Introduction
Autophagy is an important process in eukaryocytes
that enables the recycling of intracellular materials. During the
autophagic process, certain damaged proteins or organelles are
enclosed by double-membrane autophagic vacuoles and then degraded
in lysosomes (animals) or in vacuoles (yeasts and plants) for
reuse. Similar to apoptosis and cell aging, autophagy is a
biological phenomenon of great importance, involved in numerous
processes in the growth of an organism (1). Excessive or impaired autophagy may
cause diseases, including cancer (2), pathogen infections (3), neurodegenerative diseases (4) and aging (5). Thus, the regulation of autophagy has
become a focal point of investigation. In recent years, mTOR kinase
(6), type I PI3K/Akt (7), AMPK (8), P53 (9), DAPK (10), endoplasmic reticulum annexins
(11), Ire-1 (12), IP3R (13), GTPase (14), Erk1/2 (15), ceramide (16) and calcium (17), have all been observed to be
involved in the regulation of autophagy. Molecular biology methods,
including gene and protein analysis, have achieved marked success
in studies of autophagy regulation; however, these methods are
restricted in clinical practice. An approach targeting protein
kinases may soon translate laboratory findings into the clinic,
since an increasing number of newly developed kinase inhibitors are
able to inhibit corresponding kinases with the most direct targets
(18). The present study mined
particular kinases regulating autophagy and predicted the kinase
inhibitors that may regulate autophagy, which may provide benefits
in the clinical application of autophagic regulation.
Materials and methods
Mining and screening of
autophagy-associated kinases
Firstly, information regarding human protein kinases
was collected from the Kinasource protein kinase database
(http://kinasource.co.uk/Database/substrates.html)
and an Excel file listing 511 protein kinases was produced. Using
the literature-mining approach, Perl language software (www.perl.com) was used to write a literature-mining
program. The retrieved keyword ‘autophagy’ and the ‘protein kinase’
Excel file were then imported to the ActivePerl 5.16.2 software
(ActiveState Software Inc., Vancouver, BC, Canada) and the
literature information was obtained from the National Library of
Medicine’s PubMed database (http://www.ncbi.nlm.nih.gov/pubmed/). The scope of the
search included the titles and abstracts of all articles in the
PubMed database. Valuable articles were identified and the false
ones that were not related to autophagy were ruled out by manual
screening.
Secondly, autophagy-associated gene ontology (GO)
terms were searched for in the Gene Ontology Database (http://www.geneontology.org/), defining the species as
Homo sapiens in all databases and screening out the human
autophagy annotation (GO:0006914), and an ‘autophagy-associated
genes’ Excel file was generated. Documents in PubMed were searched
following the importation of the ‘autophagy-associated genes’ and
‘protein kinase’ files to the ActivePerl software, and
autophagy-associated kinases concerning the corresponding
autophagy-associated genes were screened out. Subsequently, the
documents were checked by manual screening to verify whether the
detected genes were associated with autophagy.
The kinases remaining after the intersection of the
two abovementioned methods were thus proposed as candidates of the
autophagy-associated kinases.
Enrichment analysis of
autophagy-associated kinases
Database for Annotation, Visualization and
Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/) software was used for
Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis.
The DAVID was opened and the kinase genes were submitted, with the
whole human genome selected as the background. Subsequently,
‘Functional Annotation Tool’ served as the analytical tool and the
results of the KEGG enrichment analysis were obtained and the
pathways with P≤0.05 were selected.
Regulatory network construction for
autophagy-associated kinases
The corresponding signaling pathways of the specific
kinases in the KEGG database (http://www.genome.jp/kegg/pathway.html) were
identified and an Excel file was set up, which contained each
kinase and the corresponding KEGG signaling pathways. The file was
imported to Cytoscape (version 2.6.3; http://www.cytoscape.org), with the interaction type
defined as ‘default interaction’, the source interaction as
‘kinase’, and target interaction as ‘KEGG signaling pathway’. The
terms ‘layout’, ‘cytoscape layout’ and ‘spring embedded’ were then
selected. A regulatory network was constructed and the kinases were
connected using the same signaling pathway.
Prediction of kinase inhibitors that
regulate autophagy
An Excel file of 194 kinase inhibitors was set up as
determined by a database (http://www.selleckbio.com/servlet/DownloadServlet?fileName=Selleck-Kinase-Inhibitor-Library.xlsx).
The ‘autophagy-associated kinases’ and ‘kinase inhibitors’ files
were imported to the ActivePerl software and screened. The
regulatory network was constructed using Cytoscape.
The texts associated with ‘autophagy’ and ‘candidate
kinase inhibitor’ were extracted from the PubMed database, and the
current research focus was differentiated from uninvestigated
kinase inhibitors in the screened documents.
Results
Mining and screening of
autophagy-associated kinases
Firstly, through text mining of ‘autophagy’ and
‘protein kinase’ terms in PubMed, 398 preliminary articles were
identified and retrieved. Following manual screening, 32 articles
were found to not be concerned with autophagy and were ruled out
(false rate 18.09%). In total, 62 kinases were obtained through
analyzing the remaining 366 articles.
Secondly, mining for the ‘Autophagy-Associated GO
Term’ in the Gene Ontology Database identified 56 genes with the
human ‘Autophagy’ annotation (GO:0006914). Among these, five genes
were annotated with ‘Macroautophagy’ (GO:0016236), 10 genes with
‘Negative regulation of autophagy’ (GO:0010507), seven genes with
‘Negative regulation of macroautophagy’ (GO:0016242), seven genes
with ‘Positive regulation of autophagy’ (GO:0010508), 10 genes with
‘Positive regulation of macroautophagy’ (GO:0016239)and 16 genes
with ‘Regulation of autophagy’ (GO:0010506). A total of 1,362
preliminary articles were retrieved in PubMed by importing the
‘autophagy-associated gene’ and ‘protein kinase’ files, and 1,026
articles remained following manual screening (false rate 24.67%).
In total, 146 kinases were obtained through analyzing these
articles (Fig. 1).
Using the intersection of the two abovementioned
methods, 54 candidate autophagy-associated kinases were identified
(Table I).
 | Table IAutophagy-associated kinases detected
by text mining and bioinformatic screening. |
Table I
Autophagy-associated kinases detected
by text mining and bioinformatic screening.
| Group | Family | Kinase symbol | Description | Publication
PMIDs |
|---|
Serine/threonine
protein kinases:
AGC group | DMPK | CDC | cell division cycle
42 | 22038398, 21695150,
20944112, 19788417, 19185564, 17172425, 16702956 |
| AKT | AKT | v-akt murine thymoma
viral oncogene homolog 1 | 22996802, 22989521,
22931788, 22924465, 22902833, 22892762, 22874552, 22722716,
22694815, 22535956, 21964073, 21297943, 20857426, 20629079,
20347887, 16007218, 21297943 |
| DMPK | DMPK | dystrophia
myotonica-protein kinase | 20797447,
19946639 |
| NDR | NDR | serine/threonine
kinase 38 | 19450522 |
| RSK | RSK | ribosomal protein S6
kinase | 22017876, 19185734,
18809440, 17631443 |
| SGK | SGK | serum/glucocorticoid
regulated kinase 1 | 22125061 |
| CAMK group | CAMKL | AMPK | AMP-activated protein
kinase | 21871960, 21871561,
21406597, 21390063, 20876807, 20103647 |
| CHK | choline kinase
alpha | 22858649, 21829721,
20805228, 20368736, 22499945, 21616090, 20838898, 20805228,
19672857, 17172425 |
| PASK | PAS domain containing
serine/threonine kinase | 18391949 |
| PIM | PIM | pimples | 17476689 |
| PKD | PKD | protein kinase
D1 | 21270095,
19863783 |
| DAPK | DAPK | death-associated
protein kinase 1 | 23071514, 22874566,
22699507, 21416260, 21167819, 19267229, 18806760, 21508686 |
| Trio | Trio | trio Rho guanine
nucleotide exchange factor | 22833093,
21350583 |
| MAPK | MAPK | map kinase | 22579651, 18614532,
18614532, 15131264, 22665475, 19370058 |
| CK1 group | CK1 | CK1 | casein kinase 1 | 21829721,
19943845 |
| CMGC group | RCK | MAK | male germ
cell-associated kinase | 22935150, 21641547,
20639397, 20580051, 20200163, 19144925 |
| MOK | MOK protein
kinase | 19066463,
17555713 |
| CDK | CDK | Cyclin-dependent
kinase 4 | 22858649, 21668989,
20513426, 19185564, 22896656, 22716215, 22517431, 22496786,
21898405, 21668989, 21376716, 21297943, 20946021, 20630104 |
| STE group | STE20 | LOK | lymphocyte-oriented
kinase | 22966490, 21757721,
21584293, 19074891 |
| MINK | minK-like
protein | 15840653, 14500369,
10637306, 7215561 |
| NRK | neurospecific
receptor kinase | 22496408, 22146761,
22089196, 20013139, 16885413, 11262416 |
| PAK | p21 protein
(Cdc42/Rac)-activated kinase 1 | 21678067,
21678067 |
| Other | NIK | NCK interacting
kinase | 19723111, 17671417,
17563756 |
| TKL group | RAF | BRAF | v-raf murine
sarcoma viral oncogene homolog B1 | 20459375,
20182446 |
| LRRK | LRRK | leucine-rich repeat
kinase 2 | 22988880, 22988879,
22988877, 22988862, 22931754, 22927213, 22914360, 22908196,
22773119, 22649237 |
| Other | ILK | integrin-linked
kinase | 22348065,
21823616 |
| Others | WEE | WEE | Wee1-like protein
kinase | 22858649 |
| VPS15 | PIK | phosphatidyl
inositol kinase (PIK-A) | 20818396, 20643123,
18957027, 20548331 |
| PDHK | PDK | Pyruvate
dehydrogenase kinase | 22901154, 22305748,
21724622, 20352258, 20007520, 19010909, 12453151 |
| Tyrosine protein
kinases | Abl | ABL | c-abl oncogene 1,
non-receptor tyrosine kinase | 22898871 22898604
22765290 22641616 22407228 22395361 22340716 22202070 21944254
21903901 |
| Csk | CSK | c-src tyrosine
kinase | 21214405 18368623
16903144 16903142 15325586 12416391 |
| Alk | ALK | anaplastic lymphoma
receptor tyrosine kinase | 21324370
21134980 |
| EGFR | EGFR | epidermal growth
factor receptor | 23029387 22954701
22882626 22881126 22771957 22753156 22740881 22638807 22521643
22492316 |
| Eph | EPHA | epoxide hydrolase
EphA | 21503576 |
| EPHB | epoxide hydrolase
ephB | 21503576
20046096 |
| Src | LCK | lymphocyte-specific
protein tyrosine kinase | 21611191
16886061 |
| FYN | FYN oncogene
related to SRC, FGR, YES | 20814235 |
| SRC | v-src sarcoma
(Schmidt-Ruppin A-2) viral oncogene homolog (avian) | 22911754, 22896656,
22732841, 22673740, 22575674, 22430150, 22342344, 22340716,
22246348, 22193164 |
| FGR | Gardner-Rasheed
feline sarcoma viral (v-fgr) oncogene homolog | 23039021 |
| YES | v-yes-1 Yamaguchi
sarcoma viral oncogene homolog 1 | 16461361 |
| TYK | tyrosine kinase
2 | 21300624 |
| JAK | JAK | Janus kinase 2 | 22065112, 21300624,
21088407, 20930550 20592027, 20008137, 19997066, 19817673,
19453248, 21930714 |
| IGF | insulin-like growth
factor binding protein 3 | 23071566, 22973545,
21850397, 21659463 20409077, 19680556 |
| ROS | c-ros oncogene 1,
receptor tyrosine kinase | 23079083, 23076967,
23071780, 23071110, 23059970, 23056433, 23052483, 23046998
23041169, 23028492 |
| FGFR | FGFR | fibroblast growth
factor receptor 3 | 22207710 |
| PDGFR | FLT | fms-related
tyrosine kinase 1 | 23056267, 22689683,
22095133 |
| KIT | v-kit
Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | 23071514, 22905730,
22833529, 22801786, 21943129, 21479886, 21339326, 20809291,
20660757, 19567133 |
| FAK | FAK | focal adhesion
kinase | 22732841, 22333033,
22138575, 22132975, 21807966, 21775823, 21764854, 21525242,
21460634, 21350583 |
| Axl | MER |
methylenetetrahydromethanopterin
reductase | 22860116, 21829707,
19273585 |
| Met | MET | met
proto-oncogene(hepatocyte growth factor receptor) | 22673163, 22569095,
22525272, 22466960, 21927636, 21743956, 21433059, 21340000,
21135502, 20736296 |
| Musk | MUSK | muscle, skeletal,
receptor tyrosine kinase | 22191394 |
| Syk | SYK | spleen tyrosine
kinase | 22902620 |
| Tec | TEC | tec protein
tyrosine kinase | 19915056 |
| Ack | TNK | tyrosine kinase,
non-receptor, 1 | 22191060 |
Enrichment analysis of
autophagy-associated kinases
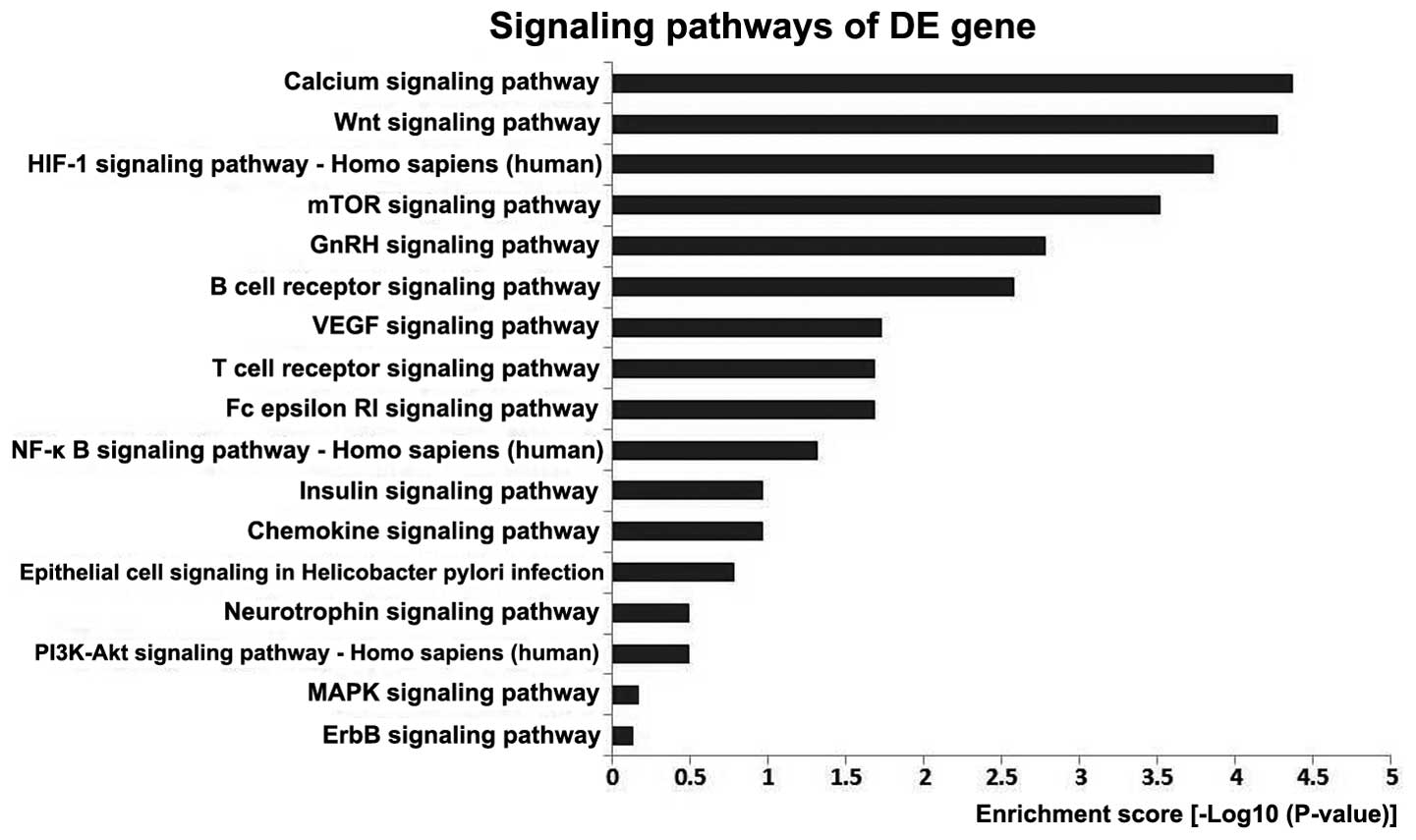
The enrichment analysis of the 54 kinases is shown
in Fig. 2, and indicates that the
functions of these kinases are mainly enriched in pathways such as
the calcium, Wnt, HIF-1 and mTOR signaling pathways.
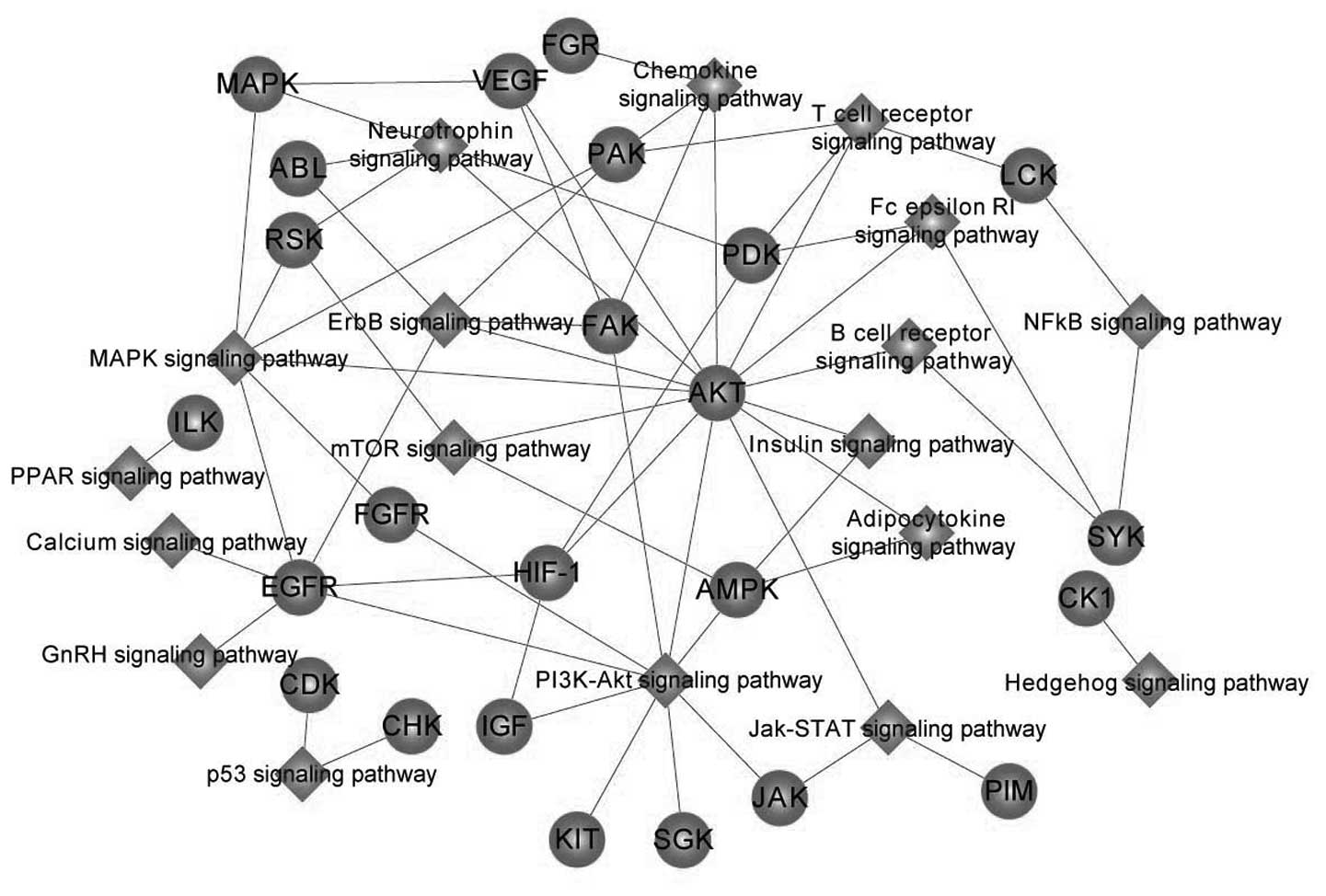
Regulatory network of
autophagy-associated kinases
With the further use of the KEGG database, the
kinases of the same KEGG signaling pathway were connected to
generate a visual network and 24 nodes signifying protein kinases
of the KEGG signaling pathways were identified (See Fig. 3).
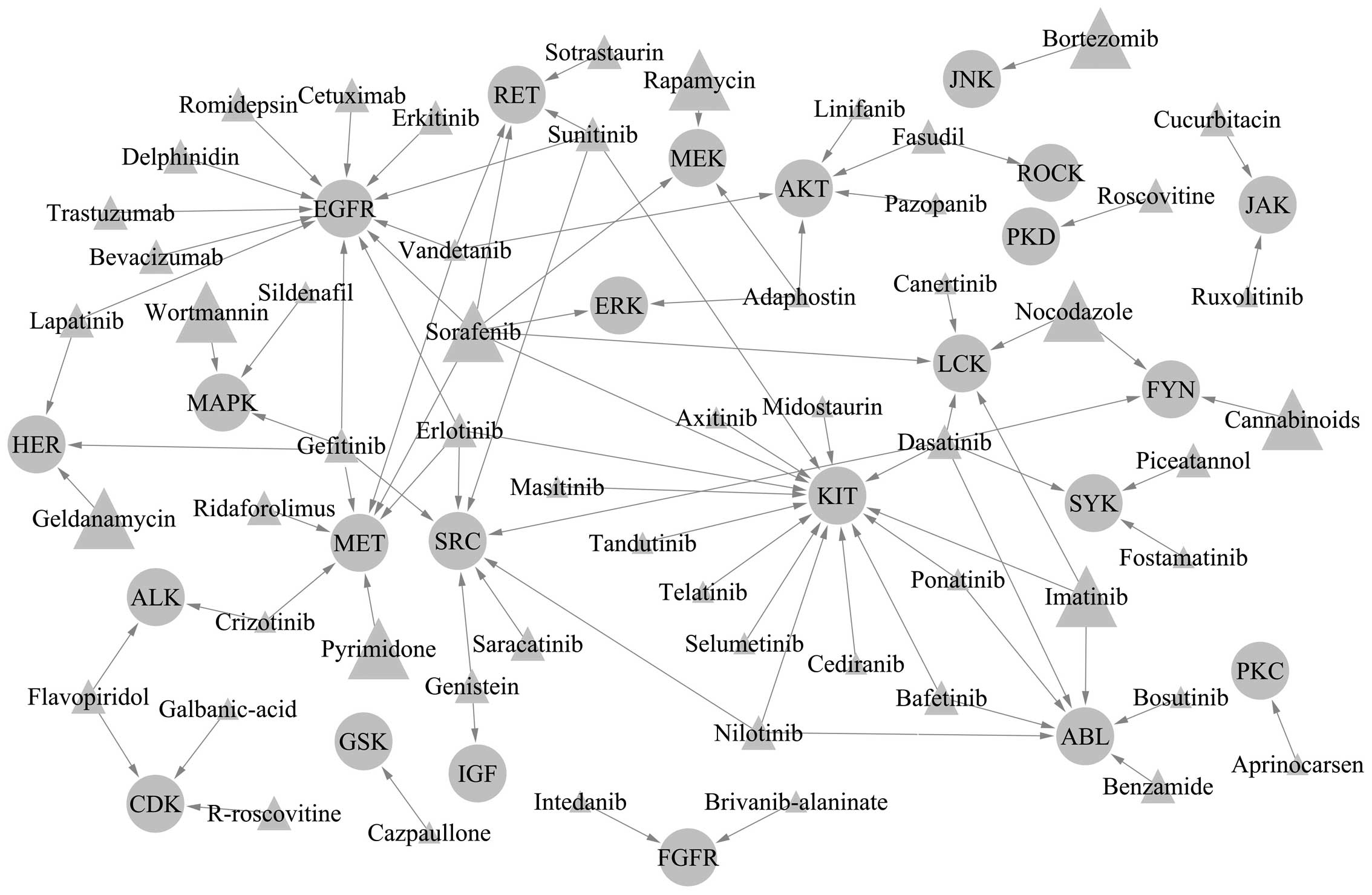
Predicted kinase inhibitors that regulate
autophagy
Through text mining, 176 preliminary articles were
retrieved, with 21 false articles (11.93%) ruled out following
manual screening. The remaining 155 articles indicated that, among
the 54 kinases shown in Table I,
24 have specific kinase inhibitors that regulate the corresponding
functions; a total of 56 kinase inhibitors were found to be
involved in the regulation of these 24 kinases. A network was
constructed as determined by these regulatory associations
(Fig. 4).
The associations between these ‘Kinase Inhibitors’
and ‘Autophagy’ were determined through further text mining and 267
articles were screened out. A total of nine of these kinase
inhibitors (wortmannin, sorafenib, geldanamycin, nocodazole,
pyrimidone, rapamycin, bortezomib, cannabinoids and imatinib) had
been widely reported in autophagy regulation research, 23 kinase
inhibitors had been seldom reported and 24 had never been reported
(Fig. 4).
Discussion
Autophagy regulates both cell survival and cell
death. However, the detailed underlying mechanisms require further
investigation. In the near future, preventing and controlling
diseases or promoting health through the regulation of autophagy
may be possible. Identifying drugs that target autophagy may be of
great importance in the treatment of numerous diseases. However,
whether autophagy may be accurately regulated requires further
analysis.
The development of bioinformatics in the
post-genomic era provides novel hypotheses in gene-to-drug
research. The concept of literature-based discovery was introduced
by Swanson in 1986 (19), and the
study indicated that abundant undiscovered public knowledge is
implied in non-interactive literature sets of articles. The
underlying discovery method is determined by the following
principle: Certain associations between two complementary natural
language text passages are largely of the form ‘A causes B’ and ‘B
causes C’. From this, it may be deduced that A and C are linked by
B, irrespective of the meaning of A or C. However, studies may not
have been published concerning a possible connection between A and
C, although such association, if validated, may be of scientific
importance. This allows for the generation of the hypotheses, such
as ‘Fish oil may be used for treatment of Reynaud’s disease’.
Currently, text mining of biomedical literature has generated a
great deal of interest in biomedical research. For example, Li
et al (20) detected 1,500
drug addiction-related genes using text mining; 396 closely related
genes were screened through meta-analysis that identified 18
enriched signaling pathways. The authors then established an
addiction-related gene database and a molecular network
interactions diagram. Gajendran et al (21) identified two novel bone
biology-related genes, S100A12 and FYN, through text mining and
bioinformatic analysis. These two studies suggest that potential
information in the literature may be identified through text mining
and bioinformatic analysis. The present study, targeted at kinases,
used kinase inhibitors as the intervention to determine kinase
inhibitors that regulate autophagy, with the aim of more rapid
translation between laboratory molecular biology results and
clinical studies.
In the present study, 54 autophagy-associated
kinases were obtained through bioinformatic analysis, indicating
that these kinases may be used as novel targets in autophagy
regulation research. The KEGG signaling pathway enrichment analysis
determining the functions of these kinases found that these kinases
are mainly enriched in certain signaling pathways.
One study observed that calcium and calcium
signaling pathways are important in autophagic regulation (22). Calcium, involved in transmembrane
signal transduction, acting as the second messenger in cytoplasm,
is of great importance in the regulation of cell survival and
apoptosis. The autophagy induced by increases in the levels of
calcium in the cytoplasm has been reported to be completed by
calcium/calmodulin-dependent protein kinase kinase β-mediated AMPK
activation and mTOR inhibition (23). Grotemeier et al (24) revealed that calcium induces
autophagy independently, without calcium-mediated AMPK activation
or complete mTOR inhibition. The fact that the increase in the
levels of calcium in the cytoplasm induce autophagy and hinder the
degradation of autophagic vacuoles indicates that calcium induces
autophagy independently without the aid of other medications. DAPK
is a calcium/calmodulin serine/threonine kinase, acting as a tumor
suppressor gene, promoting cell apoptosis and autophagy. Beclin-1
is a protein with a BH3 domain. In general, Beclin-1 combines with
the BH3 domain of the apoptosis protein bcl-2/bcl-XL inhibitor and
DAPK phosphorylates the BH3 Thr119 site in Beclin-1, resulting in
the dissociation of Beclin-1 and Bcl-XL, inducing autophagy
(25). All studies verified that
the calcium signaling pathway regulates autophagy, which is in
accordance with the present study.
Protein kinases, also termed protein phosphokinases,
are associated with almost all cell functions. However, in
particular, kinases are important in signaling between cells and
the execution of complicated cell functions, including cell
division. Thus, kinase over-activity may result in the development
of certain types of cancer (26).
The key role of kinases in controlling cell behavior has been
widely investigated as a target in the treatment of numerous other
diseases, including diabetes (27), osteoporosis (28), inflammation (29) and ophthalmopathy (30).
Techniques such as gene transfection or RNA
intervention have been shown to be distinctive and effective in the
laboratory setting for regulating kinases as targets of autophagy
research, but these methods have a number of problems when
translated into clinical practice. However, various emerging kinase
inhibitors available on the market have been observed to be
effective in regulating kinase activity (31). Thus, introducing kinase inhibitors
into switching autophagy may have more practical applications, and
this may speed up the translation between laboratory results and
clinical application, in compliance with the concept of
translational medicine.
Kinase inhibitors, compounds that inhibit protein
kinase activity, have been commonly analyzed in recent new drug
studies and an increasing number of drugs have emerged. In recent
years, a number of large pharmaceutical companies have developed
novel drugs with the primary goal of targeting kinases (32). In total more than 30 kinase
inhibitors have undergone clinical studies and been approved by the
Food and Drug Administration of the USA. First generation products,
such as imatinib (33), gefitinib
(34) and erlotinib (35), have been shown to be effective in
inhibiting the activity of the targeted kinases. Second generation
AbL kinase-targeted drugs, including dasatinib, have been
subsequently developed (36).
Third generation Aurora kinase inhibitors remain under
investigation (37). This
describes only a fraction of current kinomic studies, as a great
number of novel compounds are being developed.
The present study identified nine kinase inhibitors
(wortmannin, sorafenib, geldanamycin, nocodazole, pyrimidone,
rapamycin, bortezomib, cannabinoids and imatinib) that have been
widely reported in autophagy regulation research. Kinase inhibitors
have previously been shown to regulate autophagy.
However, other kinase inhibitors that regulate
autophagy were also identified in the present study. Among the 56
kinase inhibitors derived from the bioinformatic analysis, 24
(crizotinib, galbanic-acid, cazpaullone, intedanib,
brivanib-alaninate, sotrastaurin, linifanib, midostaurin,
ponatinib, fostamatinib, cediranib, bosutinib, aprinocarsen,
pazopanib, adaphostin, canertinib, ruxolitinib, axitinib,
telatinib, selumetinib, vandetanib, tandutinib, sildenafil and
masitinib) were not found to have been previously reported with
regard to autophagy regulation in PubMed. This indicates that
introducing these kinases into autophagy regulation analysis in
later studies may produce important benefits in the future.
References
|
1
|
Zou Y, Wang Q, Li B, Xie B and Wang W:
Temozolomide induces autophagy via ATM-AMPK-ULK1 pathways in
glioma. Mol Med Rep. 10:411–416. 2014.PubMed/NCBI
|
|
2
|
Wang W, Fan H, Zhou Y, et al: Knockdown of
autophagy-related gene BECLIN1 promotes cell growth and inhibits
apoptosis in the A549 human lung cancer cell line. Mol Med Rep.
7:1501–1505. 2013.PubMed/NCBI
|
|
3
|
Richards AL and Jackson WT: Intracellular
vesicle acidification promotes maturation of infectious poliovirus
particles. PLoS Pathog. 8:e10030462012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kingwell K: Genetics: Mutations in
autophagy gene cause a rare and severe neurodegenerative disease.
Nat Rev Neurol. 9:1822013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Salminen A, Ojala J, Kaarniranta K and
Kauppinen A: Mitochondrial dysfunction and oxidative stress
activate inflammasomes: impact on the aging process and age-related
diseases. Cell Mol Life Sci. 69:2999–3013. 2012. View Article : Google Scholar
|
|
6
|
Zou Z, Yuan Z, Zhang Q, et al: Aurora
kinase A inhibition-induced autophagy triggers drug resistance in
breast cancer cells. Autophagy. 8:1798–1810. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen A, Xiong LJ, Tong Y and Mao M:
Neuroprotective effect of brain-derived neurotrophic factor
mediated by autophagy through the PI3K/Akt/mTOR pathway. Mol Med
Rep. 8:1011–1016. 2013.PubMed/NCBI
|
|
8
|
Mack HI, Zheng B, Asara JM and Thomas SM:
AMPK-dependent phosphorylation of ULK1 regulates ATG9 localization.
Autophagy. 8:1197–1214. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhao YG, Zhao H, Miao L, et al: The
p53-induced gene Ei24 is an essential component of the basal
autophagy pathway. J Biol Chem. 287:42053–42063. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gandesiri M, Chakilam S, Ivanovska J, et
al: DAPK plays an important role in panobinostat-induced autophagy
and commits cells to apoptosis under autophagy deficient
conditions. Apoptosis. 17:1300–1315. 2012. View Article : Google Scholar
|
|
11
|
Yadunandam AK, Yoon JS, Seong YA, Oh CW
and Kim GD: Prospective impact of 5-FU in the induction of
endoplasmic reticulum stress, modulation of GRP78 expression and
autophagy in Sk-Hep1 cells. Int J Oncol. 41:1036–1042.
2012.PubMed/NCBI
|
|
12
|
Roy A and Kolattukudy PE: Monocyte
chemotactic protein-induced protein (MCPIP) promotes inflammatory
angiogenesis via sequential induction of oxidative stress,
endoplasmic reticulum stress and autophagy. Cell Signal.
24:2123–2131. 2012. View Article : Google Scholar
|
|
13
|
Criollo A, Maiuri MC, Tasdemir E, et al:
Regulation of autophagy by the inositol trisphosphate receptor.
Cell Death Differ. 14:1029–1039. 2007.PubMed/NCBI
|
|
14
|
Lipatova Z, Belogortseva N, Zhang XQ, et
al: Regulation of selective autophagy onset by a Ypt/Rab GTPase
module. Proc Natl Acad Sci USA. 109:6981–6986. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cea M, Cagnetta A, Fulciniti M, et al:
Targeting NAD+ salvage pathway induces autophagy in
multiple myeloma cells via mTORC1 and extracellular
signal-regulated kinase (ERK1/2) inhibition. Blood. 120:3519–3529.
2012.PubMed/NCBI
|
|
16
|
Meijer AJ and Codogno P: Signalling and
autophagy regulation in health, aging and disease. Mol Aspects Med.
27:411–425. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Crawford SE, Hyser JM, Utama B and Estes
MK: Autophagy hijacked through viroporin-activated
calcium/calmodulin-dependent kinase kinase-β signaling is required
for rotavirus replication. Proc Natl Acad Sci USA. 109:E3405–E3413.
2012.PubMed/NCBI
|
|
18
|
Zhang K, Wang X and Wang H: Effect and
mechanism of Src tyrosine kinase inhibitor sunitinib on the
drug-resistance reversal of human A549/DDP cisplatin-resistant lung
cancer cell line. Mol Med Rep. 10:2065–2072. 2014.PubMed/NCBI
|
|
19
|
Swanson DR: Fish oil, Raynaud’s syndrome,
and undiscovered public knowledge. Perspect Biol Med. 30:7–18.
1986.
|
|
20
|
Li CY, Mao X and Wei L: Genes and (common)
pathways underlying drug addiction. PLoS Comput Biol. 4:e22008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gajendran VK, Lin JR and Fyhire DP: An
application of bioinformatics and text mining to the discovery of
novel genes related to bone biology. Bone. 40:1378–1388. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cárdenas C and Foskett JK: Mitochondrial
Ca(2+) signals in autophagy. Cell Calcium. 52:44–51.
2012.PubMed/NCBI
|
|
23
|
Høyer-Hansen M, Bastholm L, Szyniarowski
P, et al: Control of macroautophagy by calcium,
calmodulin-dependent kinase kinase-beta, and Bcl-2. Mol Cell.
25:193–205. 2007.PubMed/NCBI
|
|
24
|
Grotemeier A, Alers S, Pfisterer SG, et
al: AMPK-independent induction of autophagy by cytosolic
Ca2+ increase. Cell Signal. 22:914–925. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zalckvar E, Berissi H, Mizrachy L, et al:
DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1
promotes dissociation of beclin 1 from Bcl-XL and induction of
autophagy. EMBO Rep. 10:285–292. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lu W, Qu JJ, Li BL, et al: Overexpression
of p21-activated kinase 1 promotes endometrial cancer progression.
Oncol Rep. 29:1547–1555. 2013.PubMed/NCBI
|
|
27
|
Sajan MP, Nimal S, Mastorides S, et al:
Correction of metabolic abnormalities in a rodent model of obesity,
metabolic syndrome, and type 2 diabetes mellitus by inhibitors of
hepatic protein kinase C-ι. Metabolism. 61:459–469. 2012.PubMed/NCBI
|
|
28
|
Liu Y, Berendsen AD, Jia S, et al:
Intracellular VEGF regulates the balance between osteoblast and
adipocyte differentiation. J Clin Invest. 122:3101–3113. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kim SB, Kang OH, Joung DK, et al:
Anti-inflammatory effects of tectroside on UVB-induced HaCaT cells.
Int J Mol Med. 31:1471–1476. 2013.PubMed/NCBI
|
|
30
|
Murakami Y, Matsumoto H, Roh M, et al:
Receptor interacting protein kinase mediates necrotic cone but not
rod cell death in a mouse model of inherited degeneration. Proc
Natl Acad Sci USA. 109:14598–14603. 2012. View Article : Google Scholar
|
|
31
|
Marech I, Patruno R, Zizzo N, et al:
Masitinib (AB1010), from canine tumor model to human clinical
development: where we are? Crit Rev Oncol Hematol. 91:98–111. 2014.
View Article : Google Scholar
|
|
32
|
Ferrè F, Palmeri A and Helmer-Citterich M:
Computational methods for analysis and inference of
kinase/inhibitor relationships. Front Genet. 30:1962014.PubMed/NCBI
|
|
33
|
Niu CC, Zhao C, Yang ZD, et al:
Downregulation of γ-catenin inhibits CML cell growth and
potentiates the response of CML cells to imatinib through β-catenin
inhibition. Int J Mol Med. 31:453–458. 2013.
|
|
34
|
Wu J, Min R, Wu M and Chen W: Gefitinib
induces mitochondrial-dependent apoptosis in Saccharomyces
cerevisiae. Mol Med Rep. 4:357–362. 2011.PubMed/NCBI
|
|
35
|
Guttman-Yassky E, Mita A, De Jonge M, et
al: Characterisation of the cutaneous pathology in non-small cell
lung cancer (NSCLC) patients treated with the EGFR tyrosine kinase
inhibitor erlotinib. Eur J Cancer. 46:2010–2019. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Song Z, Lu P, Furman RR, et al: Activities
of SYK and PLCgamma2 predict apoptotic response of CLL cells to SRC
tyrosine kinase inhibitor dasatinib. Clin Cancer Res. 16:587–599.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li Y, Tang K, Zhang H, et al: Function of
Aurora kinase A in Taxol-resistant breast cancer and its
correlation with P-gp. Mol Med Rep. 4:739–746. 2011.PubMed/NCBI
|