Introduction
Non-small cell lung cancer (NSCLC) is the most
common type of lung cancer and is a leading cause of lung
cancer-associated mortality worldwide (1–3).
Patients with NSCLC are mainly treated using platinum-based
chemotherapy, including cisplatin (diaminedichloroplatinum, DDP)
treatment (4). However, the
continuous infusion or multiple administration of DDP often results
in the development of drug resistance, leading to treatment failure
(5). Therefore, it is necessary to
elucidate the molecular mechanisms underlying DDP resistance in
NSCLC. A greater understanding of these mechanisms may aid the
identification of novel therapeutic targets for attenuating DDP
resistance.
MicroRNAs (miRNAs), small, noncoding regulatory RNAs
of 21–25 nucleotides, are critical regulators of
post-transcriptional gene expression (6). Alterations in miRNA expression have
been demonstrated to be involved in regulating drug resistance in
various tumors. A role of miR-451 in regulating resistance of MCF-7
breast cancer cells to chemotherapeutic drug doxorubicin has been
reported (7). miR-215 expression
has been demonstrated to result in osteosarcoma and colon cancer
cell-chemoresistance to methotrexate and Tomudex (8). miRNA-106a (miR-106a) belongs to the
miR-17 family and has been reported to confer DDP resistance to
esophageal adenocarcinoma cells (9). Huh et al (10) demonstrated that dysregulation of
miR-106a expression confers paclitaxel resistance to ovarian cancer
cells. A study suggested that anti-miR-106a (specific to miR-106a)
induced A549-cell apoptosis and increased the sensitivity of A549
cells to anti-cancer drugs (11).
However, the effects of miR-106a and the mechanism through which
miR-106a influences DDP-induced apoptosis in NSCLC remain to be
elucidated.
The adenosine triphosphatase-binding cassette (ABC)
transporter family of genes comprises multi-drug
resistance-associated genes, including 48 members classified into
seven subfamilies (ABCA-ABCG) in humans (12). The majority of these transporters,
including ABCB1, ABCC2 and ABCG2, have been well studied in their
capacity for influencing cancer-drug resistance (13–17).
The activity of these transporters may therefore influence the
efficacy of chemotherapeutic drugs in tumor treatment. Although
miR-106a may have an important role in conferring DDP-resistance
(18), the mechanisms through
which miR-106a regulates ABC transporter proteins and modulates
DDP-induced apoptosis in NSCLC cells remain to be elucidated. The
present study therefore aimed to elucidate the mechanism by which
miR-106a interacts with ABCA1 and modulates DDP resistance in NSCLC
A549 cells. The results may indicate a potential therapeutic target
for the treatment of NSCLC patients with DDP-resistance.
Materials and methods
NSCLC A549 and A549/DDP cell culture
A549 cells and A549/DDP cells (Guangzhou Zixiutang
Biotechnology Co, Ltd., Guangzhou, China) were cultured in
RPMI-1640 medium (Gibco-BRL, Invitrogen Life Technologies,
Carlsbad, CA, USA), supplemented with 10% fetal bovine serum (FBS)
and 1% PS (100 U/ml penicillin, 100 μg/ml streptomycin; DingGuo,
Beijing, China) and maintained in a humidified incubator with 5%
CO2 at 37°C. To maintain the DDP-resistant phenotype,
DDP was added to the culture media with a final concentration of 1
mg/ml for A549/DDP cells. A549/DDP cells were cultured for one week
in DDP-free medium prior to their experimental use.
Cell viability assay
A549 and A549/DDP cells were plated in 96-well
plates at a density of 5,000 cells/well. Eight hours following
transfection, cells were treated with various doses of DDP (0.125,
0.25, 0.5, 1.0, 2.0, 4.0 and 8.0 mg/ml; Dinghui Pharmaceutical
Factory, Hunan, China) in combination with MTT and were incubated
for 4 h at 37°C. The cells were subsequently agitated with MTT
solvent on an orbital shaker for 10 min in the dark. The absorbance
at 570 nm was measured using an automatic microplate reader (Ani
Labsystems, Ltd. Oy, Vantaa, Finland).
Reverse transcription quantitative
polymerase chain reaction (RT-qPCR) analysis of mRNA
expression
Total RNA was extracted using RNAiso Plus (Takara
Bio, Inc., Otsu, Japan) according to the manufacturer’s
instructions. cDNA was synthesized using the RevertAid First-Strand
cDNA Synthesis kit (Fermentas, Thermo Fisher Scientific,
Pittsburgh, PA, USA) according to the manufacturer’s instructions.
Following the reverse transcription reaction, qPCR was conducted
using the ABI 7300 PCR System (Applied Biosystems Life
Technologies, Foster City, CA, USA), using U6 and β-actin as
endogenous controls for data normalization. Primers were as
follows: β-actin forward, 5′-AATCTGGCACCACACCTTCT-3′ and reverse,
5′-AGCACAGCCTGGATAGCAAC-3′; ABCA1 forward,
5′-TCTCCAGAGCCAACCTGGCAGCA-3′ and reverse,
5′-CCACAGGAGACAGCAGGCTAGCGA-3′.
Transfection of miR-106a mimic, inhibitor
and short interfering (si)RNA-ABCA1
The miR-106a mimic, inhibitor anti-miR-106a and
relative control mimic were purchased from GenePharma (Shanghai,
China). The sequences were as follows: miR-106a mimic,
5′-GAUGGACGUGACAUUCGUGAAAA-3′ and anti-miR-106a,
5′-CUACCUGCACUGUAAGCACUUUU-3′. siRNA-ABCA1 (si-ABCA1) was also
purchased from GenePharma. The sequences used in the present study
were as follows: siR sense, 5′-GGAUGUACAACGAACAGUACA-3′ and
antisense, 5′-UACUGUUCGUUGUACAUCCAG-3′ (12). In a transfection assay, the cells
were transfected with 50 nM miR-106a mimic, anti-miR-106a or
si-ABCA1 and siRNA control using Lipofectamine® 2000
reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) according
to the manufacturer’s instructions. The medium was replaced with
fresh RPMI-1640 (DingGuo) containing 10% FBS (DingGuo) and
antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin) 6 h
post-transfection. For qPCR and western blot analysis, the cells
were collected following an additional 48 h of incubation. Three
independent experiments were performed.
Dual luciferase activity assay
The 3′-untranslated region(UTR) of human
ABCA1 cDNA containing the putative target site for miR-106a
was chemically synthesized and inserted into the pGL3-control
vector, downstream of the luciferase gene (Promega, Madison, WI,
USA). Cells were plated at 2×105 cells/well in 24-well
plates. Subsequently, pGL3-ABCA1–3′-UTR and 80 ng Renilla
luciferase-herpes simplex virus-thymidine kinase control reporter
vector (Promega) were transfected in combination with miR-106a
mimic, anti-miR-106a or controls using Lipofectamine®
2000 (Invitrogen Life Technologies) according to the manufacturer’s
instructions. Luciferase activity was measured using the Dual
Luciferase Reporter Assay system (Promega). Firefly luciferase
activity was normalized to Renilla luciferase activity for
each transfected well.
Western blot analysis
To obtain whole cell lysates, the cell pellet was
resuspended in whole cell lysis buffer (10 mM Tris at pH 7.4, 1 mM
NaF, 1 mM Na3VO4, 1 mM phenylmethanesulfonyl
fluoride, 0.1 % sodium dodecyl sulfate, 1% Triton X-100 and the
protease inhibitor cocktail; Nanjing chemistry company, Nanjing,
China) and was subsequently lysed by freezing-thawing followed by
sonication. The first antibody was rabbit polyclonal anti-ABCA1
(EMD Millipore, Billerica, MA, USA; 1:100 dilution) and
anti-β-actin antibody (Abcam, San Francisco, CA, USA; 1:1,000
dilution). The secondary antibody was goat anti-rabbit
immunoglobulin G, conjugated with horseradish peroxidase at a
dilution of 1:1,000. The bound antibodies were detected using
Enhanced Chemiluminescence Plus Western Blotting Detection system
(GE Healthcare, Little Chalfont, UK). β-actin was used as an
internal control to normalize ABCA1 expression levels.
Representative images from one of three independent experiments are
exhibited. The blotting band intensity was quantified using Image-J
software (http://imagej.nih.gov/ij/).
Annexin V and propidium iodide (PI)
staining
Enumeration of apoptotic cells was performed using
fluorescein isothiocyanate (FITC)-conjugated Annexin V and PI (BD
Pharmingen, San Diego, CA, USA). Cells were washed twice in cold 1×
PBS and resuspended in Annexin V-binding buffer (BD Pharmingen) at
a concentration of 3×106 cells/ml. This suspension (100
μl) was stained with 5 μl Annexin V-FITC and 5 μl PI. The cells
were gently vortexed and incubated for 15 min at room temperature
in the dark. Following the addition of 400 μl binding buffer to
each tube, cells were analyzed by flow cytometry (Cytomics FC500;
Beckman Coulter, Miami, FL, USA).
Statistical analysis
Values are presented as the mean + standard
deviation and experiments were repeated three times. The difference
was determined by two-tailed Student’s t-tests using GraphPad 5.0
software (GraphPad Software, Inc., La Jolla, CA, USA). P<0.05
was considered to indicate a statistically significant difference
between values.
Results
miR-106a is upregulated in DDP-resistant
A549/DDP cells and involved in conferring DDP-resistance to A549
cells
Survival curves of the A549 and A549/DDP cells in
response to various doses of DDP were detected (Fig. 1A). The results revealed that the
A549/DDP cell lines exhibited greater resistance to DDP compared
with that of the A549 cell line. Subsequently, the involvement of
miR-106a in DDP-resistant A549/DDP cells was investigated using
RT-qPCR. The results indicated that miR-106a expression levels were
higher in A549/DDP cells than in A549 cells (Fig. 1B), which suggested that miR-106a
may be associated with DDP resistance in NSCLC cells, which was in
agreement with a previous study (10). Furthermore, in order to investigate
the association between miR-106a and DDP resistance in A549 cells,
the effect of overexpression of miR-106a on A549 cells was
evaluated. RT-qPCR revealed that transfection with the miR-106a
mimic significantly increased miR-106a mRNA expression levels,
which suggested that miR-106a was efficiently transfected into the
cells (Fig. 1C and D). The
miR-106a mimic-transfected A549 cells had a significantly higher
survival rate than that of the negative control (miR-control)
group. The effect of miR-106a repression on A549/DDP cells was also
tested. The relative results (Fig. 1E
and F) indicated that the A549/DDP cells transfected with
anti-miR-106a had a significantly lower survival rate than that of
the control group. These results demonstrated that miR-106a confers
DDP-resistance in A549 cells and knockdown of miR-106a was able to
moderately sensitize A549/DDP cells to DDP.
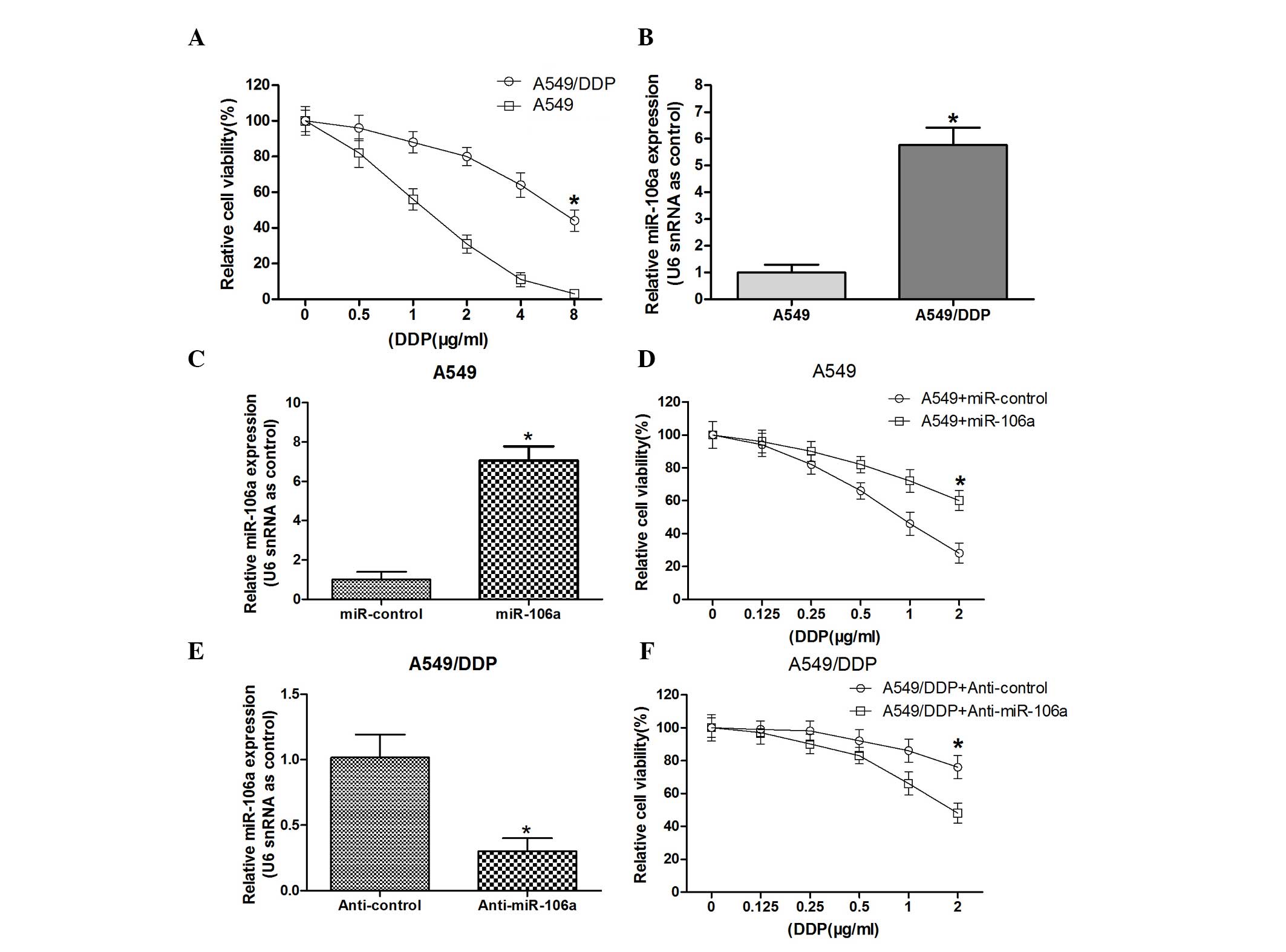 | Figure 1miR-106a is upregulated in
DDP-resistant A549/DDP cells and involved in conferring DDP
resistance to A549 cells. (A) Survival curves of A549/DDP and A549
cells. Cells were treated with various doses of DDP (0.125, 0.25,
0.5, 1.0, 2.0, 4.0 and 8.0 mg/ml). Following 48 h of incubation,
cell viability was measured by MTT assay. (B) RT-qPCR analysis of
miR-106a mRNA expression levels in A549 and A549/DDP cells. U6
snRNA was used as a loading control. (C and E) qRT-PCR indicated
that miR-106a expression levels were significantly increased or
decreased in the cells transfected with the miR-106a mimic or
anti-miR-106a in A549 cells or A549/DDP cells, respectively. (D and
F) Transfected A549 cells or A549/DDP cells were incubated with
various doses of DDP (0.125, 0.25, 0.5, 1.0 and 2.0 mg/ml) for 48 h
prior to MTT assay to determine cell viability.
*P<0.05. DDP, cisplatin; RT-qPCR, reverse
transcription quantitative polymerase chain reaction; snRNA, small
nuclear RNA; miR, micro RNA; mRNA, messenger RNA. |
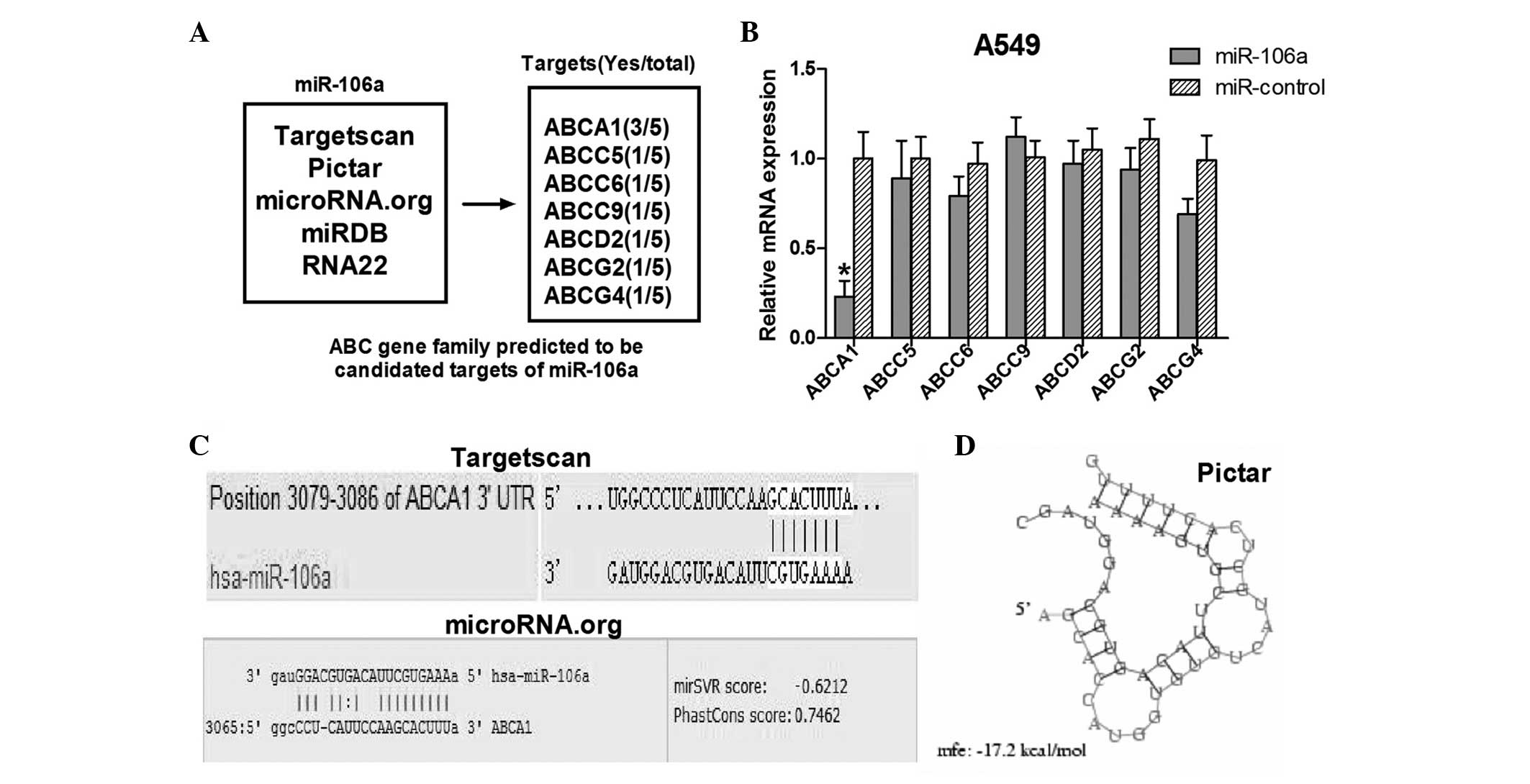
ABCA1 is a candidate target gene of
miR-106a
As miR-106a has a pivotal function in conferring DDP
resistance of NSCLC A549 cells, it is important to explore the
target genes of miR-106a involved in the DDP-resistance mechanism.
To determine which ABC transporter genes may be regulated by
miR-106a, six miRNA target prediction algorithms were utilized in
order to identify the target genes of miR-106a. A list of seven ABC
transporter genes was identified, including ABCA1, ABCC5, ABCC6,
ABCC9, ABCD2, ABCG2 and ABCG4 (Fig. 2A). Among these genes, ABCA1
had the greatest frequency (three of the five algorithms predicted
that the 3′UTR of ABCA1 contained the putative binding site
of miR-106a). A549 cells were subsequently transfected with
miR-106a and RT-qPCR was performed in order to detect the
expression levels of the seven candidate target genes of miR-106a.
Of the potential target genes identified, ABCA1 was
selected, as it was demonstrated to have the lowest expression
levels in the transfected A549 cells (Fig. 2B). The alignments of miR-106a with
the ABCA1 3′UTR insert predicted by Targetscan, Pictar and
microRNA.org software are illustrated in Fig. 2C and D.
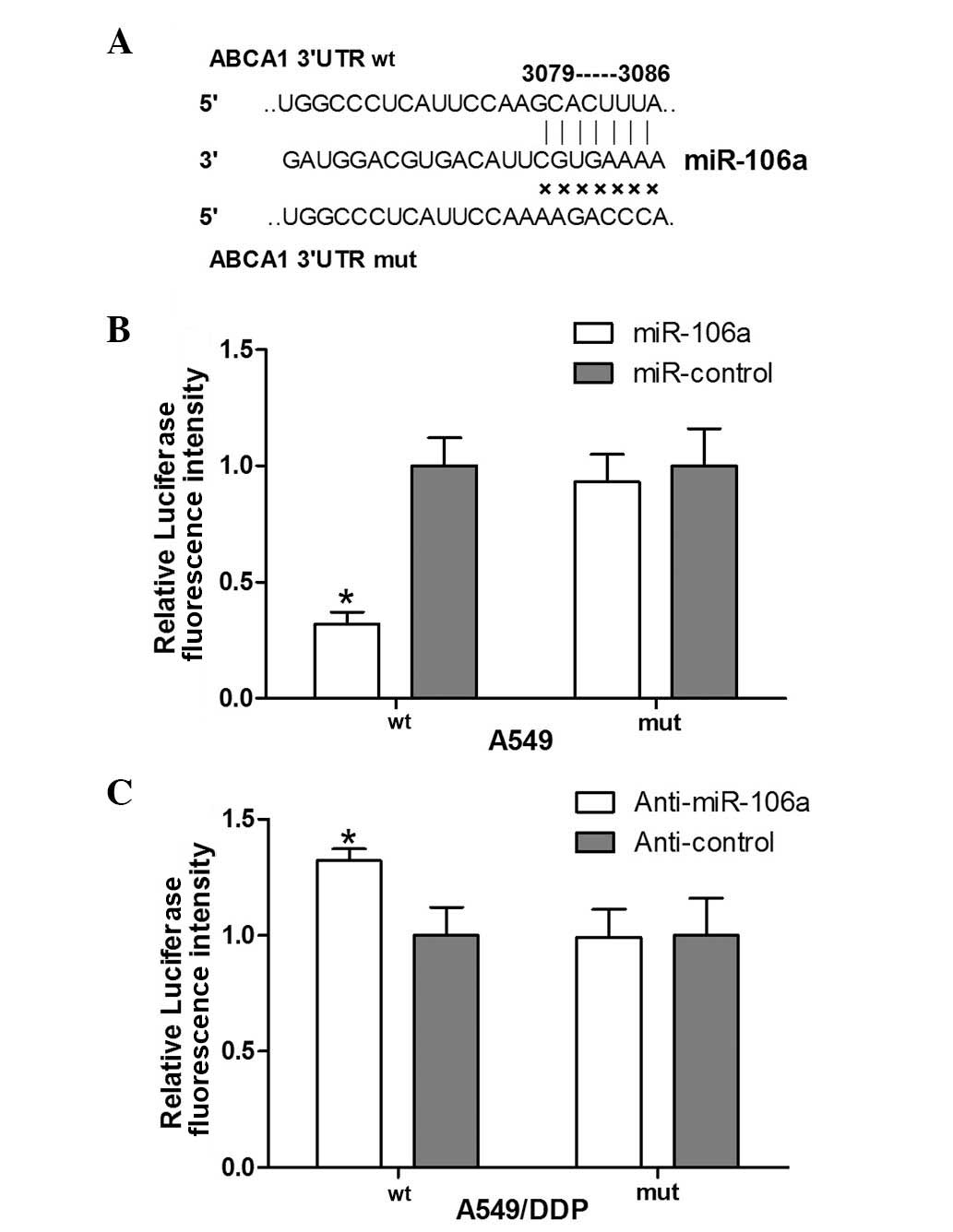
ABCA1 is a direct target of miR-106a
To determine whether miR-106a directly regulates
ABCA1 by binding to the 3′UTR of the target gene, a
luciferase reporter assay was performed. Wild-type and mutant ABCA1
3′UTR sequences which contained the miR-106a binding site were
constructed and inserted into pGL3 vectors (Fig. 3A). The reporter construct and
miR-106a mimics were transfected into the DDP-sensitive cell line
(A549). Concurrently, the reporter construct and anti-miR-106a were
transfected into the DDP-resistant A549/DDP cell line. When
miR-106a was overexpressed in A549 cells, luciferase expression
levels were significantly lower than those in the miR-control
group. However, the luciferase intensity in the mutant ABCA1
3′UTR cells was unaffected by miR-106a transfection (Fig. 3B). In analogy, repression of
miR-106a caused an increase in luciferase levels in DPP-resistant
cells, while mutant cells were unaffected (Fig. 3C). It was therefore concluded that
ABCA1 was a direct target of miR-106a.
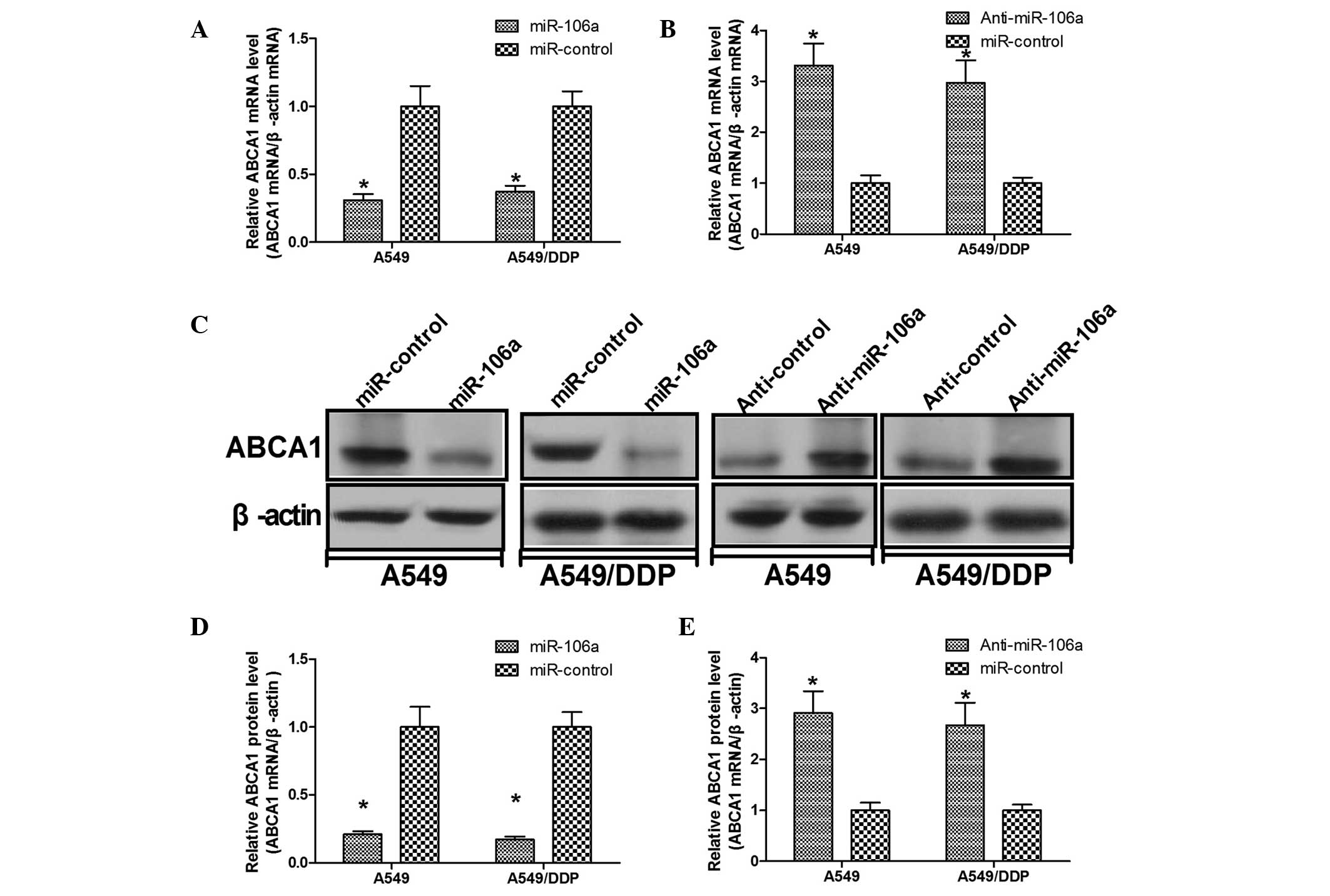
miR-106a downregulates ABCA1 mRNA and
protein expression levels
RT-qPCR and western blotting assays were performed
in order to determine the regulation of endogenous ABCA1
expression levels by miR-106a. The results demonstrated that
overexpression of miR-106a significantly decreased ABCA1 mRNA and
protein expression levels in A549 and A549/DDP cells (P<0.05;
Fig. 4A, C and D). Simultaneously,
repression of miR-106a significantly increased ABCA1 mRNA and
protein expression levels in A549 and A549/DDP cells (P<0.05;
Fig. 4B, C and E). These results
suggested that miR-106a negatively regulated ABCA1
expression in A549 and A549/DDP cells.
ABCA1 is an important signaling molecule
in miR-106a-regulated DDP-resistance in A549/DDP cells
The results of the present study demonstrated that
miR-106a overexpression induced DDP-resistance, miR-106a knockdown
rescued DDP sensitivity and miR-106a was able to directly target
ABCA1, a membrane transporter involved in drug uptake. To determine
whether ABCA1 had a key role in miR-106a-regulated DDP
resistance, ABCA1 siRNA or si-negative control (NC) was transfected
into anti-miR-106a transfected A549/DDP cells and the cell survival
rate under various concentrations of DDP was evaluated. Western
blot analysis indicated that ABCA1 siRNA effectively reduced the
protein expression levels of ABCA1 (Fig. 5A). ABCA1 knockdown significantly
increased the survival rate of the A549/DDP cells transfected with
anti-miR-106a compared with that of the si-NC group (Fig. 5B), which suggested that miR-106a
modulated DDP resistance in A549/DDP cells by targeting ABCA1. To
confirm this mechanism, an additional rescue assay was performed
using flow cytometry. As indicated in Fig. 5C and D, ABCA1 knockdown
significantly decreased the apoptotic rate of A549/DDP cells
transfected with anti-miR-106a compared with that of the si-NC
group.
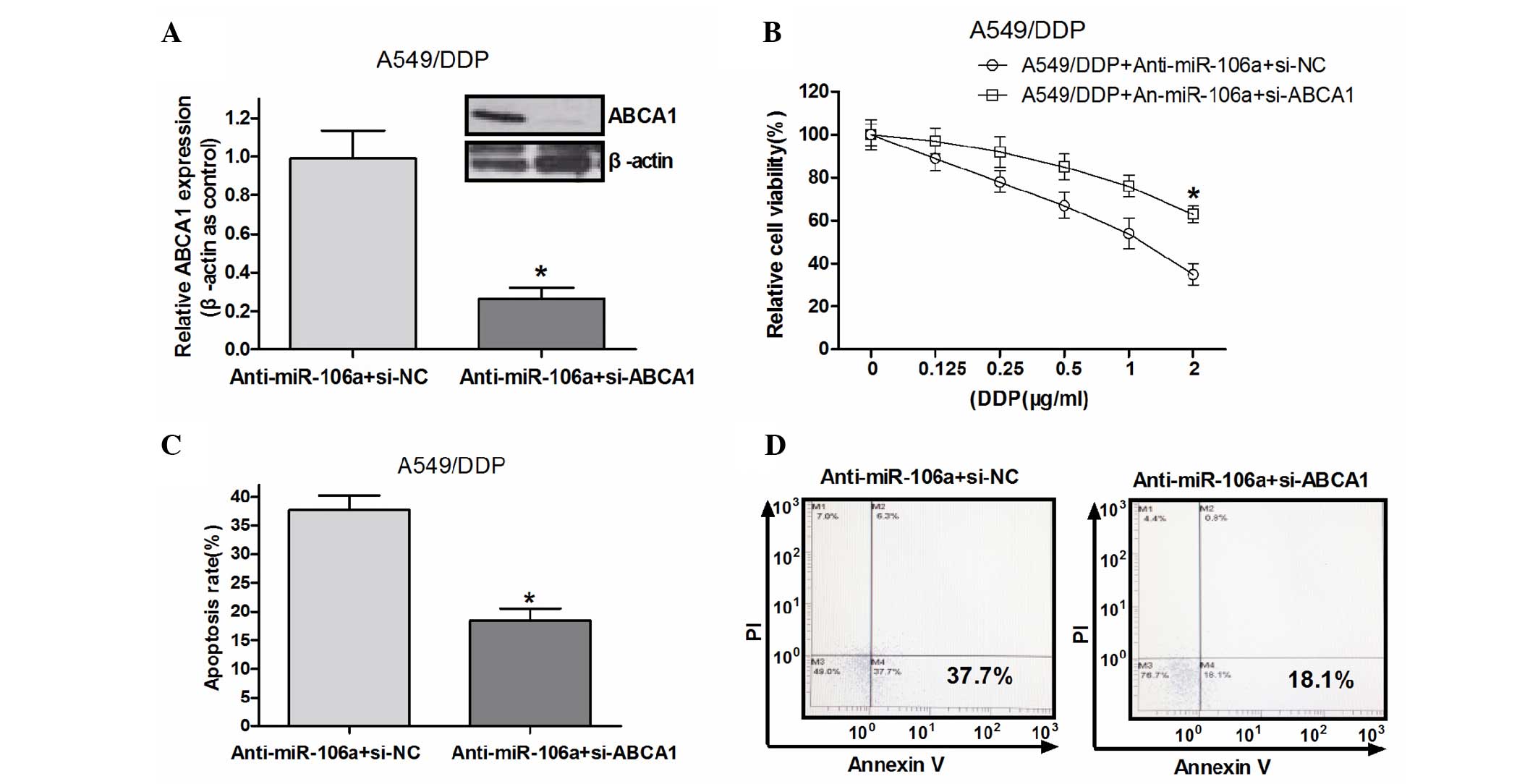 | Figure 5ABCA1 has an important role in
miR-106a-regulated DDP-resistance in A549/DDP cells. (A) ABCA1
protein levels in miR-106a inhibitor-transfected A549/DDP cells
following treatment with ABCA1 siRNA or siRNA-NC. (B) A549/DDP
cells were treated with various doses of DDP (0.125, 0.25, 0.5, 1.0
and 2.0 mg/ml) 48 h post-transfection. Cell viability was
determined by MTT assay. (C) miR-106a-inhibited A549/DDP cells were
treated with ABCA1 siRNA or a siRNA-NC, 48 h following
transfection. Apoptotic rate was determined by flow cytometry. (D)
Representative figures. *P<0.05 compared with NC.
mRNA, messenger RNA; siRNA, short interfering RNA; miR, micro RNA;
DDP, cisplatin; PI, propidium iodide; ABCA1, adenosine
triphosphatase-binding cassette sub-family A, member 1; NC,
negative control. |
Discussion
Chemoresistance is one of the most significant
obstacles in the successful treatment of lung cancer (5). Extensive studies have demonstrated
that miRNAs may act as regulators of chemosensitivity in addition
to regulating oncogenes or tumor-suppressor genes in various types
of human cancer (19,20). With respect to lung cancer, miR-31
was reported to inhibit cisplatin-induced cell apoptosis by
regulating the drug transporter ABCB9 (21). A recent study indicated that the
upregulation of miR-92b influenced the cisplatin-chemosensitivity
phenotype of lung cancer cells by targeting PTEN (22). Another study demonstrated that
miR-495 enhanced the sensitivity of NSCLC cells to platinum by
modulation of copper-transporting P-type adenosine triphosphatase A
(23). However, few studies have
investigated the role of miR-106a and its target genes in lung
cancer-cell drug resistance, although its association with cancer
has been well characterized. In the present study, the expression
levels of miR-106a in DDP-resistant A549 cells (A549/DDP) were
detected by RT-qPCR and the results indicated that the upregulation
of miR-106a expression levels were associated with DDP resistance.
It was also confirmed that the modulation of miR-106a in A549/DDP
cells resensitized them to DDP.
The ABC gene families have been well characterized
in their association with drug or multi-drug resistance in various
types of human cancers, including P-glycoprotein (ABCB1)
(24,25). In the present study, three
computational algorithms were used to predict putative targets of
miR-106a. These algorithms identified seven members of the ABC gene
family which had putative target sites in their 3′UTR. ABCA1
was validated as the most significant target of miR-106a in lung
cancer cell lines by reporter-gene assay and western blot analysis.
In several cell lines, functional targets of miR-106a have been
verified, including SLC2A3, p130 and FASTK (26–28).
The present study, for the first time, suggests that miR-106a
directly targeted the ABCA1 3′UTR and regulated its mRNA and
protein expression levels.
ABCA1 is a member of the ABC transporter
family and has an important role in cellular and body metabolism.
It has been reported that cancer-specific ABCA1
hypermethylation and loss of protein expression result in high
intracellular cholesterol levels and therefore contribute to the
creation of an environment conducive to tumor progression (29). Another study demonstrated an
anti-cancer function of the cholesterol exporter ABCA1 in human
cancer cells and reported that reconstitution of ABCA1
expression inhibited tumor formation (30). ABCA1 has also been
demonstrated to have important roles in resistance to curcumin in
M14 melanoma cells (31). These
reports support the results of the present study in suggesting that
ABCA1, as an important ABC gene transporter, mediated
miR-106a-induced DDP resistance in A549 cells. The present study
additionally established a rescue assay to confirm this hypothesis.
miR-106a-repressed A549/DDP cells were transfected with si-ABCA1,
which demonstrated that the viability of A549/DDP cells was
recovered in the cells in the si-ABCA1 group compared with those in
the control group. Furthermore, the high apoptotic rate of A549/DDP
cells caused by miR-106a knockdown was attenuated by si-ABCA1.
These results associated miR-106a, which has an anti-apoptotic
role, and ABCA1, which acts as a tumor suppressor, with DDP
resistance in A549 cells. It was therefore hypothesized that
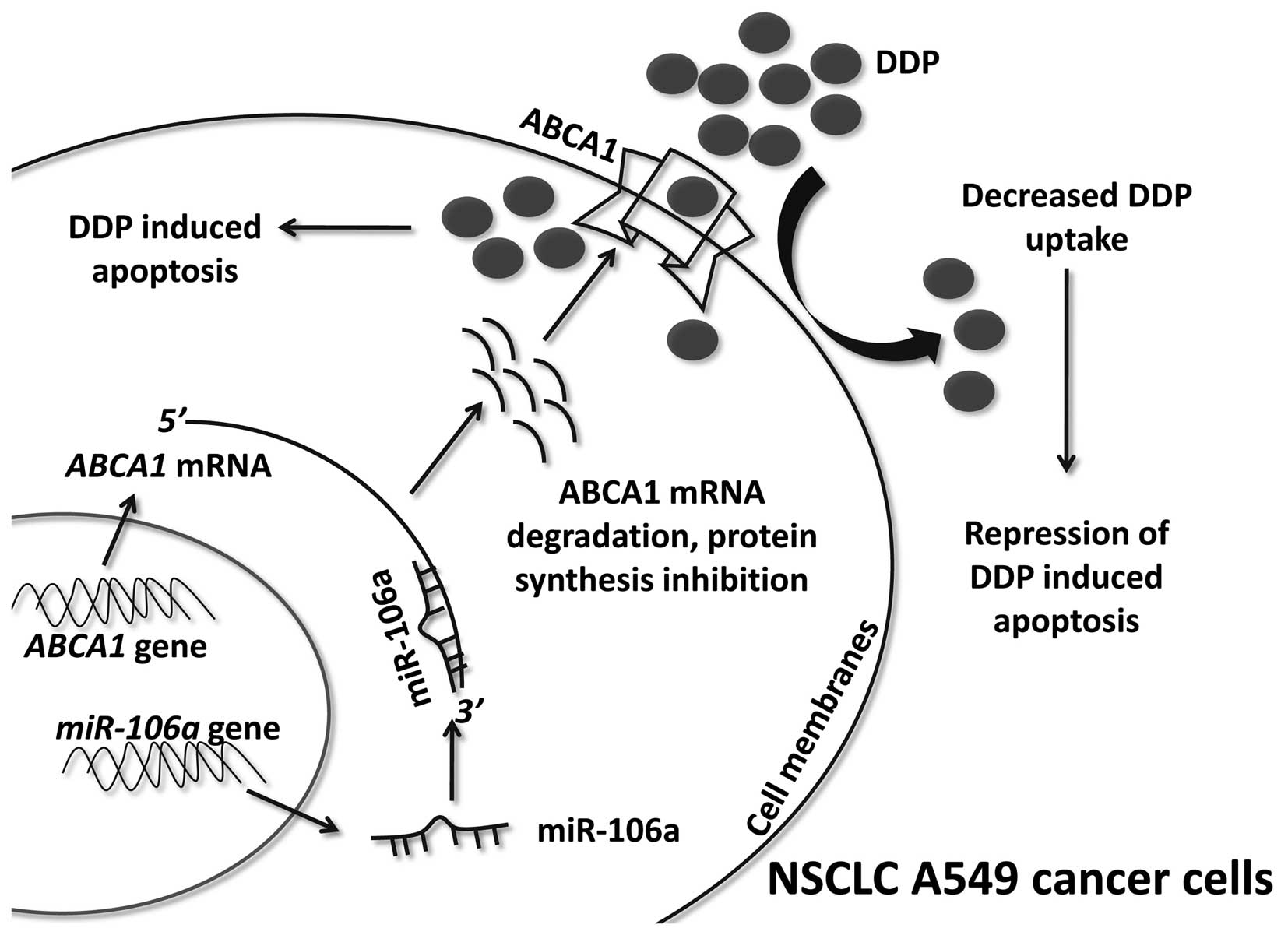
miR-106a was upregulated in a DDP-resistant NSCLC cell line
compared with a corresponding DDP-sensitive NSCLC cell line
(A549/DDP compared to A549). miR-106a upregulation suppressed ABCA1
mRNA and protein expression levels. Thus, the inhibition of
ABCA1 resulted in decreased DDP uptake in A549 cells. These
factors contributed to the miR-106a-induced DDP resistance of A549
cells (Fig. 6).
Further study will reveal that miR-106a is an
anti-apoptotic factor in other NSCLC cell lines. The present study
provided the first evidence, to the best of our knowledge, that
miR-106a has an important role in conferring DDP-resistance by
targeting ABCA1 in A549 cell lines and thus exhibits
anti-apoptotic properties. The present study highlighted the
potentially important role of miR-106a in the development of DDP
resistance and suggested that miR-106a may serve as a biomarker
which may be used to predict patient response to DDP in NSCLC.
References
|
1
|
Siegel R, DeSantis C, Virgo K, Stein K,
Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S, et al:
Cancer treatment and survivorship statistics, 2012. CA Cancer J
Clin. 62:220–241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Center MM, DeSantis C and Ward
EM: Global patterns of cancer incidence and mortality rates and
trends. Cancer Epidemiol Biomarkers Prev. 19:1893–1907. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Saika K and Machii R: Cancer mortality
attributable to tobacco by region based on the WHO Global Report.
Jpn J Clin Oncol. 42:771–772. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stinchcombe TE and West HL: Maintenance
therapy in non-small-cell lung cancer. Lancet. 374:1398–1400. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chang A: Chemotherapy, chemoresistance and
the changing treatment landscape for NSCLC. Lung cancer. 71:3–10.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kovalchuk O, Filkowski J, Meservy J,
Ilnytskyy Y, Tryndyak VP, Chekhun VF and Pogribny IP: Involvement
of microRNA-451 in resistance of the MCF-7 breast cancer cells to
chemotherapeutic drug doxorubicin. Mol Cancer Ther. 7:2152–2159.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Song B, Wang Y, Titmus MA, Botchkina G,
Formentini A, Kornmann M and Ju J: Molecular mechanism of
chemoresistance by miR-215 in osteosarcoma and colon cancer cells.
Mol Cancer. 9:962010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hummel R, Hussey DJ and Haier J:
MicroRNAs: predictors and modifiers of chemo- and radiotherapy in
different tumour types. Eur J Cancer. 46:298–311. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huh JH, Kim TH, Kim K, Song JA, Jung YJ,
Jeong JY, Lee MJ, Kim YK, Lee DH and An HJ: Dysregulation of
miR-106a and miR-591 confers paclitaxel resistance to ovarian
cancer. Br J Cancer. 109:452–461. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang PY, Li YJ, Zhang S, Li ZL, Yue Z, Xie
N and Xie SY: Regulating A549 cells growth by ASO inhibiting miRNA
expression. Mol Cell Biochem. 339:163–171. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Locher KP: Structure and mechanism of ABC
transporters. Curr Opin Struct Biol. 14:426–431. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Iwasaki H, Okabe T, Takara K, Yoshida Y,
Hanashiro K and Oku H: Down-regulation of lipids transporter ABCA1
increases the cytotoxicity of nitidine. Cancer Chemother Pharmacol.
66:953–959. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Borst P, Evers R, Kool M and Wijnholds J:
A family of drug transporters: the multidrug resistance-associated
proteins. J Natl Cancer Inst. 92:1295–1302. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gottesman MM, Fojo T and Bates SE:
Multidrug resistance in cancer: role of ATP-dependent transporters.
Nat Rev Cancer. 2:48–58. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cole SP, Bhardwaj G, Gerlach JH, Mackie
JE, Grant CE, Almquist KC, Stewart AJ, Kurz EU, Duncan AM and
Deeley RG: Overexpression of a transporter gene in a
multidrug-resistant human lung cancer cell line. Science.
258:1650–1654. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Deeley RG and Cole SP: Function, evolution
and structure of multidrug resistance protein (MRP). Semin Cancer
Biol. 8:193–204. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fang Y, Shen H, Li H, et al: miR-106a
confers cisplatin resistance by regulating PTEN/Akt pathway in
gastric cancer cells. Acta Biochim Biophys Sin (Shanghai).
2013.45(11): 963–72
|
|
19
|
Gong C, Yao Y, Wang Y, Liu B, Wu W, Chen
J, Su F, Yao H and Song E: Up-regulation of miR-21 mediates
resistance to trastuzumab therapy for breast cancer. J Biol Chem.
286:19127–19137. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Suh SS, Yoo JY, Nuovo GJ, Jeon YJ, Kim S,
Lee TJ, Kim T, Bakàcs A, Alder H, Kaur B, et al: MicroRNAs/TP53
feedback circuitry in glioblastoma multiforme. Proc Natl Acad Sci
USA. 109:5316–5321. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dong Z, Zhong Z, Yang L, Wang S and Gong
Z: MicroRNA-31 inhibits cisplatin-induced apoptosis in non-small
cell lung cancer cells by regulating the drug transporter ABCB9.
Cancer Lett. 343:249–257. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li Y, Li L, Guan Y, Liu X, Meng Q and Guo
Q: MiR-92b regulates the cell growth, cisplatin chemosensitivity of
A549 non small cell lung cancer cell line and target PTEN. Biochem
Biophys Res Commun. 440:604–610. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Song L, Li Y, Li W, Wu S and Li Z: miR-495
enhances the sensitivity of non-small cell lung cancer cells to
platinum by modulation of copper-transporting P-type adenosine
triphosphatase A (ATP7A). J Cell Biochem. 115:1234–1242. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gillet JP, Efferth T and Remacle J:
Chemotherapy-induced resistance by ATP-binding cassette transporter
genes. Biochim Biophys Acta. 1775:237–262. 2007.PubMed/NCBI
|
|
25
|
Yamagishi T, Sahni S, Sharp DM, Arvind A,
Jansson PJ and Richardson DR: P-glycoprotein mediates drug
resistance via a novel mechanism involving lysosomal sequestration.
J Biol Chem. 288:31761–31771. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dai DW, Lu Q, Wang LX, Zhao WY, Cao YQ, Li
YN, Han GS, Liu JM and Yue ZJ: Decreased miR-106a inhibits glioma
cell glucose uptake and proliferation by targeting SLC2A3 in GBM.
BMC Cancer. 13:4782013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu Z, Gersbach E, Zhang X, Xu X, Dong R,
Lee P, Liu J, Kong B, Shao C and Wei JJ: miR-106a represses the Rb
tumor suppressor p130 to regulate cellular proliferation and
differentiation in high-grade serous ovarian carcinoma. Mol Cancer
Res. 11:1314–1325. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhi F, Zhou G, Shao N, Xia X, Shi Y, Wang
Q, Zhang Y, Wang R, Xue L, Wang S, et al: miR-106a-5p inhibits the
proliferation and migration of astrocytoma cells and promotes
apoptosis by targeting FASTK. PLoS One. 8:e723902013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lee BH, Taylor MG, Robinet P, et al:
Dysregulation of cholesterol homeostasis in human prostate cancer
through loss of ABCA1. Cancer Res. 73:1211–1218. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Smith B and Land H: Anticancer activity of
the cholesterol exporter ABCA1 gene. Cell Rep. 2:580–590. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bachmeier BE, Iancu CM, Killian PH,
Kronski E, Mirisola V, Angelini G, Jochum M, Nerlich AG and Pfeffer
U: Overexpression of the ATP binding cassette gene ABCA1 determines
resistance to Curcumin in M14 melanoma cells. Mol Cancer.
8:1292009. View Article : Google Scholar : PubMed/NCBI
|