Introduction
Hepatitis B virus (HBV) infection is a severe
worldwide health concern. HBV is a DNA virus with a rapid rate of
mutation. Based on the heterogeneity of the HBV nucleotide
sequence, the HBV strains are divided into eight genotypes, A to H,
with a characteristic geographical distribution (1,2). For
example, genotypes A and D are mainly found in Europe, Africa and
the Americas (3), whereas
genotypes B and C predominate in Asia (4). In the present study, genotype B is
mainly observed in South China and genotype C is mainly found in
North China (5–7). Numerous studies have indicated that
the HBV genotypes are intrinsically linked to the severity of liver
disease in acute and chronic HBV infections. B-type HBV is often
associated with mild liver diseases (8,9),
whereas C-type HBV typically results in severe liver diseases,
including cirrhosis and liver cancer (10,11).
The HBV genotypes have been proven to be responsive to antiviral
therapy (12). Therefore, an assay
for the identification of the HBV genotypes is of significance in
improving the prediction of prognosis and the determination of the
optimal treatment regimen for liver disease caused by HBV
infection.
Conventional methods to determine the HBV genotype
(13,14) are labor-intensive, inaccurate or
expensive. For instance, INNO-LiPA is a simple method suitable for
the analysis of small volumes; however, it is not cost-effective
for efficient high-throughput analysis in a routine clinical
diagnostic setting (15). HBV DNA
sequencing is currently regarded as the gold standard for
genotyping despite its low sensitivity for the evaluation of mixed
infections and the requirement for interpretation of complex peak
patterns.
In the present study, a SNaPshot assay was developed
based on the polymerase chain reaction (PCR) amplification using
fluorescent-marked terminators as well as capillary electrophoresis
to simultaneously analyze the four common HBV genotypes (A-D). The
SNaPshot technique was selected in order to effectively overcome
the shortcomings of the other methods and for the development of an
effective and feasible method of clinical analysis capable of
providing improved prognosis prediction, treatment determination
and patient monitoring.
Materials and methods
Samples and HBV DNA extraction
The samples of four HBV genotypes (A-D) plasmids and
123 clinical serum samples available in our laboratory were
subjected to the multiplex SNaPshot assay. All reagents were
originally obtained from Shanghai Sangon Inc. (Shanghai, China).
Blood samples were drawn from 123 patients with an HBV-positive
infection at The Second Affiliated Hospital of Chongqing Medical
University (Chongqing, China) by venipuncture. The serum samples
were obtained from whole blood which was allowed to clot and were
then centrifuged and the serum samples were stored at −80°C prior
to testing. The HBV DNA was extracted using a viral DNA extraction
kit (Shanghai HuaShun Inc., Shanghai, China) according to the
manufacturer’s instructions. All the procedures were in compliance
with the Helsinki Declaration. The donors were included in the
study following written informed consent. The study protocol was
approved by the Ethics Committee of the Chongqing Medical
University (Chongquing, China; reference number: CQMU 2010–25).
HBV nested-PCR primers and SNaPshot
probes design
Nested-PCR and SNaPshot probes designed for the
present study are shown in Table I
and II. The HBV nested-PCR
primers were designed using Primer 3 software (http://frodo.wi.mit.edu) according to the base
sequences of the conserved regions of various HBV genotypes
(nt260–700 and nt310–610, respectively). Four pairs of SNaPshot
probes used for the detection of HBV types A-D were designed to
anneal with the sense strand immediately adjacent to the specific
site. Each SNaPshot probe was synthesized with a different length
of a poly(dT) tail to allow for separation of the SNaPshot products
on the basis of size.
 | Table IPrimer sequences used in the HBV PCR
protocol. |
Table I
Primer sequences used in the HBV PCR
protocol.
| Name | Sequences |
|---|
| P1:HBV 260F |
5′-CTCGTGGTGGACTTCTCTCA-3′ |
| P2:HBV 310F |
5′-GGCCAAAATTCGCAGTCCC-3′ |
| P3:HBV 610R |
5′-GATGATGGGATGGGAATACA-3′ |
| P4:HBV 700R |
5′-CGAACCACTGAACAAATGGCA-3′ |
 | Table IIProbe sequences and concentration used
in the HBV SNaPshot protocol. |
Table II
Probe sequences and concentration used
in the HBV SNaPshot protocol.
| No. | Name | Genotype | Sequences | Concentration,
μM | Production length,
bp |
|---|
| 1 | Probe1 | HBV A |
5′-GCTGCTATGCCTCATCTTCTT-3′ | 1.5 | 22 |
| 2 | Probe2 | HBV B |
5′-TTTTTTTGGCCAAAATTCGCAGTCCC-3′ | 3.0 | 27 |
| 3 | Probe3 | HBV C |
5′-TTTTTTTTTTTATGTTGCCCGTTTGTCCTCTA-3′ | 0.75 | 33 |
| 4 | Probe4 | HBV D |
5′-TTTTTTTTTTTTTTTTTTTTGTACAGCAACAGAGGGA-3′ | 3.0 | 39 |
HBV DNA PCR amplification
Amplification was performed in a volume of 25 μl
containing 0.5 μl of 10 μM of each primer (P1 and P4), 12.5 μl
master mix (Takara, Dalian, China), 10 ng template DNA dissolved in
0.5 μl solvent and 11 μl ddH2O. The PCR reaction
conditions were as follows: 94°C for 4 min, 35 cycles of 94°C for
30 sec, 56°C for 40 sec and 72°C for 50 sec, and a final step at
72°C for 10 min. In total, 3 μl PCR product mixture was analyzed by
1.5% agarose gel electrophoresis. A volume of 0.5 μl negative PCR
products was nested in P2 and P3 at PCR reaction conditions of 94°C
for 4 min, 35 cycles of 94°C for 30 sec, 50°C for 30 sec and 72°C
for 40 sec, followed by 72°C for 10 min.
HBV PCR product purification
Subsequent to PCR, 15 μl PCR product mixture was
treated for SNaPshot analysis. Treatment was conducted with 5 units
SAP (shrimp alkaline phosphatase, Applied Biosystems, Grand
Island, NY, USA) and two units Exo I (exonuclease I, Applied
Biosystems) incubated at 37°C for 1 h, followed by incubation at
75°C for 15 min to remove excess deoxyribonucleotide triphosphates
(dNTPs) and primers. The samples were then stored at 4°C. The PCR
products for the sequencing reactions were purified further using
ethanol/NaAc and stored at 4°C.
SNaPshot analysis
SNaPshot assays were performed using a SNaPshot
multiplex kit (Applied Biosystems). The reactions were performed in
a total reaction volume of 10 μl containing 1 μl purified PCR
product (5–10 ng), 5 μl SNaPshot ready multiplex mix (Applied
Biosystems), 1 μl mixture of each of the four SNaPshot single base
extension probe mixtures (final concentrations of each probe are
shown in Table II) and 3 μl
ddH2O. Thermal cycling was performed under the following
conditions: 25 cycles of 10 sec each at 96°C, 10 sec at 50°C and 30
sec at 60°C. Labeled extension products were treated further with 1
unit SAP for 1 h at 37°C and 15 min at 75°C, then mixed with
0.5 μl extension products with 0.5 μl Genescan-120 LIZ size
standard and 9 μl Hi-Di formamide (Applied Biosystems) and
denatured at 95°C for 5 min. The products were then immediately
placed in an ice bath for 5 min and relocated through a 3100
sequencing analyzer using POP-6 polymer electrophoresis (Orbita).
The fluorescence signal was analyzed with Gene Mapper 3.5 software
(Applied Biosystems). The samples were sequenced simultaneously
with P1.
Sequencing
The PCR products for sequencing reactions were
electrophoresed on an ABI 3100 sequencing analyzer using POP-6
polymer (Orbita) with P1.
Sensitivity analysis
Two approaches were used to determine the
sensitivity of the SNaPshot method. The first one involved serial
dilutions of the B-type plasmid DNA
(1×100–1×108 copies/ml) in HBV-negative serum
and the second one, 10 ng template DNA containing the B-type
plasmid DNA at 0, 5, 10, 20, 50, 80, 90, 95 and 100% in the C-type
plasmid.
Specificity and accuracy analysis
The HBV genotypes, A-D, plasmids and 123
HBV-positive serum samples were assayed using the SNaPshot method
and DNA direct sequencing. Where the results of SNaPshot and
sequencing were inconsistent, the PCR products were cloned into the
pMD18-T vector (Takara Bio, Inc., Shiga, Japan) according to
manufacturer’s instructions and 20 clones of each sample were
selected for further sequencing.
Statistical analysis
The statistical package SPSS 11.5 (SPSS, Inc.,
Chicago, IL, USA) was employed for data analysis. P≤0.05 was
considered to indicate a statistically significant difference.
Results
Strategies for HBV genotyping
The strategies for detection of the HBV genotypes,
A-D, are presented in Fig. 1. HBV
DNA templates were amplified by PCR or nested-PCR, followed by a
SNaPshot single base extension and electrophoresis. Probe 1
recognized the HBV locus 436, while probes 2, 3 and 4 recognized
the HBV loci 320, 482 and 555, respectively. The single base
extension of these four probes produced 22, 27, 33 and 39 bp
fragments, respectively (Table
II).
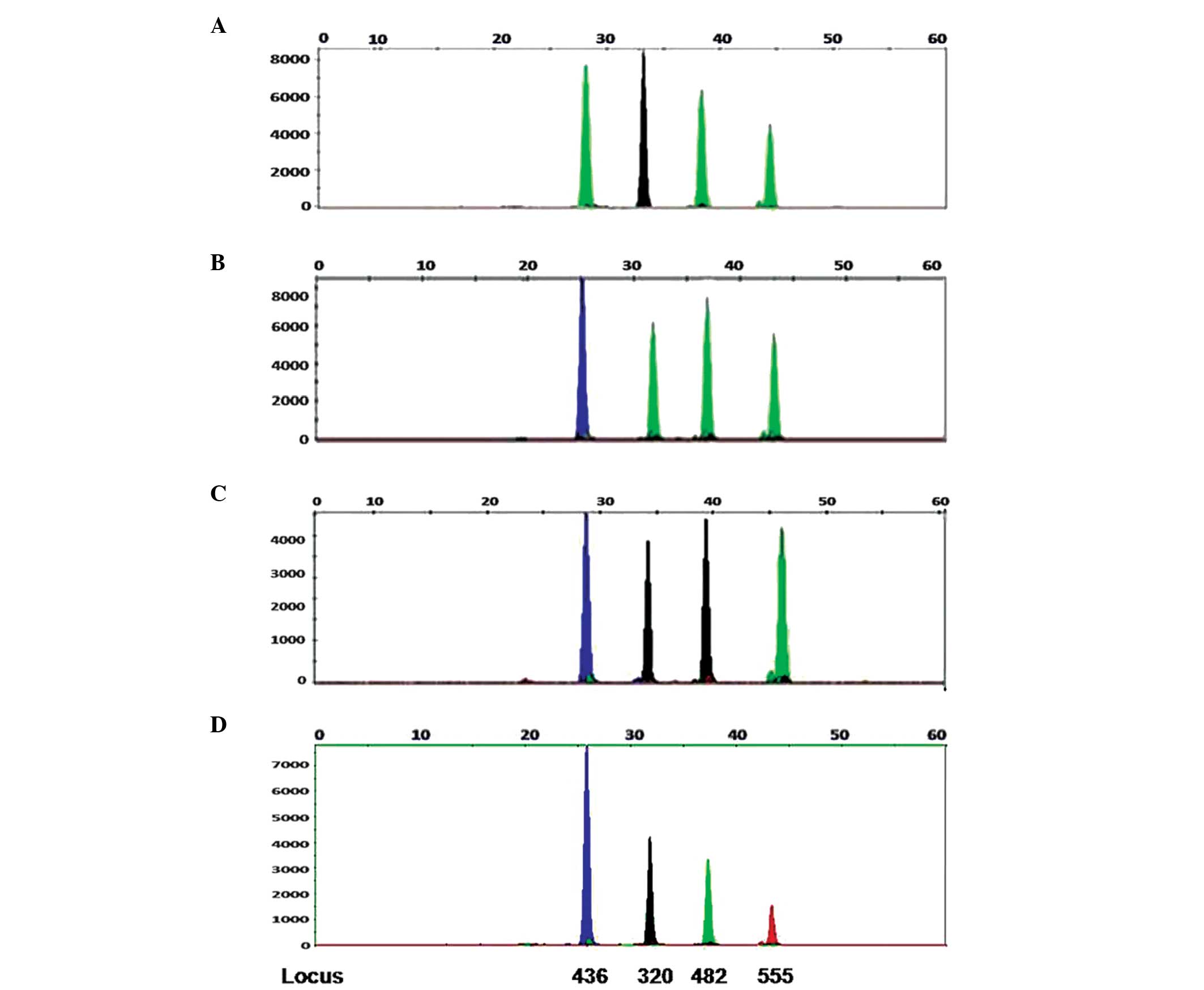 | Figure 1Strategies and specificity of the HBV
A–D plasmids detected by the SNaPshot assay. SNaPshot results
obtained from the analysis of the HBV plasmid. The HBV DNA
templates were amplified by polymerase chain reaction, single base
extension and electrophoresis. Probe 1 recognized the HBV locus
436, while probes 2, 3 and 4 recognized the HBV locus 320, 482 and
555, respectively. The x-axis represents the size (bp) of the probe
pair with the incorporated nucleotides, while the y-axis
corresponds to the relative fluorescence units of the peak. The
probe mix included 1.5 μM probe A, 3.0 μM probe B, 0.75 μM probe C
and 3.0 μM probe D. A–D correspond to the HBV genotypes, A–D,
plasmids. Red refers to T, blue refers to G, green refers to A and
black refers to C. Genotypes A, B, C and D correspond to ACAA,
GAAA, GCCA and GCAT, respectively. |
HBV DNA amplification
HBV DNA PCR amplification appeared to be a major
problem of the SNaPshot analysis. Two sets of primers for HBV PCR
were designed to amplify the distinct fragments, 440 and 300 bp.
Four DNA plasmids and 111 samples were effectively amplified using
the outer primers. In total, 12 samples that were effectively
amplified using the nested primers were subsequently quantified
using a HBV nucleic acid detection kit (Zhongshan Da an Gene Co.,
Ltd., Guangzhou, China). The minimum concentration was
2.14×102 IU/ml.
SNaPshot analysis optimization
The probe length (21, 26, 32 and 38 bp), the
concentration (1.5, 3.0, 0.75 and 3.0 μM; Table II), and the template concentration
were optimized to ensure consistency in the height and position of
the peaks representing the four types of extension products. A
significant linear regression correlation with the peak (r=0.995)
was identified at the DNA template concentrations ranging from
2.5–10 ng/μl.
Specificity analysis
This novel multiplex SNaPshot method was validated
by analysis of the four types of HBV genotypes (A-D) and the
HBV-negative patient samples. The predicted results were obtained
for the four plasmid types. Non-specific signals were observed and
HBV-negative patient samples had no signal. The results indicated
the detection of an HBV A-type for ACAA, a B-type for GAAA, a
C-type for GCCA, and a D-type for GCAT. There were two different
extension bases between the two different HBV types. Overall, this
method demonstrated good specificity (Fig. 1).
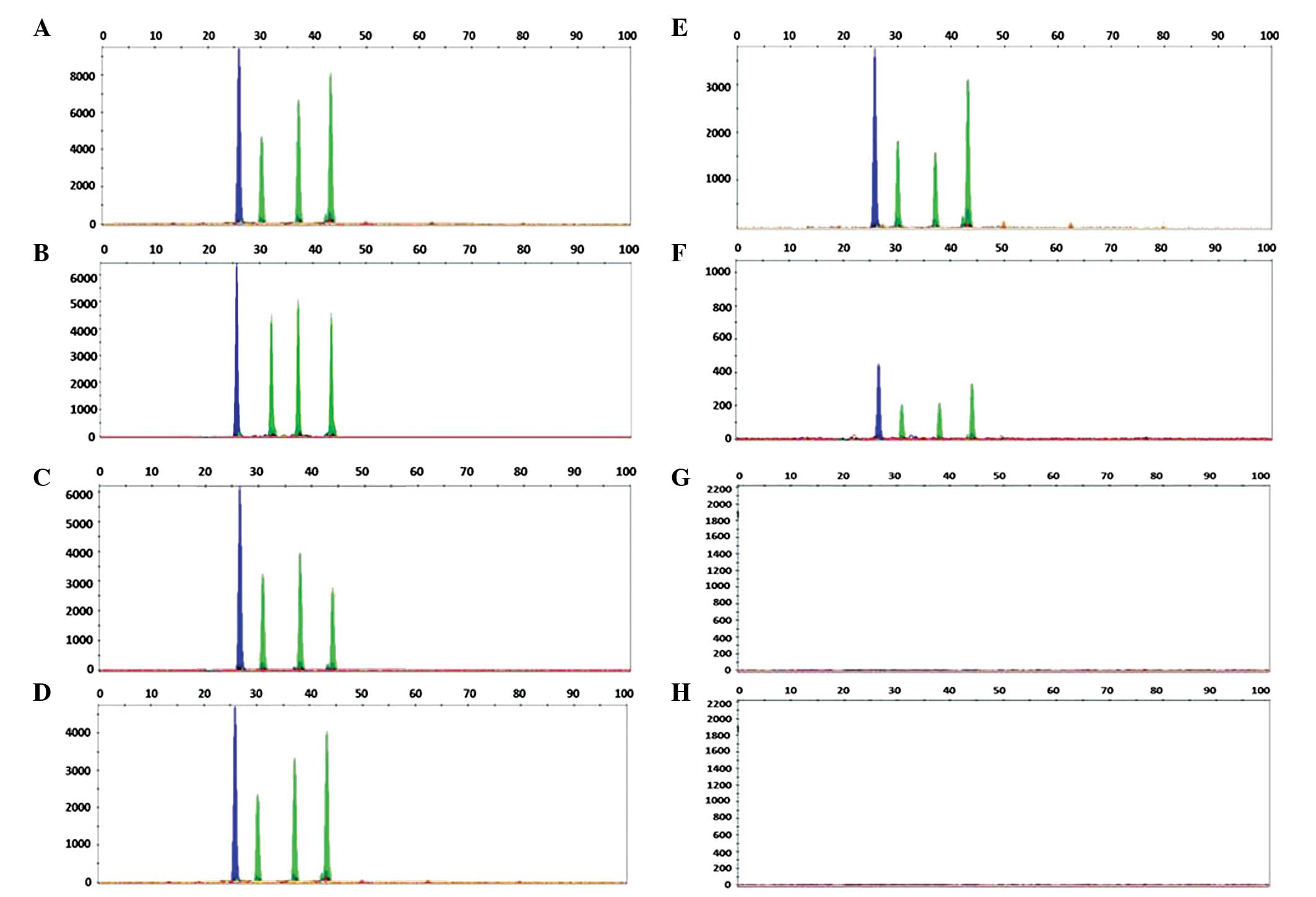
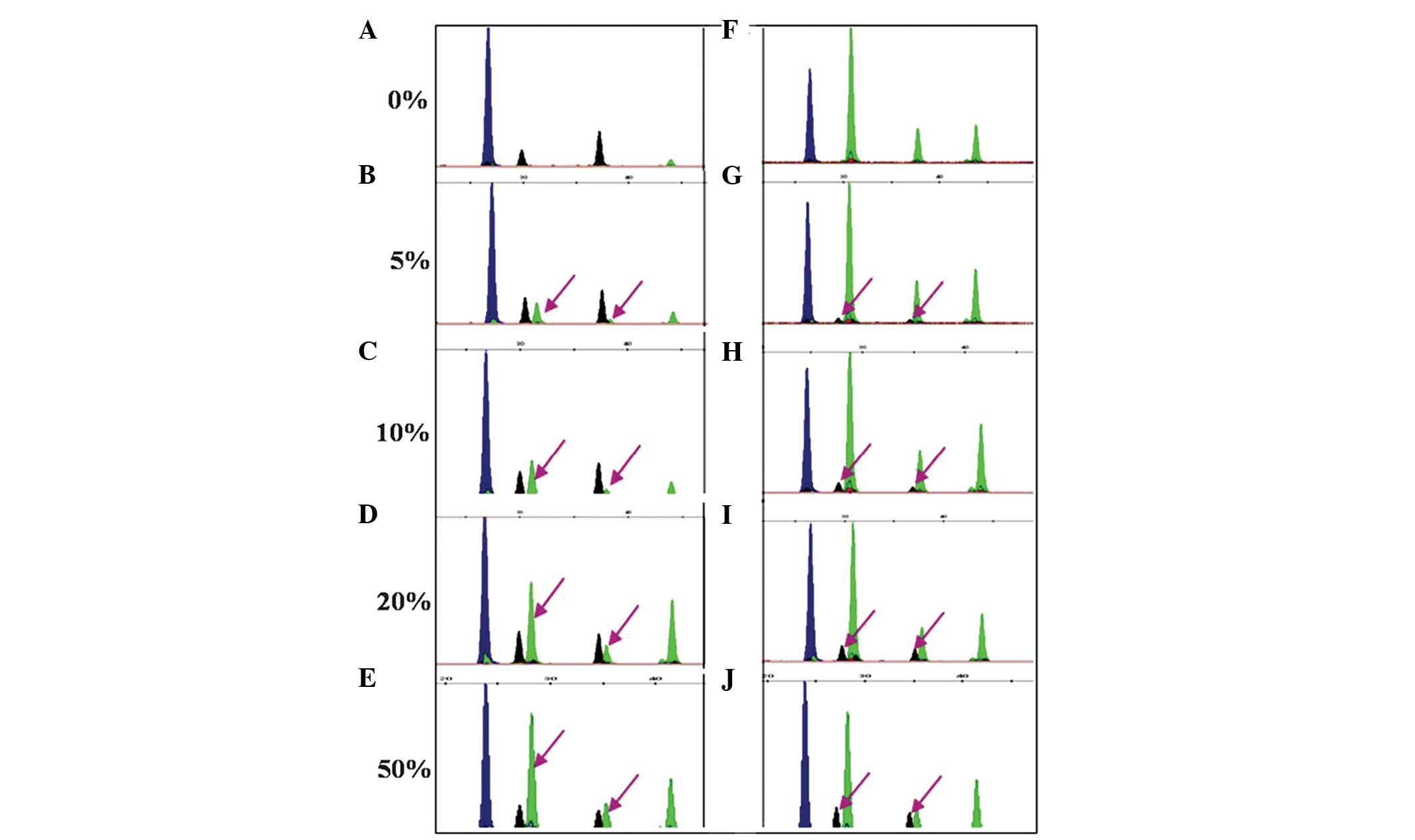
Sensitivity analysis
The sensitivity of the SNaPshot assay was assessed
by analysis of the B-type plasmid diluted in HBV-negative serum and
mixed samples containing the B-type with the C-type plasmids at
varying concentrations or proportions. A correct analysis of the
HBV genotype was achieved up to a viral load of 1×103
copies/ml. Simultaneously, successful detection of BC mixed samples
was achieved up to a 5% level. The results are shown in Fig. 2 and 3.
Comparison of the SNaPshot assay and DNA
sequencing
The SNaPshot assay was evaluated using clinical
serum samples from 123 patients identified with an HBV infection.
All the samples were analyzed by the SNaPshot assay and DNA
sequencing. Discrepancies in the results between the two methods
were confirmed by subclone sequencing. Out of the 123 samples, 80
cases of B-type, 29 cases of C-type, nine cases of BC-type, four
cases of BD mixed infection and one case of BCD-type were
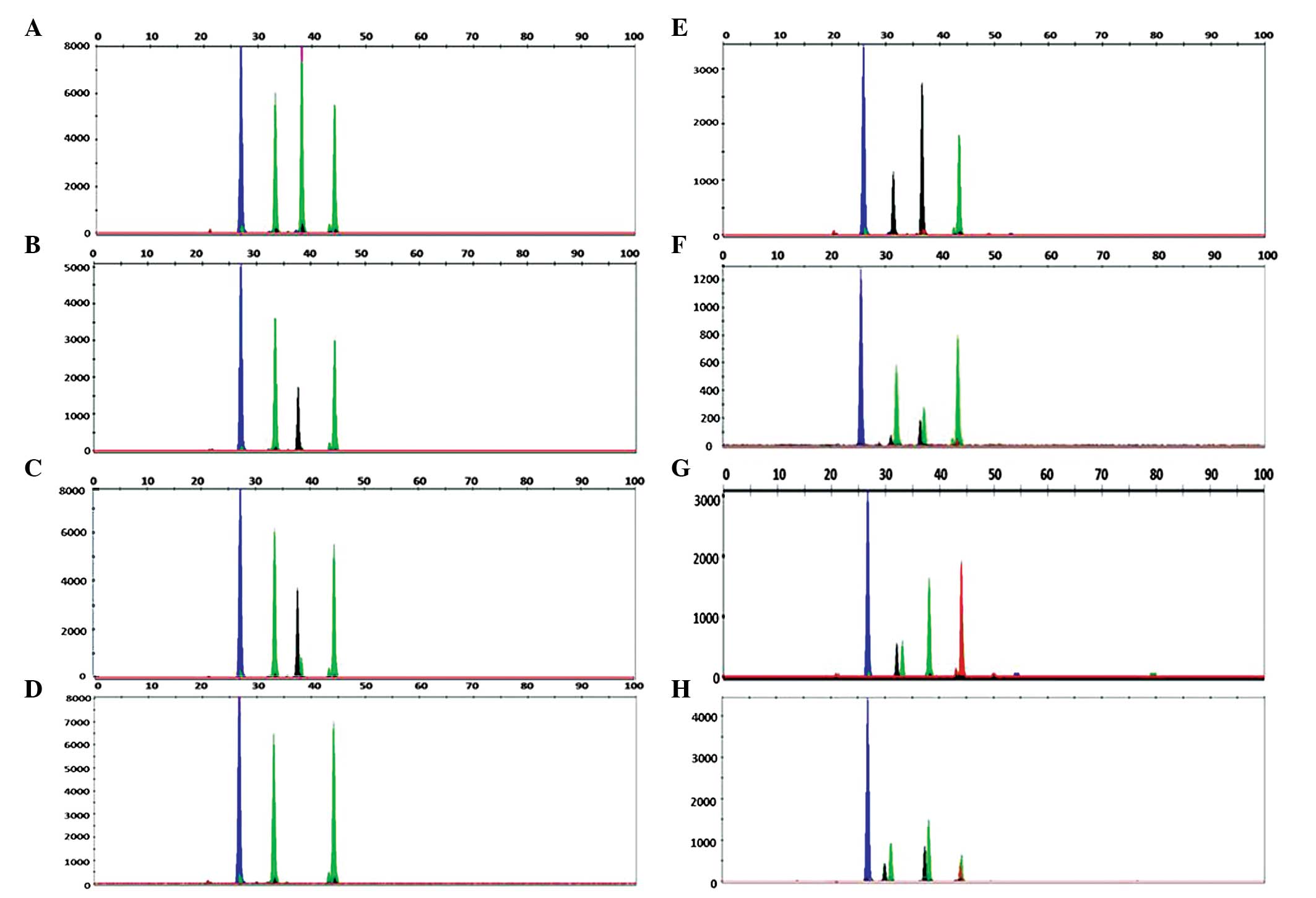
identified using the SNaPshot assay. In the detection process, four
types of electrophoresis patterns were observed in the B-type
samples, which included GAAA, GACA, GA (CA) A and GA-A. Fig. 4A–D show typical results from
samples assayed by SNaPshot. DNA sequencing analysis revealed two
base mutations in the GA-A samples differing in the no. 3 extension
probe sequences. A comparison of the results obtained using the
SNaPshot assay and DNA sequencing demonstrated 100% (123/123)
sensitivity and validity of the SNaPshot assay. However,
inconsistent results were obtained for 12 samples. Clone sequencing
analysis of 20 randomly selected clones for each of these 12
samples was subsequently performed and the results were found to be
fully consistent with the SNaPshot assay analysis (12/12; Table III; Fig. 4E–H). The consistency of the
SNaPshot assay with DNA sequencing and clone sequencing assays was
90.24 (111/123) and 100% (12/12), respectively. The accuracy of the
SNaPshot assay for detection of mixed HBV genotype infections was
found to be equivalent to or higher than that of direct DNA
sequencing [100% (14/14) vs. 14.29% (2/14); P=5.983×10−6
according to Fisher’s exact test].
 | Table IIIResults of serum samples detection by
the SNaPshot and sequencing assay. |
Table III
Results of serum samples detection by
the SNaPshot and sequencing assay.
| SNaPshot assay |
|---|
|
|
|---|
| Tests and
results | B-type | C-type | D-type | BC-type | BD-type | BCD-type |
|---|
| DNA sequencing,
n=123 |
| B-type | 80 | | | 5a | 2a | 1a |
| C-type | | 29 | | 3a | | |
| D-type | | | | | 1a | |
| BC-type | | | | 1 | | |
| BD-type | | | | | 1 | |
| BCD-type | | | | | | |
| Clone sequencing,
n=12 |
| B-type | | | | | | |
| C-type | | | | | | |
| D-type | | | | | | |
| BC-type | | | | 8 | | |
| BD-type | | | | | 3 | |
| BCD-type | | | | | | 1 |
Discussion
In the present study, a SNaPshot assay was
established for the simultaneous detection of the four most common
HBV genotypes in clinical specimens. The methodology employed for
this assay is also regarded as a short-sequencing technology. The
reaction cocktails consisted of a polymerase, four
fluorescent-labeled dNTPs and extension probes. The position of the
nucleotide base at the termination site is associated with the
genotype, and this is readily detectable by differential
fluorescence analysis. As many as ten SNP polymorphic loci are able
to be readily detected in a single run experiment.
The SNaPshot assay described in the present study
utilized four colors of fluorescence labeling (green, red, blue and
black) to allow single-peak fluorescence waveform identification of
the four bases (A, T, G and C) with 3100 gene sequencing. The four
HBV genotypes, A-D, were differentiated based on the simultaneous
appearance of these four differential fluorescent labels: Green
(A), red (T), blue (G) and black (C), for ACAA, GAAA, GCCA and
GCAT, respectively. Mixed infections containing different HBV
genotypes produced distinct bimodal distributions in separate
locations of the electropherogram reflecting the number of
different genotypes detected. Therefore, electropherograms
generated from each species demonstrated that the genotypes
examined, A-D, were able to be clearly identified and
differentiated from one another using this novel SNaPshot
assay.
The detection level of the SNaPshot assay based on
nesting PCR was 1×103 copies/ml. The detection level of
BC-mixed infections was 5%. In the present study, 123 HBV isolates
from clinical specimens (HBV DNA ≥ 2.14×102 IU/ml) were
analyzed using the multiplex SNaPshot assay. The test sensitivity
of the standard plasmids and clinical samples was 100%. The
incidence of B-type, C-type, BC-type, B-type and BCD-type was shown
to be 65.04, 23.58, 7.32, 3.25 and 0.81%, respectively, using the
SNaPshot assay. An analysis of the sequences of the GA-A
electropherogram demonstrated that the no. 3 SNaPshot probes were
not extended as a single base for the last base mutations (A-G).
However, this did not significantly affect the determination of the
HBV B-type. A comparison of the SNaPshot assay and DNA sequencing
methods revealed that there were five cases of BC-, two cases of
BD- and one case of BCD-type among the eight B-type samples; three
cases of BC-type among the three C-type samples and one case of
BD-type among the D-type samples all detected by DNA sequencing.
The mixing ratio of the 12 clinical samples was lower than 20% and
the DNA copies were below the test level.
Similar results were obtained by the SNaPshot and
the cloning sequence analysis. The SNaPshot analysis demonstrated
that the HBV B-genotype was predominant in the Chongqing region,
followed by the HBV C-type. A small amount of the BC-, BD- and
BCD-type infections were also identified in the Chongqing region,
although the BCD-type infections were the least common. The results
obtained using the SNaPshot assay were consistent with the DNA
sequencing results and the DNA cloning sequence analysis in the
detection of different HBV genotypes in the high-copy DNA samples
(111/111) and the small number of mixed infection in the serum
samples (12/12). The sensitivity of the assay was 100% (123/123)
and the accuracy of detection of mixed types was higher compared
with that of direct DNA sequencing [100% (14/14) vs. 14.29% (2/14);
P=5.983×10−6].
The results of the present study demonstrate that
the SNaPshot assay offers a rapid, robust and highly reliable
alternative to traditional DNA sequencing methods for the
identification of the HBV genotypes A-D. Although the complete DNA
sequence can be detected by HBV genome sequencing, the test results
are unclear, providing complex background signals with more
numerous peaks and lower detection sensitivity for the mixed
infections due to the presence of residual primers.
In conclusion, the present study showed that the
SNaPshot method is able to be used for the simultaneous analysis of
the four common HBV genotypes. The sensitivity and accuracy of this
technique was shown to be higher with regard to a single product,
simple peak shapes and no background noise compared with the
sequencing method. The SNaPshot method is limited by its potential
for the detection of known genotypes only. However, HBV genotype
research is advancing and the gene sequence of the virus is now
recognized worldwide. Thus, the present study provides the basis of
a novel technique that is suitable for further development in
clinical HBV genotyping.
Acknowledgements
The present study was financially supported by the
863 projects of the Ministry of Science and Technology of the
People’s Republic of China (grant no. 2008AA02Z424) and the
National Youth Foundation (grant no. KJ100309). The authors would
like to thank Dr Hu Jieli for providing the HBV A-D plasmids and
Lai Yuqi Bachelor and Dr Chen Feilan for revising the
manuscript.
References
|
1
|
Norder H, Couroucé AM and Magnius LO:
Complete genomes, phylogenetic relatedness, and structural proteins
of six strains of the hepatitis B virus, four of which represent
two new genotypes. Virology. 198:489–503. 1994. View Article : Google Scholar
|
|
2
|
Okamoto H, Tsuda F, Sakugawa H, et al:
Typing hepatitis B virus by homology in nucleotide sequence:
comparison of surface antigen subtypes. J Gen Virol. 69(Pt 10):
2575–2583. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kaya S, Cetin ES, Aridogan BC, Onal S and
Demirci M: Distribution of hepatitis B virus (HBV) genotypes among
HBV carriers in Isparta. Iran Biomed J. 11:59–63. 2007.PubMed/NCBI
|
|
4
|
Duong TN, Horiike N, Michitaka K, et al:
Comparison of genotypes C and D of the hepatitis B virus in Japan:
a clinical and molecular biological study. J Med Virol. 72:551–557.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pujol FH, Navas MC, Hainaut P and Chemin
I: Worldwide genetic diversity of HBV genotypes and risk of
hepatocellular carcinoma. Cancer Lett. 286:80–88. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang Z, Liu Z, Zeng G, et al: A new
intertype recombinant between genotypes C and D of hepatitis B
virus identified in China. J Gen Virol. 86:985–990. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lu J, Gong W, Cheng H, et al: Detection of
HBV genotypes of tumor tissues and serum by a fluorescence
polarization assay in north-western China’s hepatocellular
carcinoma patients. Virol J. 8:3622011.PubMed/NCBI
|
|
8
|
Liu J, Li Y, Chen T, et al: The
distribution of HBV genotypes and clinical significance in familial
clustering in an infected population with unfavorable prognosis.
Arch Virol. 153:2157–2161. 2008. View Article : Google Scholar
|
|
9
|
Tseng TC and Kao JH: HBV genotype and
clinical outcome of chronic hepatitis B: facts and puzzles.
Gastroenterology. 134:1272–1273. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sánchez-Tapias JM, Costa J, Mas A,
Bruguera M and Rodes J: Influence of hepatitis B virus genotype on
the long-term outcome of chronic hepatitis B in western patients.
Gastroenterology. 123:1848–1856. 2002.
|
|
11
|
Kao JH and Chen DS: HBV genotypes and
outcome of HBV infection. Hepatology. 41:2162005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Batoctoy KS, Tseng TC, Kao JH, Quiza FE,
Garcia LH Sr and Lao-Tan J: HBV/A and HBV/C genotype predominance
among patients with chronic hepatitis B virus infection in Cebu
City, Philippines. Hepatol Int. 5:774–781. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ali MM, Hasan F, Ahmad S and Al-Nakib W:
Comparative evaluation of INNO-LiPA HBV assay, direct DNA
sequencing and subtractive PCR-RFLP for genotyping of clinical HBV
isolates. Virol J. 7:1112010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Thibault V, Pichoud C, Mullen C, et al:
Characterization of a new sensitive PCR assay for quantification of
viral DNA isolated from patients with hepatitis B virus infections.
J Clin Microbiol. 45:3948–3953. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mercier M, Laperche S, Girault A, Sureau C
and Servant-Delmas A: Overestimation of incidence of hepatitis B
virus mixed-genotype infections by use of the new line probe
INNO-LiPA genotyping assay. J Clin Microbiol. 49:1154–1156. 2011.
View Article : Google Scholar : PubMed/NCBI
|