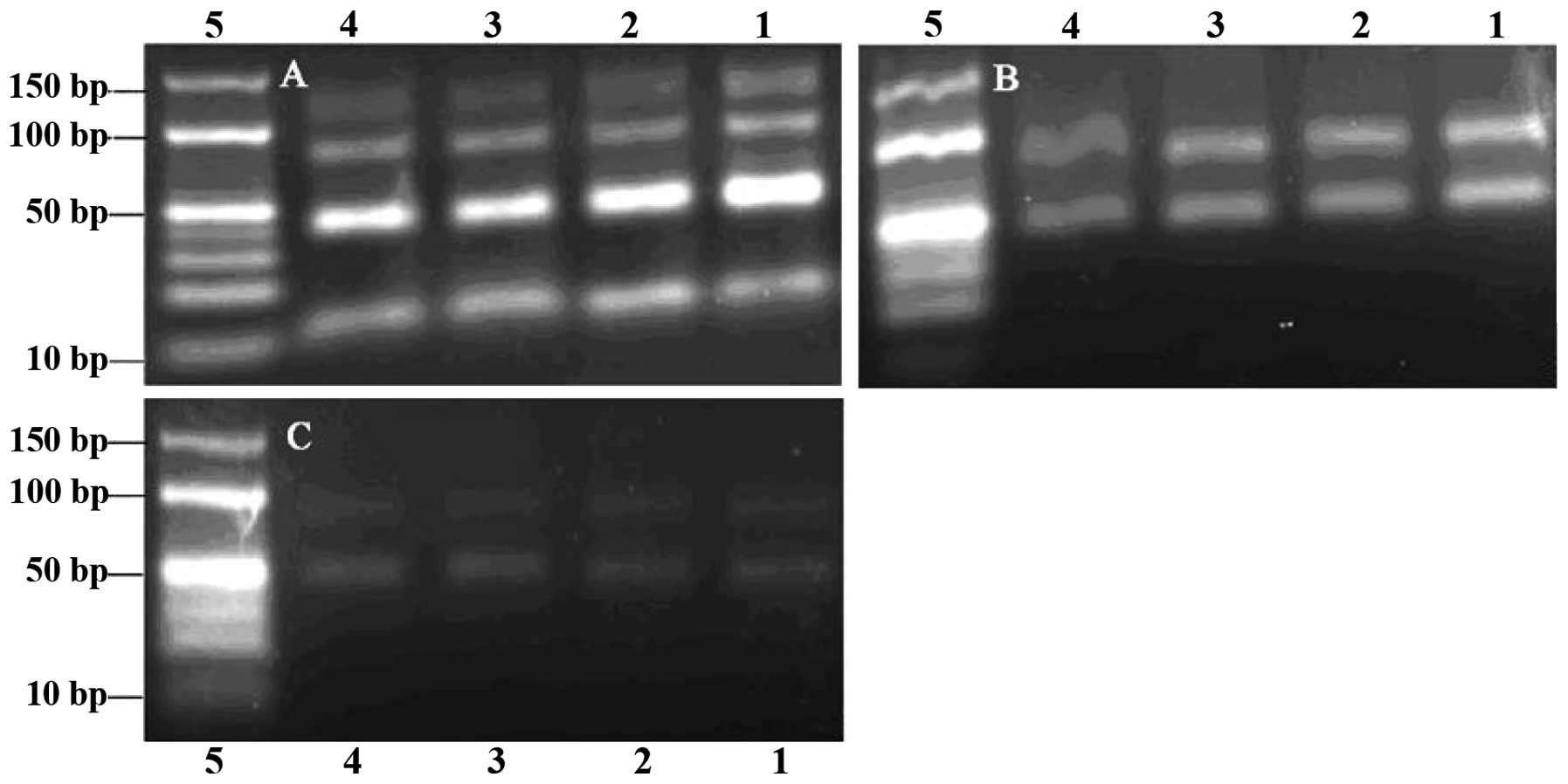
In the agarose electrophoresis gels, the 78-bp ssDNA
fragment migrated between the 40- and 50-bp bands of the dsDNA
ladder, and the 78-bp dsDNA near the 80-bp band. PCR products were
successfully obtained in symmetric PCR reactions when the number of
heating cycles was 5–24 (Fig. 1A and
B), while more PCR by-products were observed when this number
was >28 (Fig. 1C). Visible
bands were obtained from the PCR reactions performed with annealing
temperatures 53.3–65.3°C, while when this temperature was >60°C,
the PCR product quantities began to decrease (Fig. 1C). Since the selection would fail
if an extremely high number of PCR by-products were formed, and
because we aimed to obtain a high number of specific PCR products,
we chose an annealing temperature of 60°C and 24 heating cycles as
the most appropriate conditions, in order to obtain the maximum
number of specific PCR products with a minimal number of PCR
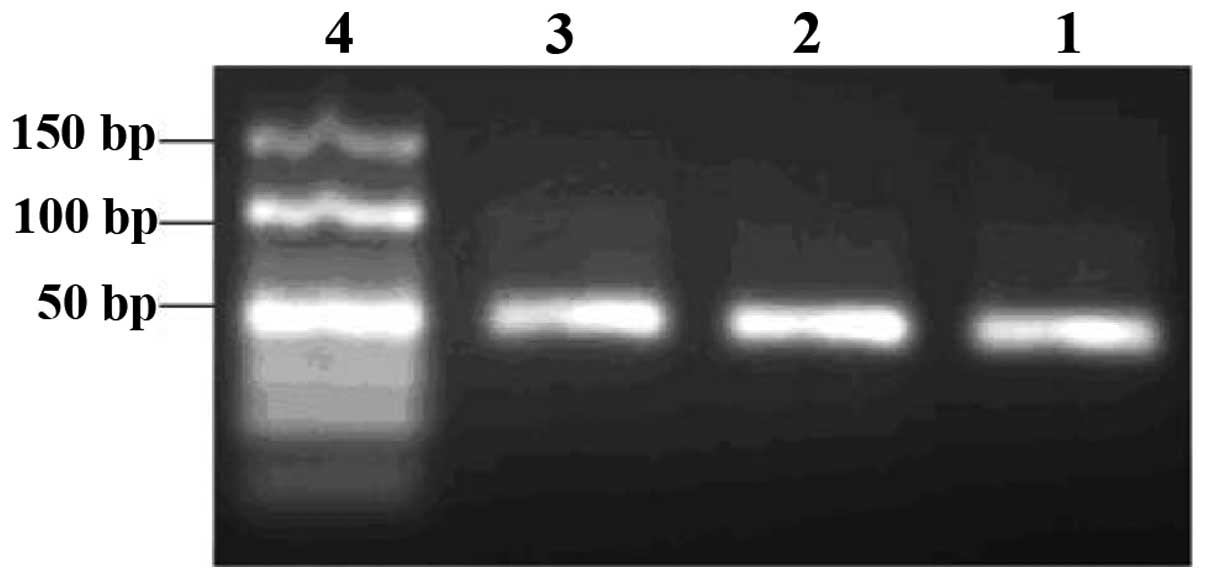
by-products formed. Similarly, based on the gel examination of
products from the indirect asymmetric PCR (ssDNA was amplified by
symmetric PCR to dsDNA and then dsDNA was amplified by asymmetric
PCR to ssDNA; Fig. 2), we chose a
100:1 proportion for the forward and reverse primers, and 40
heating cycles. The direct asymmetric PCR amplification (ssDNA was
amplified by asymmetric PCR to ssDNA) gave only non-specific
by-products and not ssDNA.
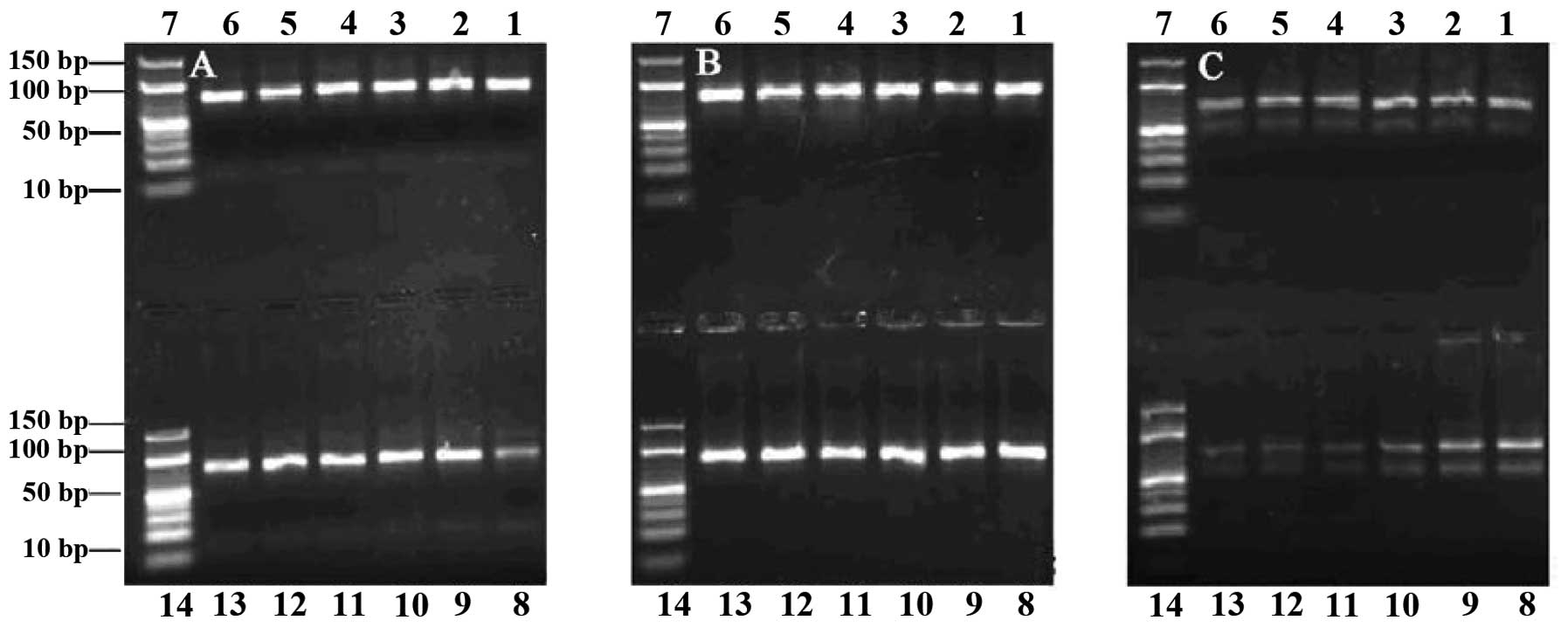 | Figure 1Agarose gel (3%) electrophoresis
analysis of polymerase chain reaction (PCR) products. Number of
heating cycles: (A) 5, (B) 24 and (C) 28. Lanes 1–6 and 8–13,
products from PCR performed with annealing temperatures 53.3, 53.6,
54.4, 55.7, 57.1, 58.6, 60.0, 61.4, 62.9, 64.2, 65.0 and 65.3°C,
respectively. Lanes 7 and 14, DNA ladder. |
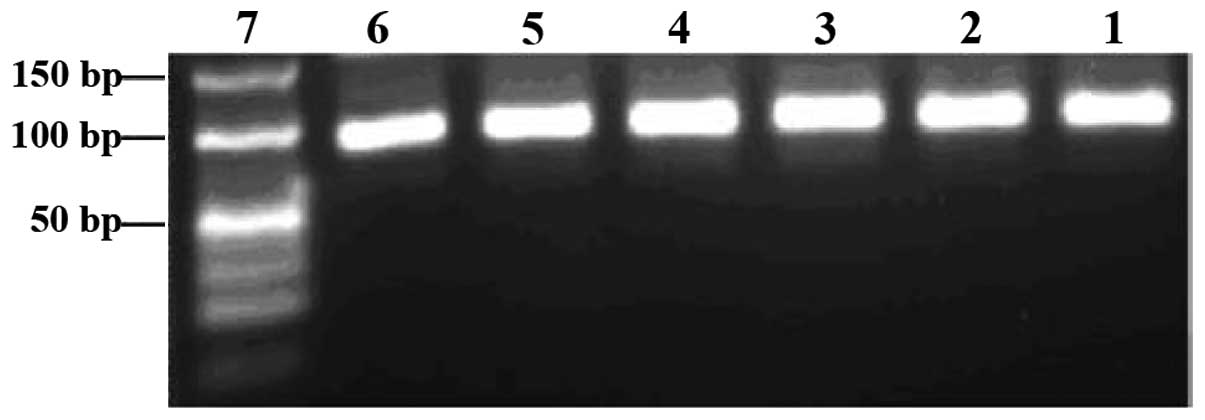
In vitro selection
To isolate aptamers that specifically bind to Ras
protein, we utilized Ras protein as a selection target. Sterile
dsH2O was used as a negative target control. The random
ssDNA library was used to screen ligands that bind to Ras protein.
Different pools of aptamers that bind to Ras with high affinity and
specificity were successfully obtained after 11 rounds of selection
by SELEX. Each pool of aptamers was examined before and after PCR
amplification on 3% agarose gels. The aptamer pools migrated
between the 40- and 50-bp bands of the dsDNA ladder, and their PCR
products near the 80-bp band of the dsDNA ladder (Figs. 3 and 4).
Binding rates and affinities between the
aptamer pools and Ras protein
The binding rates and affinities between the aptamer
pools and the Ras protein were calculated. The results showed that
the binding rates increased gradually from 2.4 to 34.5% along the
process of the in vitro selection. The binding affinities,
as evidenced by measured OD, increased gradually from 0.220 to
1.080. Kd was calculated based on the equation described in
‘Materials and methods’; results are shown in Table II. No further improvement in the
binding rates and affinities was observed after 11 rounds of
selection. The 11th aptamer pool showed the highest binding rate
(34.5%) and the highest binding affinity with an OD at 1.080 and a
18.3 nM Kd (Table II).
 | Table IIRas binding rates (%), optical density
(OD) and equilibrium dissociation constant (Kd; in nM) for aptamer
pools from different rounds of selection. |
Table II
Ras binding rates (%), optical density
(OD) and equilibrium dissociation constant (Kd; in nM) for aptamer
pools from different rounds of selection.
| Round | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
|---|
| Rate | 2.4 | 4.0 | 9.5 | 11.5 | 18.7 | 19.1 | 24.4 | 29.4 | 34.3 | 33.9 | 34.5 |
| OD | 0.220 | 0.358 | 0.445 | 0.498 | 0.638 | 0.702 | 0.819 | 0.917 | 1.012 | 0.998 | 1.080 |
| Kd | 51.5 | 48.6 | 47.1 | 47.4 | 39.4 | 39.9 | 32.6 | 24.8 | 19.8 | 19.6 | 18.3 |
Cloning and sequencing of selected
aptamers
The aptamer pool from the 11th round of selection
was cloned. Plasmid DNA was isolated from individual clones,
purified and analyzed by sequencing. A total of 50 individual
aptamers was thus obtained. The constant regions of these sequences
were all identical to the sequence initially designed. There were
45 sequences comprising 35 base pairs in the random region as
initially designed, and 5 mutated sequences, among which 4 deletion
mutations (one base pair deleted in three aptamers and two base
pairs in one) and 1 insertion mutation (one base pair). Two of the
50 sequences were identical to each other.
Aptamer sequence analysis
Following rejection of the 5 mutated sequences, the
45 retained sequences were grouped into 10 major families based on
their primary structures, analyzed by DNAMAN 5.29 software.
Conserved primary structure motifs were observed in aptamers Ra1 to
Ra44 grouped in families 1 to 9. Only the sequence of aptamer Ra45,
family 10, was not associated with those of the other aptamers
(Table III). The secondary
structure analysis revealed different conformations among aptamers,
but of a single type: stem-loop structure. These stem-loop
structures were located at 5′-, 3′-fixed or random sequences, and
consisted of 5′-, 3′-fixed, or fixed and random sequences. The most
conserved sequences were part of the stem-loop structure with
angles.
 | Table IIISequence of aptamers of different
families, grouped based on the presence of common motifs (in
bold). |
Table III
Sequence of aptamers of different
families, grouped based on the presence of common motifs (in
bold).
Binding affinity between individual
aptamers and Ras protein
A total of 45 biotin-labeled individual aptamers was
synthesized based on their respective sequences. The binding
affinities between individual aptamers and Ras were measured by
ELISA. The OD of the complexes ranged between 0.459 and 1.213 and
their Kd between 48.3 and 15.3 nM (Table IV). Aptamers Ra1 and Ra2 had the
highest affinity with an OD at 1.213 and a 15.3 nM Kd. Aptamer Ra27
had the lowest affinity with an OD at 0.459 and a 48.3 nM Kd. To
explore whether the binding affinity of aptamers to the Ras protein
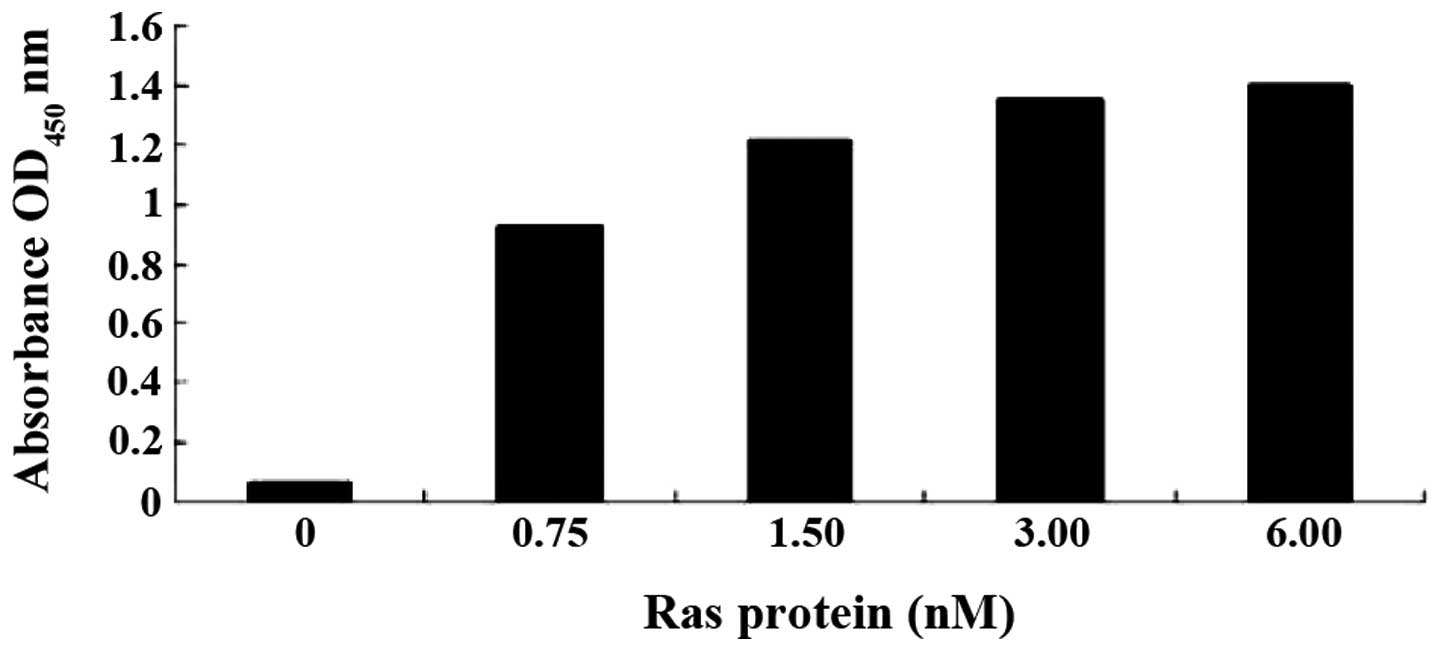
is dose-dependent, a series of mixtures with a fixed concentration
of aptamer (100 nM) and increasing concentrations of Ras (0, 0.75,
1.50, 3.00 and 6.00 nM) was prepared for ELISA assay. OD
measurements showed that the affinities of Ra1 (Fig. 5) or Ra2 (data not shown) aptamer
binding to the Ras protein are dependent on the aptamer dose.
 | Table IVOptical density (OD) and equilibrium
dissociation constant (Kd; in nM)) values of different
aptamers. |
Table IV
Optical density (OD) and equilibrium
dissociation constant (Kd; in nM)) values of different
aptamers.
| Aptamer | Ra1 | Ra1-mut | Ra27 | Ra27-mut |
|---|
| OD | 1.213 | 0.715 | 0.459 | 0.621 |
| Kd | 15.3 | 27.8 | 48.3 | 34.2 |
The data from binding affinity estimations were
subsequently mapped to the primary and secondary structures of the
corresponding aptamers. We found that most aptamers with high
binding affinity also had a high number of small stem-loops, while
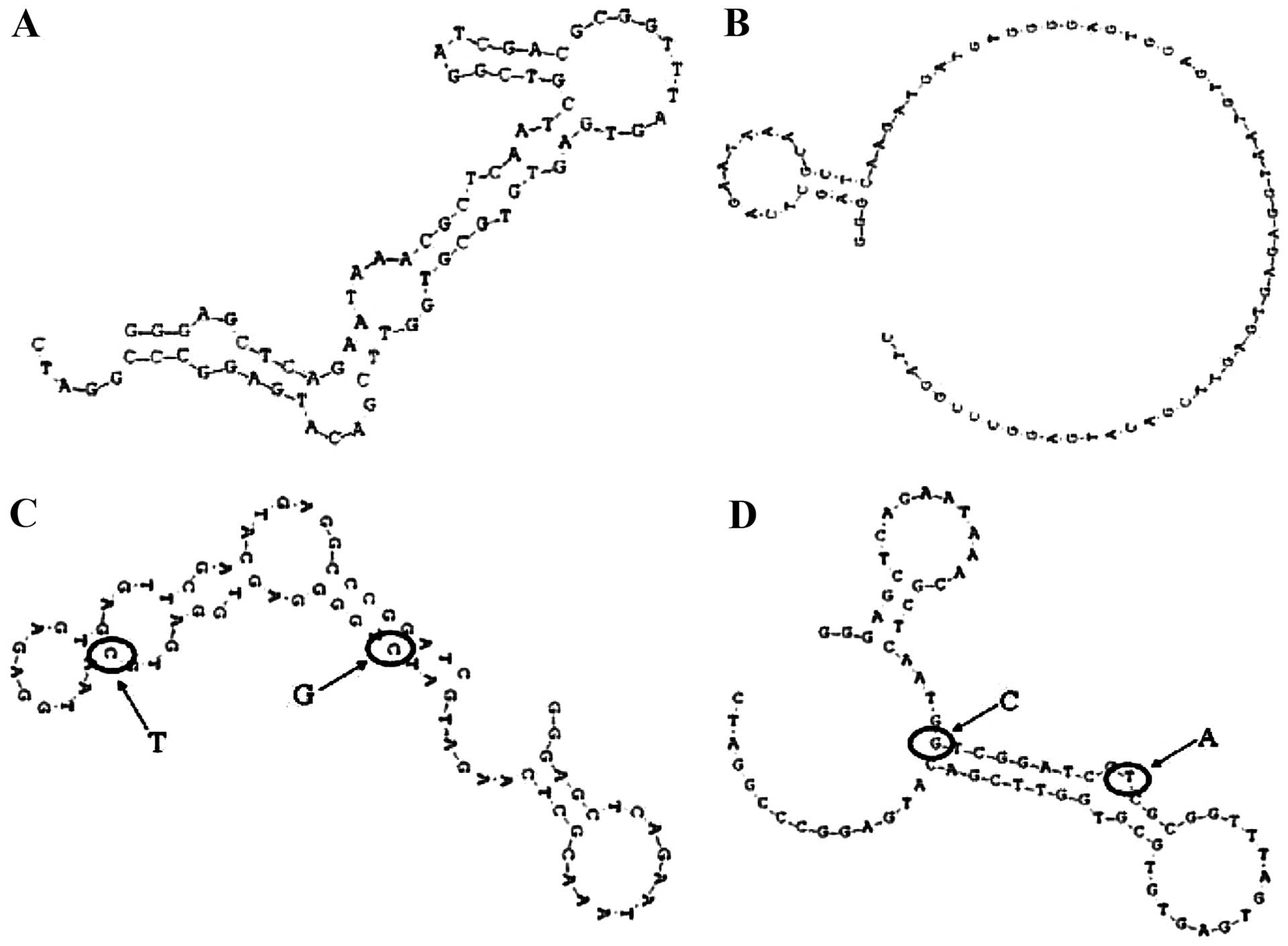
those with low affinity had fewer small stem-loops (Fig. 6). We therefore hypothesized that
the terminal loop of a stem-loop structure may be the site of
binding to the target Ras protein. To test this hypothesis,
aptamers Ra1-mut and Ra27-mut were synthesized, in which two bases
in the terminal loop of the stem-loop structure of Ra1 and Ra27
were mutated (Fig. 6C and D). The
Ra1- and Ra27-mut affinities of binding to the Ras protein were
then measured. The OD of the Ra1-mut aptamer was markedly decreased
compared to the original aptamer Ra1, while the opposite trend was
observed for aptamer Ra27-mut. Thus, the terminal loop of a
stem-loop structure might be the site of binding to the target Ras
protein.
Discussion
Restenosis after coronary surgery or angioplasty
remains an issue that needs to be solved (6–8).
In-stent restenosis (ISR) occurs in 15–50% of patients after
bare-metal stent implantation and still constitutes one of the most
common adverse events (9).
Although the use of drug-eluting stents has significantly reduced
ISR incidence rates, ISR persists as an unresolved clinical issue,
especially for patients presenting highly complex lesions (10,11).
A previous study showed that the proliferation and
migration of VSMCs play a very important role in restenosis
(12). Ras protein can activate
numerous signaling pathway proteins, such as MAPKs and PI3K/Akt, to
induce VSMC proliferation and migration (13). Using the SELEX technique, aptamers
that bind with high affinity and specificity to a wide range of
selected molecular targets can be efficiently isolated from large
randomized ssDNA libraries. Aptamers can be highly potent
antagonists of specific protein-protein interactions (14). Mutations in the KRAS gene
are required for the early occurrence and maintenance of
tumorigenesis and are the most frequently found mutations in
numerous types of human malignant diseases. Jeong et al
(15) identified and characterized
an RNA aptamer that specifically binds to the mutant KRAS protein,
which could be useful as a ligand for specific therapy and
diagnostic applications for cancer types promoted by mutations in
this protein. RNA aptamers with affinity for the Ras-binding domain
(RBD) of Raf-1 were also isolated from a degenerate pool by in
vitro selection. These aptamers efficiently inhibited the
interaction of Ras with the Raf-1 RBD, and inhibited Ras-induced
Raf-1 activation in a cell-free system (16). The Ras superfamily comprises 50
related proteins bearing a GTP-binding domain involved in signal
transduction. There are three types of Ras proteins in mammalian
cells, H-Ras, N-Ras and K-Ras. The most common of these is H-Ras.
Activated H-Ras protein is considered critical for cell
proliferation and differentiation (16). Therefore, we attempted to obtain
high-affinity ssDNA aptamers recognizing the H-Ras protein by SELEX
as a means to treat restenosis occurring after coronary surgery or
angioplasty.
In our study, Ras protein was first coated in Nunc
96-well plates, blocked using BSA and then the ssDNA library was
added along with binding buffer. The unbound ssDNA was removed and
the bound ssDNA, containing the aptamers, was eluted. Bound ssDNA
was amplified by asymmetric PCR for the next round of selection. By
repeating this process, the bound ssDNA was enriched and the
corresponding affinities increased. In the course of the in
vitro selection, we set up blank control groups in order to
eliminate false positives, i.e., ssDNA non-specifically bound to
the plates and/or to BSA. At the beginning of the selection
process, since the density of the aptamers was low, the used doses
of ssDNA and Ras were high and their proportions were low, in order
to isolate more ssDNA with high affinity. At later stages, because
the density of the aptamers had increased, the doses of ssDNA and
Ras had to be reduced and the rates had to increase. The ssDNA
molecules competitively bound to limited concentrations of the Ras
protein. Thus, high-affinity ssDNA molecules were enriched and
low-affinity ones were removed. Our results showed that the binding
rates increased from 2.4 to 34.5% along the selection process. The
binding affinities (based on OD) increased from 0.220 to 1.080,
while Kd decreased from 51.5 to 18.3 nM. No further improvement in
binding rates and affinities was observed after the 11th round of
selection. Sequences from this pool were cloned, and 50 clones were
sequenced. The results showed that aptamers with a high affinity to
Ras protein were successfully isolated.
ssDNA molecules, which fold into complex secondary
and tertiary conformations, including local regions of duplex
structures, may be distorted as a result of base mismatches,
bulges, pseudoknots and hairpins. The binding affinity of an ssDNA
is based on its primary and secondary structures. ssDNA molecules
fold, as a function of their primary structure, into complex
three-dimensional structures, providing a great diversity of
binding specificities. SELEX has been applied to a wide range of
targets including growth factors, enzymes and nucleic acids. The
aptamer NX-1838, an injectable angiogenesis inhibitor used to treat
macular degeneration-induced blindness, showed satisfactory
clinical effects (17).
Binding affinities between individual aptamers and
Ras were estimated by OD and Kd measurements, with OD ranging
between 0.459 and 1.213 and Kd between 48.3 and 15.3 nM. Aptamers
Ra1 and Ra2 showed the highest affinity with an OD of 1.213 and a
Kd of 15.3 nM. Aptamer Ra27 had the lowest affinity (OD = 0.459, Kd
= 8.3 nM). Based on primary structure analyses of these aptamers,
conserved primary structure motifs were found to be shared among
aptamers Ra1 to Ra44. Only aptamer Ra45 was not related to any of
the aptamers. The secondary structure analysis showed that the
conformations of the aptamers differed, but that all were of the
same type: stem-loop. The stem-loop was located at 5′-, 3′-fixed or
random sequences. The stem loops consisted of 5′-, 3′-fixed, or
fixed and random sequences. To investigate the intrinsic
relationship between the binding affinities and structures, we
compared the aptamers with regards to their binding affinities and
primary and secondary structures. We found that most of the
aptamers with high affinity (e.g., Ra1) had more small stem-loops,
while the low-affinity ones, such as Ra27, had fewer. The most
conserved sequences were located at stem-loops with angles. A
decrease in binding affinity was observed for the aptamer Ra1-mut
compared to Ra1, while the OD of Ra27-mut was increased compared to
Ra27. The aptamers Ra1-mut, with two mutated bases, and Ra27, had a
lower binding affinity compared to Ra1 and Ra27-mut, respectively.
The binding affinity of aptamer Ra1 to Ras was dose-dependent.
Therefore, these more small stem-loops might be the binding sites
of the aptamers to Ras, and conserved motifs within these sequences
might play an important role in binding. Li et al (18) also suggested that the stem-loops
may be the sites of protein-targeting aptamers, but it was unclear
where the conserved sequences located on their sequence. A future
study should investigate whether the function of Ras is inhibited
by the aptamers identified herein and how this affects VSMC
proliferative and migrating ability.
In summary, high-affinity ssDNA aptamers that can
bind to the Ras protein have been successfully selected. They could
serve as preventive and therapeutic agents for restenosis occurring
after coronary surgery or angioplasty in the future.
Acknowledgements
This study was supported by a grant from the
National Natural Science Foundation of China (no. 30872537).
References
|
1
|
Wan S, Shukla N, Yim A, et al: Orally
administered penicillamine is a potent inhibitor of neointimal and
medial thickening in porcine saphenous vein-carotid artery
interposition graft. J Thorac Cardiovasc Surg. 133:494–500. 2007.
View Article : Google Scholar
|
|
2
|
Liu Q, Lu Z, Zhou H, et al: The mechanical
study of vascular endothelial growth factor on the prevention of
restenosis after angioplasty. J Tongji Med Univ. 21:195–197. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jin G, Chieh-His Wu J, Li YS, et al:
Effects of active and negative mutants of Ras on rat arterial
neointima formation. J Surg Res. 94:124–132. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cerchia L, Ducongé F, Pestourie C, et al:
Neutralizing aptamers from whole-cell SELEX inhibit the RET
receptor tyrosine kinase. PLoS Biol. 3:e1232005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bunka DH and Stockley PG: Aptamers come of
age - at last. Nat Rev Microbiol. 4:588–596. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lu YG, Chen YM, Li L, Zhao RZ, Fu CH and
Yan H: Drug-eluting stents vs. intracoronary brachytherapy for
in-stent restenosis: a meta-analysis. Clin Cardiol. 34:344–351.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Reddy MA, Sahar S, Villeneuve LM, Lanting
L and Natarajan R: Role of Src tyrosine kinase in the atherogenic
effects of the 12/15-lipoxygenase pathway in vascular smooth muscle
cells. Arterioscler Thromb Vasc Biol. 29:387–393. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Alfonso F: Interventions for drug-eluting
stent restenosis - to cut, or not to cut: is that the question?
Circ J. 74:1796–1797. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kastrati A, Mehilli J, Dirschinger J,
Pache J, Ulm K, Schuhlen H, Seyfarth M, Schmitt C, Blasini R,
Neumann FJ and Schomig A: Restenosis after coronary placement of
various stent types. Am J Cardiol. 87:34–39. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liistro F, Fineschi M, Angioli P,
Sinicropi G, Falsini G, Gori T, Ducci K, Bravi A and Bolognese L:
Effectiveness and safety of sirolimus stent implantation for
coronary in-stent restenosis: the TRUE (Tuscany Registry of
Sirolimus for Unselected In-Stent Restenosis) registry. J Am Coll
Cardiol. 48:270–275. 2006. View Article : Google Scholar
|
|
11
|
Aminian A, Kabir T and Eeckhout E:
Treatment of drug-eluting stent restenosis: an emerging challenge.
Catheter Cardiovasc Interv. 74:108–116. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kenagy RD, Fukai N, Min SK, Jalikis F,
Kohler TR and Clowes AW: Proliferative capacity of vein graft
smooth muscle cells and fibroblasts in vitro correlates with graft
stenosis. J Vasc Surg. 49:1282–1288. 2009. View Article : Google Scholar
|
|
13
|
Tuerk C and Gold L: Systematic evolution
of ligands by exponential enrichment: RNA 1igands to bacteriophage
T4 DNA polymerase. Science. 249:505–510. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jeong S, Han SR, Lee YJ, Kim JH and Lee
SW: Identification of RNA aptamer specific to mutant KRAS protein.
Oligonucleotides. 20:155–161. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kimoto M, Shirouzu M, Mizutani S, Koide H,
Kaziro Y, Hirao I and Yokoyama S: Anti-(Raf-1) RNA aptamers that
inhibit Ras-induced Raf-1 activation. Eur J Biochem. 269:697–704.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang JX, Tang W, Shi LP, et al:
Investigation of the immunosuppressive activity of artemether on
T-cell activation and proliferation. Br J Pharmacol. 150:652–661.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lee JH, Canny MD, De Erkenez A, Krilleke
D, Ng YS, Shima DT, Pardi A and Jucker F: A therapeutic aptamer
inhibits angiogenesis by specifically targeting the heparin binding
domain of VEGF165. Proc Natl Acad Sci USA. 102:18902–18907. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li ZZ, Han YW, Liu LL, Han YP, Lu Y and
Wang CX: In vitro selection of aptamers to gastric cancer cells by
SELEX. Biotechnology. 19:42–46. 2009.
|