Introduction
Mesenchymal stem cells (MSCs) were initially
identified as an adherent, fibroblast-like population obtained from
adult bone marrow (BM) by Friedenstein et al (1,2).
MSCs are able to differentiate into multiple lineages (3,4) and
are increasingly proposed as a therapeutic strategy for tissue
regeneration and repair (5).
Although BM has been the primary source of MSCs in the past
(6–8), the use of BM-derived MSCs (BM-MSCs)
is limited due to multiple factors, including the high degree of
viral exposure, potential donor morbidity, as well as significant
decreases in cell number and proliferation/differentiation capacity
associated with age (9), and the
highly invasive procedure required in order to obtain BM.
Therefore, it was important to find alternative sources to provide
MSCs. Further studies have identified additional MSC sources,
including adult synovial membranes and the fetal liver and spleen
(10–12). However, there has only been a
limited number of studies on MSCs isolated from human fetuses
(gestational age, 12–16 weeks). Comparison of fetal tissue-derived
MSCs (FT-MSCs) (13) and
adult-derived MSCs revealed that the biological activity and the
differentiative and multiplication capacity of the former were
greater than those of the latter (14). Comparison of the in vitro
and in vivo characteristics of BM-MSCs and FT-MSCs requires
analyses of their respective gene expression profiles in order to
elucidate their fundamental mechanisms, including self-renewal
during long-term expansion, differentiation into mature cells and
tissue-repair properties.
Quantitative polymerase chain reaction (qPCR) is a
commonly used technique to determine the relative change in mRNA
expression of target genes. Due to the accuracy, ease of use and
reproducibility of qPCR analysis, it is frequently used in MSC
research. However, qPCR accuracy is influenced by various external
and internal factors, including the amount of starting sample, RNA
preparation, cDNA synthesis and PCR efficiency. Therefore, it is
necessary to normalize gene expression levels by comparison to
reference genes (RGs) as internal controls (15). An ideal RG should not be influenced
by cell cycle, cell passages or experimental conditions (16); simultaneously, it should be stably
expressed in various samples (17,18).
However, to the best of our knowledge, no single RG has been
reported to be universal and completely constant. Furthermore,
increasing evidence indicated that the expression levels of
commonly used RGs vary significantly between cell types and
experimental conditions (19,20).
Thus, the selection of suitable RGs for idiographic study is a
prerequisite for any qPCR assay to obtain reliable results. The aim
of the present study was to identify and assess the stabilities and
reliabilities of ten RGs which are commonly used in BM- and FT-MSCs
for qPCR.
Ten common RGs, including 18S, ACTB,
B2M, HPRT, GAPDH, TBP, PPIA,
RPLP0, PGK1 and RPL13A (Table I)were selected for the present
study and their expression stabilities were analyzed using geNorm
(21), NormFinder (22) and BestKeeper (23) software. The present study aimed to
identify the optimal RGs for further research on BM-MSCs and
FT-MSCs.
 | Table ISummary of reference genes used in
the present study. |
Table I
Summary of reference genes used in
the present study.
| Symbol | Name | Function | Accession
number |
|---|
| 18S | 18S ribosomal
RNA | Ribosomal
subunit | NM_10098.1 |
| GAPDH |
Glyceraldehyde-3-phosphate
dehydrogenase | Enzyme in
glycolysis and nuclear functions | NM_002046 |
| RPLP0 | Ribosomal protein,
large, P0 | Structural
component of the 60S subunit of ribosomes | NM_001002.3 |
| ACTB | Beta-actin | Cytoskeletal
structural actin | NM_001101 |
| PPIA |
Peptidyl-prolylisomerase A | Accelerates the
folding of proteins | NM_021130.3 |
| PGK1 | Phosphoglycerate
kinase 1 | Glycolytic
enzyme | NM_000291.3 |
| B2M |
Beta-2-microglobulin | Component of the
MHCI molecules | NM_004048.2 |
| RPL13A | Ribosomal protein
L13a | Structural
component of the 60S ribosomal subunit | NM_012423.2 |
| HPRT | Hypoxanthine
phosphoribosyl transferase 1 | Enzyme in purine
metabolic pathway | NM_000194 |
| TBP | TATA box binding
protein | General
transcription factor | NM_003194 |
Materials and methods
MSCs
The study was approved by the Ethical Committee of
China-Japan Union Hospital, Jilin University (Changchun, China).
BM-MSCs were isolated from femur-derived bone marrow samples that
were obtained by surgical operation (China-Japan Union Hospital) on
otherwise healthy patients (aged between 18 and 43 years) following
receipt of their informed consent. FT-MSCs were obtained from Jilin
Zhongke Bio-engineering Co., Ltd. (Changchun, China). For all
experiments, pools of the various cell types were prepared by
mixing equal numbers of cells from five donors of the same passage
number. Cells were not cultured for more than four passages.
Identification of MSCs
Flow cytometric characterization of
MSCs
MSCs of passage three were labeled with the
following anti-human antibodies: CD14-phycoerythrin (PE), CD34-PE,
CD45-fluorescein isothiocyanate (FITC), CD73-PE, CD90-FITC,
CD105-peridinin chlorophyll, CD44-PE (BD Biosciences, San Jose, CA,
USA). A total of 106 labeled cells were evaluated by
flow cytometry (Beckman Coulter Fc500, Brea, CA, USA) and the data
were analyzed with CXP software (Beckman Coulter Fc500).
MSC differentiation potential
For the differentiation of MSCs into adipocytes and
osteoblasts, cells were incubated in adipogenesis differentiation
medium (StemPro® Adipogenesis Differentiation kit;
Gibco-BRL, Invitrogen Life Technologies, Carlsbad, CA, USA) and
osteogenesis differentiation medium (StemPro®
Osteogenesis Differentiation kit; Gibco-BRL), respectively
according to the manufacturer’s instructions. Adipogenic
differentiation was measured by staining cells in wells with Oil
Red O (Sigma-Aldrich, St. Louis, MO, USA) on day 21 of culture
following fixing the cells with 5% paraformaldehyde (Beyotime,
Shanghai, China) for 5 min at room temperature. Following 35 days
of incubation, osteogenic differentiation was evaluated by staining
the cells with Alizarin Red S (Alizarin S staining kit; Genmed,
Shanghai, China).
RNA isolation and cDNA synthesis
Total RNA was isolated using TRIzol reagent
(Invitrogen Life Technologies, Carlsbad, CA, USA) according to the
manufacturer’s instructions and RNA integrity was
electrophoretically verified by ethidium bromide staining. RNA
concentrations and A260/A280 nm absorbance ratios were measured
spectrophotometrically with a Synergy HT Multi-Mode microplate
reader (BioTek Instruments, Inc., Winooski, VT, USA). From 500 ng
of RNA, cDNA was synthesized using an RNA PCR kit (avian
myeloblastosis virus; TaKaRa, Dalian, China) according to the
manufacturer’s instructions. cDNA was stored at −20°C until
required.
Selection of candidate RGs and primer
design
RGs were selected for analysis based on a literature
search on the subject of RG studies in MSCs. In the present study,
ten candidate RGs were selected and were as follows: 18S,
ACTB (24,25), B2M (26,27),
HPRT, GAPDH (28–34), TBP, PPIA, RPLP0, PGK1,
RPL13A. The full name, function and accession number of the RGs
assessed in the present study are listed in Table I. RGs were selected from varying
functional classes, which significantly reduces the chance that the
genes may be co-regulated.
Primer pairs used for qPCR were designed using
primer three input (http://flypush.imgen.bcm.tmc.edu/primer/primer3_www.cgi).
The qPCR primers were synthesized by Sigma Genesys (Sigma-Aldrich)
with melting temperature (Tm) at 60±1°C. All primers
were purified by Ultrapage (Sangon, Biotech, Shanghai, China).
Primer efficiencies were determined using a 10-fold dilution series
of cDNA as templates for qPCR reactions. An approximation of PCR
efficiency was calculated using the slope of the calibration curve
according to the following equation: E=10−1/slop, where
‘slop’ represented the linear regression slope (18). Reactions were performed in
triplicate and data were analyzed by using the 2−ΔΔCt
method (10). The primer sequences
and corresponding amplicon sizes are listed in Table II.
 | Table IIPrimer sequences, product sizes and
PCR efficiency. |
Table II
Primer sequences, product sizes and
PCR efficiency.
| Gene | Primer sequences
(5′-3′) | Product size
(bp) | PCR efficiency |
|---|
| 18S |
F-GTGGAGCGATTTGTCTGGTT
R-AACGCCACTTGTCCCTCTAA | 115 | 1.90 |
| GAPDH |
F-ATGGGGAAGGTGAAGGTCG
R-GGGGTCATTGATGGCAACAATA | 108 | 1.99 |
| RPLP0 |
F-CTGGAAGTCCAACTACTTCCT
R-CATCATGGTGTTCTTGCCCAT | 160 | 2.74 |
| ACTB |
F-GAAGATCAAGATCATTGCTCCT
R-TACTCCTGCTTGCTGATCCA | 111 | 1.89 |
| PPIA |
F-TCCTGGCATCTTGTCCAT
R-TGCTGGTCTTGCCATTCCT | 179 | 2.17 |
| PGK1 |
F-GCCACTTGCTGTGCCAAATG
R-CCCAGGAAGGACTTTACCTT | 102 | 2.62 |
| B2M |
F-CTATCCAGCGTACTCCAAAG
R-GAAAGACCAGTCCTTGCTGA | 188 | 2.08 |
| RPL13A |
F-CGAGGTTGGCTGGAAGTACC
R-CTTCTCGGCCTGTTTCCGTAG | 121 | 2.00 |
| HPRT |
F-CCTGGCGTCGTGATTAGTGAT
R-AGACGTTCAGTCCTGTCCATAA | 131 | 1.78 |
| TBP |
F-GCACAGGAGCCAAGAGTGA
R-GTTGGTGGGTGAGCACAAG | 174 | 2.10 |
qPCR
qPCR was performed in 96-well plates with the ABI
PRISM 7500 Sequence Detection system (Perkin Elmer, Inc., Waltham,
MA, USA). PCR conditions were as follows: 50°C for two minutes,
95°C for 10 min followed by 40 cycles of denaturation at 95°C for
15 sec, annealing at 58°C for 15 sec and extension at 72°C for 30
sec during which fluorescence was measured. Expression levels were
recorded as cycle threshold (Ct). Data were acquired using the 7500
Software (Applied Biosystems Life Technologies, Foster City, CA,
USA). The mean Ct values of the triplicate reactions were used for
data analysis.
Data analysis
Expression stabilities of the ten RGs were assessed
via the three commonly used software programs geNorm, NormFinder
and BestKeeper. In geNorm and NormFinder, Ct values were converted
into relative quantities via the formula 2−(Ct−lowest
Ct). The raw Ct values were used directly for BestKeeper
analysis. These three programs are based on Microsoft Excel
(Microsoft Corp., Redmond, WA, USA) using various algorithms to
determine the expression stability of RGs. GeNorm calculated a gene
expression stability measure (M) and pairwise variation (V)
parameter. M is the mean pairwise variation for a given gene
compared to other tested genes and stepwise exclusion of the gene
with the highest M value followed by recalculation was performed
until the two most stable genes were left. Lower M values represent
higher expression stability. M=1.5 was used as an experimental
parameter. Above this value, the gene was considered to be
unreliable as an RG. V was calculated to determine the minimal
number of RGs required to normalize the expression of genes of
interest. V=0.15 was also used as an experimental parameter; below
this value, the number of RGs was sufficient for valid
normalization.
NormFinder computed RG stability values via an
analysis of variance-based model. Lower values indicated higher
stabilities. NormFinder was also able to compare inter- and
intra-group variations in gene stability.
BestKeeper analyzed RG stability based on the
standard deviation (SD) and coefficient of correlation (r) of all
RGs. SD values were obtained from the Ct values of each RG, and r
values were the correlation coefficient calculated using Pearson’s
pair-wise correlation analyses between each RG and the geometric
mean of the Ct values. Those genes with an SD >1.0 were
considered to be unreliable as a stable RG and the remaining genes
are ranked according to their r values.
Results
Isolation and characteristics of
MSCs
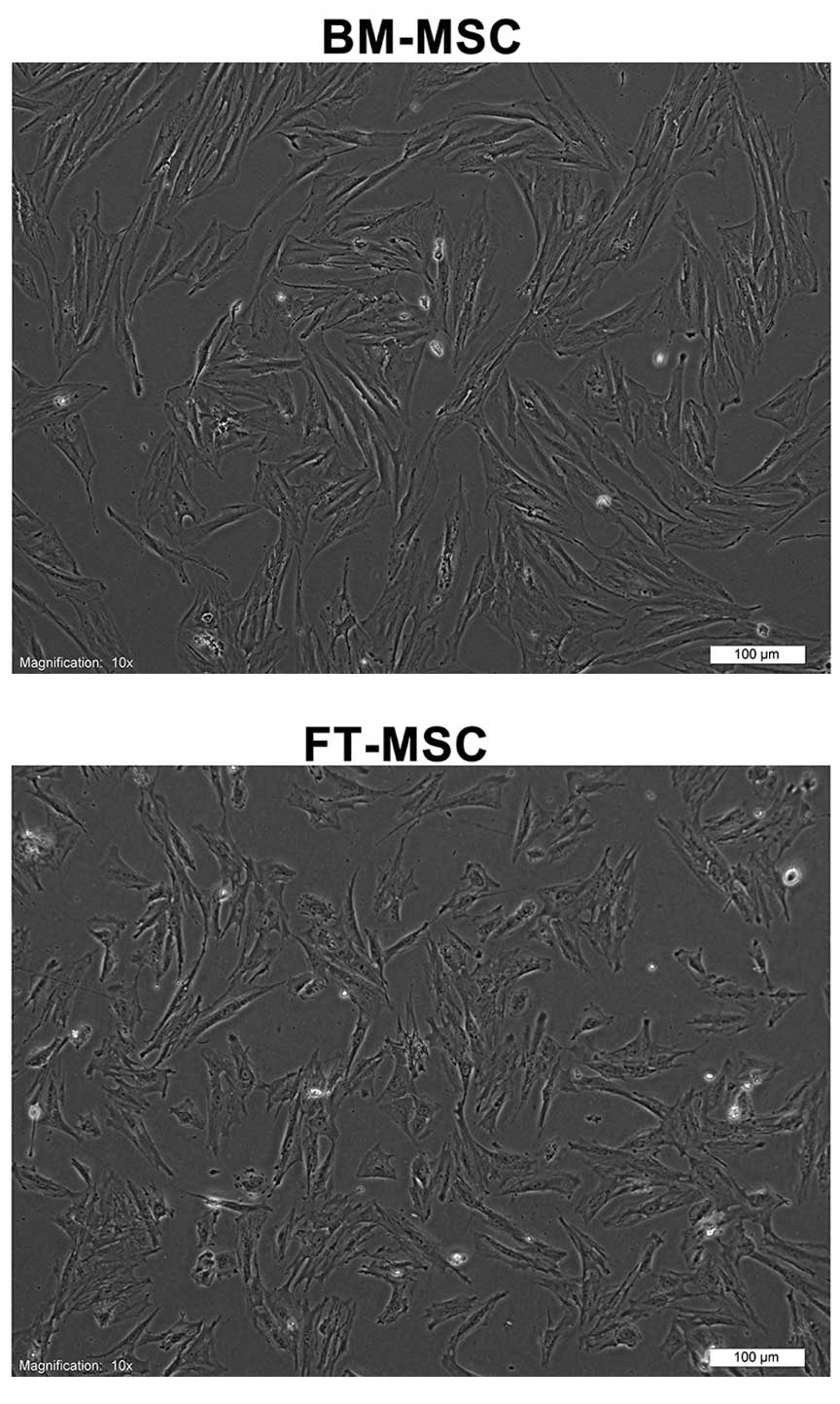
Human MSCs were obtained from human BM and FT. The
adherent cells had fibroblastic morphologies (Fig. 1). The cell-surface antigen profiles
of these cells at three passages in culture were analyzed by flow
cytometry. These cells were strongly positive for MSC-specific
surface markers, including CD44, CD73, CD90 and CD105, but were
negative for CD14, CD34 and CD45. These cells also exhibited
mesenchymal differentiation potential, as assessed by culturing
them in adipogenic and osteogenic medium (data not shown).
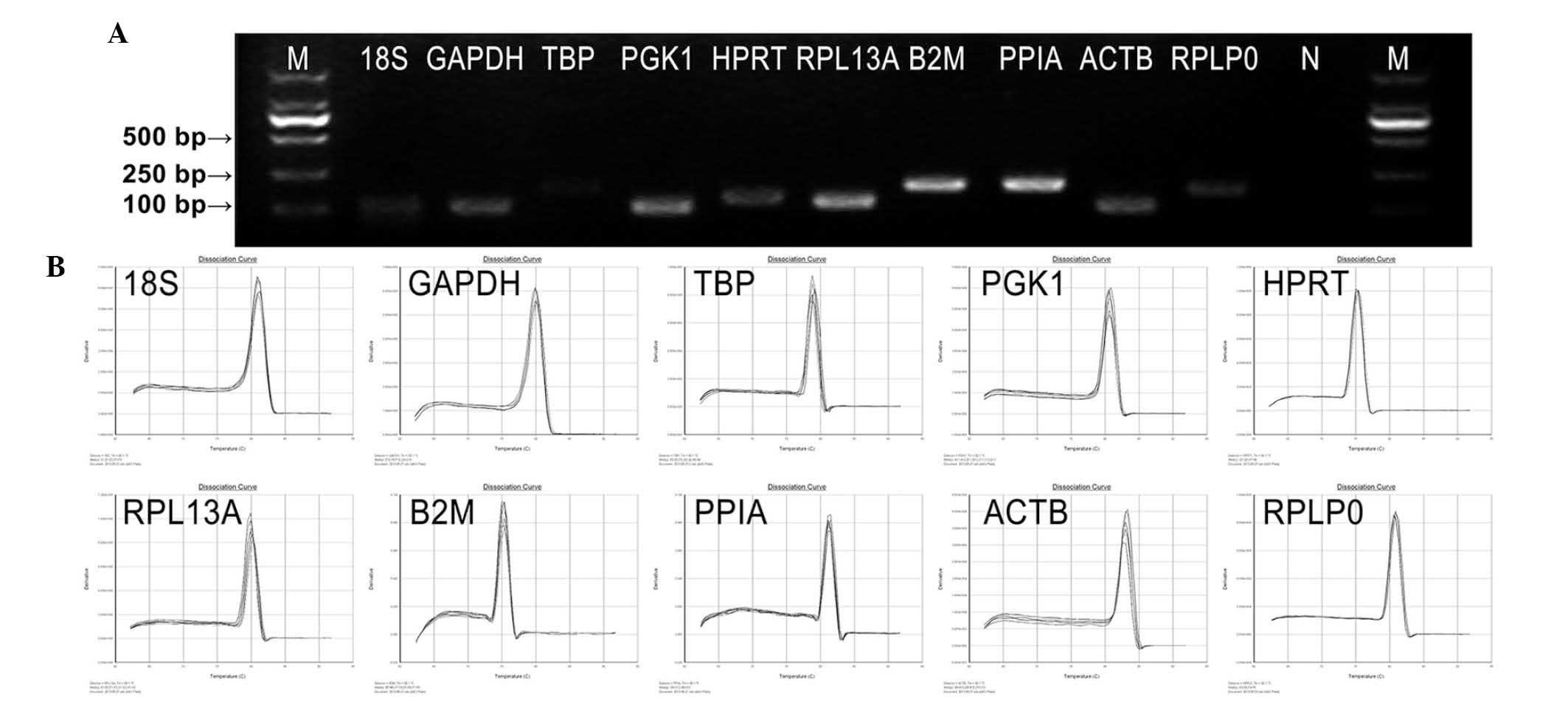
Amplification specificity and efficiency
of primers
The primer sequences, corresponding amplicon sizes
and PCR efficiencies of primers, are listed in Table II. The amplification performance
of each primer pair was tested by qPCR. Primer specificities for
qPCR were verified by dissociation curve analysis and 2% agarose
gel electrophoresis. No primer-dimers or multiple bands/peaks were
detected, confirming a single amplified band of a predicted size
(Fig. 2). All primer pairs
amplified a single PCR product of expected size, produced a slope
>-4.0, exhibited correlation coefficients (r2)
>0.97 and the corresponding qPCR efficiencies were in the range
of 1.8–2.7 obtained from a 10-fold dilution series of the template
cDNA. These data indicated that the amplification efficiencies of
the primers analyzed reached the standard requirements of
conventional qPCR.
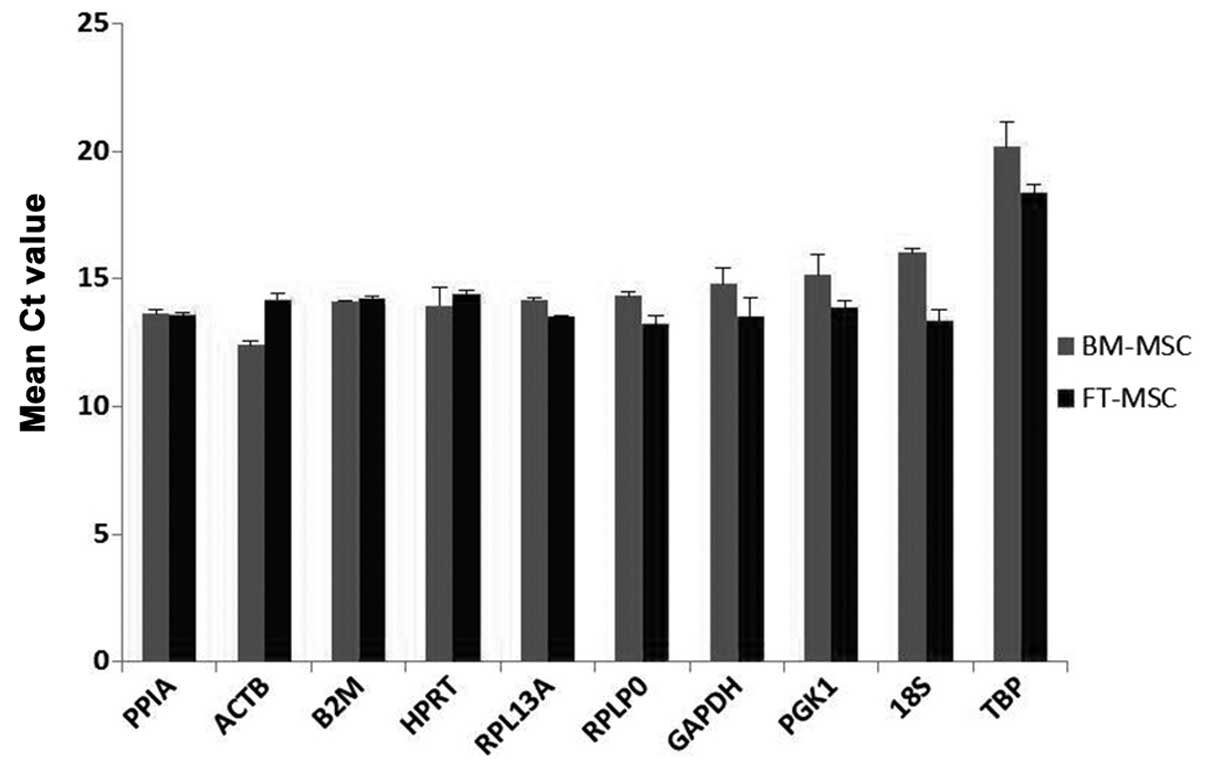
Expression levels of RGs
The expression levels of all ten RGs were analyzed
by comparison of Ct values using two biological samples with three
technical replicates. Fig. 3
exhibits the mean Ct values for each gene in two samples. All the
RGs displayed Ct values in MSCs, ranging from 12.39 for ACTB
to 20.16 for TBP. Among these RGs, ACTB (Ct
12.39–14.19) and PPIA (Ct 13.55–13.62) displayed the highest
RNA transcription levels. The lowest RNA transcription levels were
observed for TBP (Ct 18.37–20.16), followed by 18S
(Ct 13.36–16.02). The individual RGs had varying expression ranges
across the samples. Among the ten RGs in the present study,
18S had the greatest variation in its transcript expression
levels (2.7 cycles), while PPIA and B2M had
significantly lower expression variations (0.06 and 0.14 cycles,
respectively). These Ct standard deviations indicated an initial
hypothesis concerning the variability in expression. The RGs were
ranked from greatest to lowest variability in expression:
18S>TBP>ACTB>PGK1>GAPDH>RPLP0>RPL13A>HPRT>B2M>PPIA.
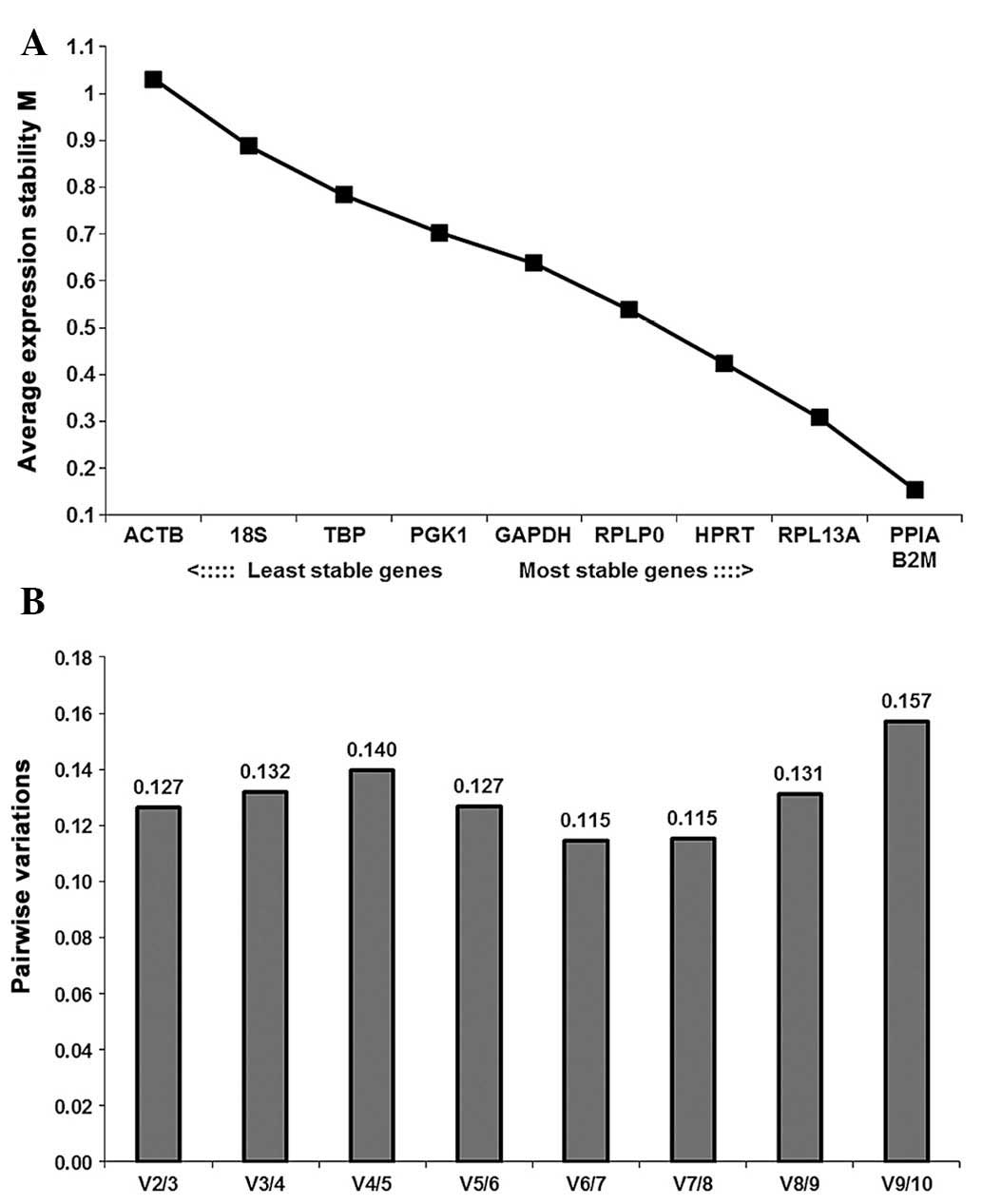
GeNorm analysis
The average stability values (M) of the ten RGs in
all samples analyzed are indicated in Fig. 4A. PPIA and B2M
(M=0.154) were identified as the most stably expressed RGs, while
ACTB (M=1.031) was the least stably expressed. These ten RGs
demonstrated inconspicuous variation between BM-MSCs and FT-MSCs as
the M-value of each gene was below the default limit of M≤1.5. The
stability ranking of the selected RGs was
B2M/PPIA>RPL13A>HPRT>RPLP0>GAPDH>PGK1>TBP>18S>ACTB.
GeNorm defines a pairwise variation of 0.15 as the cutoff value,
below which the inclusion of an additional RG is unnecessary
(28). Here, the V2/3 value (the
pairwise variation upon increasing the number of normalization
factors from two to three) was 0.127, which was below the cutoff
value; hence, the use if a combination of two RGs was sufficiently
stable for the analysis of BM-MSCs and FT-MSCs (Fig. 4B).
NormFinder analysis
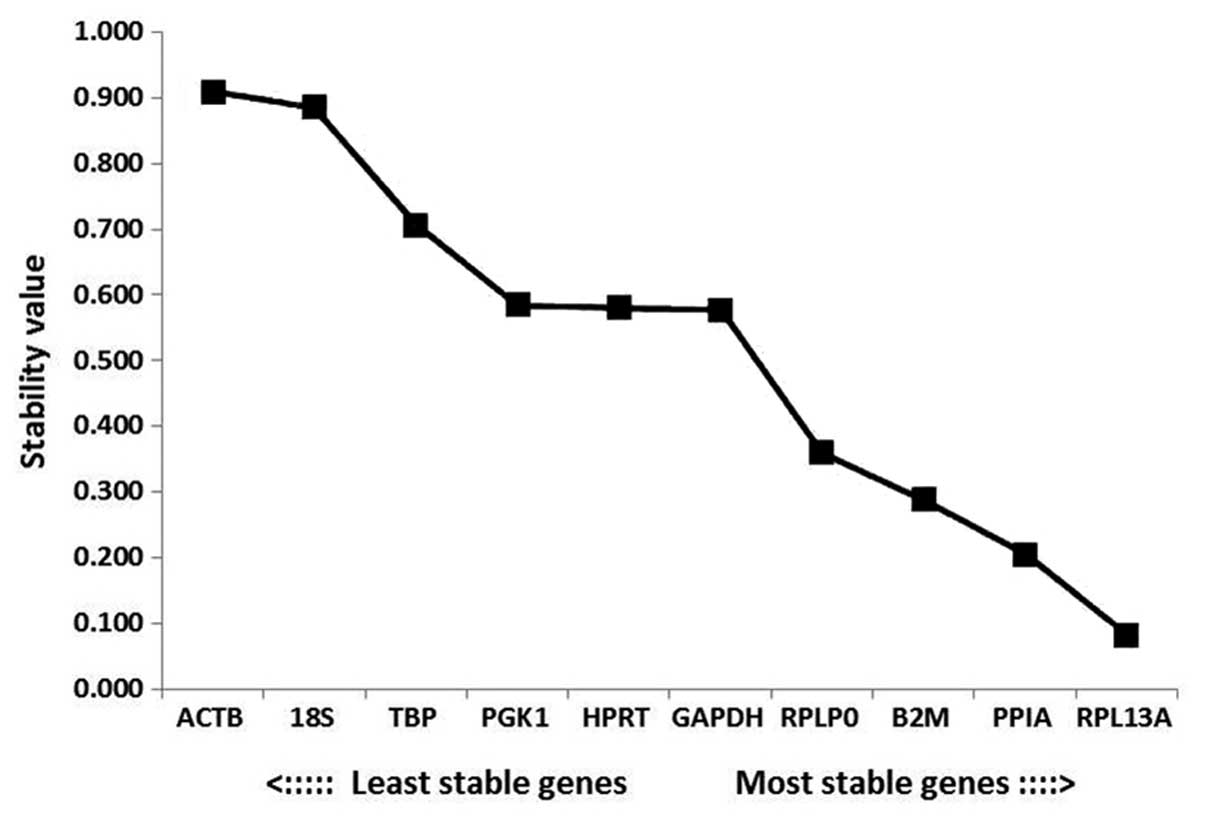
According to the outputs from the NormFinder
analysis, the most stable gene was RPL13A (stability value,
0.082), followed by PPIA and B2M. The most unstable
genes were ACTB, 18S and TBP (stability values,
0.909, 0.885 and 0.707, respectively; Fig. 5). Among the RGs analyzed in the
present study, the ranking of the stabilities according to
NormFinder software was
RPL13A>PPIA>B2M>RPLP0>GAPDH>HPRT>PGK1>TBP>18S>ACTB.
The most stable combination was PPIA and RPLP0, which
produced a stability value of 0.131.
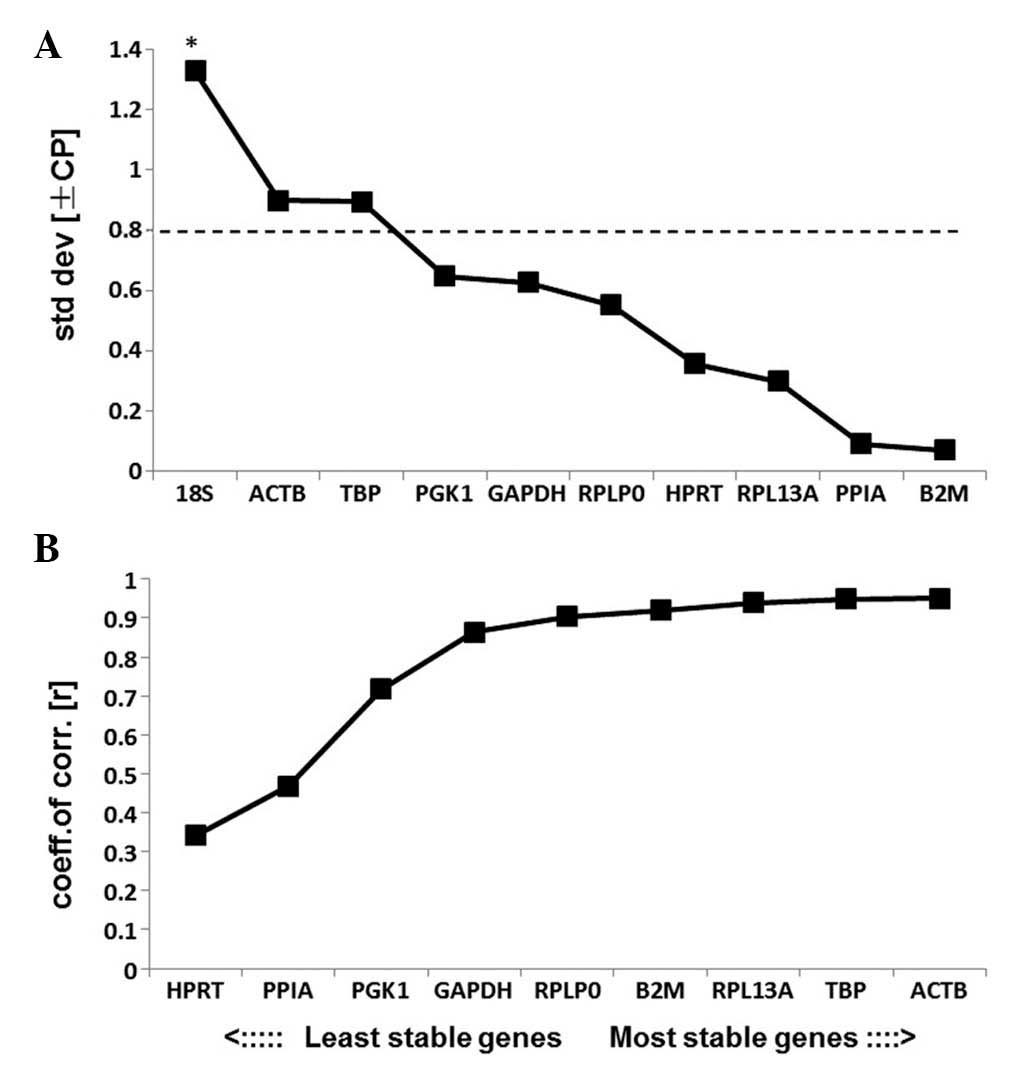
BestKeeper analysis
The results of BestKeeper analysis are displayed in
Table III. According to
BestKeeper, genes with an SD>1.0 are considered to have an
unacceptable range of variation. The results of the present study
indicated that all of the RGs analyzed, excluding 18S, had an SD
level under 1.0 and may therefore be used as credible RGs (Fig. 6A). 18S had an SD>1.0 and was
therefore eliminated from further analysis and the remaining genes
were ranked according to their coefficient of correlation (r).
HPRT was the most unstable RG and ACTB was the most
stable RG according to the results of BestKeeper analysis (Fig. 6B). The ranking of the stabilities
was
ACTB>TBP>RPL13A>B2M>RPLP0>GAPDH>
PGK1>PPIA>HPRT.
 | Table IIIStability of ten reference genes
evaluated by BestKeeper. |
Table III
Stability of ten reference genes
evaluated by BestKeeper.
| ACTB | PPIA | RPLP0 | RPL13A | HPRT | GAPDH | B2M | PGK1 | 18S | TBP |
|---|
| geo Mean [CP] | 13.25789 | 13.59014 | 13.76474 | 13.84548 | 14.15068 | 14.15517 | 14.18604 | 14.48262 | 14.6256 | 19.23467 |
| ar Mean [CP] | 13.289 | 13.5905 | 13.777 | 13.84875 | 14.157 | 14.1765 | 14.18625 | 14.50275 | 14.688 | 19.26175 |
| min [CP] | 12.279 | 13.492 | 13.005 | 13.541 | 13.446 | 13.063 | 14.109 | 13.648 | 13.048 | 18.14 |
| max [CP] | 14.343 | 13.736 | 14.452 | 14.207 | 14.487 | 15.228 | 14.3 | 15.706 | 16.127 | 20.841 |
| std dev [±CP] | 0.899 | 0.091 | 0.5515 | 0.29775 | 0.3555 | 0.6255 | 0.06975 | 0.64675 | 1.329 | 0.89475 |
| CV [%CP] | 6.764994 | 0.669585 | 4.003049 | 2.150014 | 2.511125 | 4.412232 | 0.491673 | 4.459499 | 9.048203 | 4.645217 |
| coeff. of corr.
[r] | −0.951 | 0.468 | 0.903 | 0.939 | −0.341 | 0.863 | −0.92 | 0.718 | 0.953 | 0.948 |
Comparison of software analyses
The results obtained from geNorm, NormFinder and
BestKeeper were analyzed comprehensively. GeNorm and NormFinder
indicated that PPIA, B2M and RPL13A were the most
stable RGs, whereas ACTB, 18S and TBP were the least
stable RGs. However, ACTB was ranked as the most stable gene
based on r values obtained from BestKeeper analysis. A summary of
the rankings produced by the three software programs is presented
in Table IV.
 | Table IVRanking of reference gene
stability. |
Table IV
Ranking of reference gene
stability.
| | | BestKeeper |
|---|
| | |
|
|---|
| Rank | geNorm | NormFinder | r | SD |
|---|
| 1 |
B2M,PPIA | RPL13A | ACTB | B2M |
| 2 | * | PPIA | TBP | PPIA |
| 3 | RPL13A | B2M | RPL13A | RPL13A |
| 4 | HPRT | RPLP0 | B2M | HPRT |
| 5 | RPLP0 | GAPDH | RPLP0 | RPLP0 |
| 6 | GAPDH | HPRT | GAPDH | GAPDH |
| 7 | PGK1 | PGK1 | PGK1 | PGK1 |
| 8 | TBP | TBP | PPIA | TBP |
| 9 | 18S | 18S | HPRT | ACTB |
| 10 | ACTB | ACTB | | 18S |
Discussion
RGs are required to provide a scale in qPCR
experiments. If the scale varies significantly between samples in
an experiment, it loses the function of measuring gene expression
in those samples. An ideal RG should be neither influenced nor
regulated by experimental conditions or treatments. Increasing
evidence indicated that there is no single RG that is able to be
used for multiple experiments; however, an increasing number of
studies suggested that a group of putative RGs for certain specific
experimental setups may be recommended for future studies (35–37).
MSCs, primarily derived from BM, have been examined
widely for their capacities in repairing damaged tissues (38–40).
The prospective clinical applications of BM-MSCs are varied. Their
ability to differentiate into desired mature cell types and
additional indirect mechanisms have been recognized to have
important roles in the treatment of autoimmune diseases (40). FT-MSCs, which are self-renewing and
pluripotent, are a valuable research tool and have potential for
use in regenerative medicine. Thus, fetal tissue provides a
potential substitute to BM as a source of MSCs (41). In order to make comparisons between
the mRNA expression features of two sources of MSCs, it is
important to identify a standardized, reproducible set of RGs to
normalize qPCR analysis.
To the best of our knowledge, the present study was
the first to validate the stability of RGs used for the comparison
of BM-MSCs and FT-MSCs. Expression stabilities of ten candidate
RGs, including 18S, GAPDH, RPLP0, ACTB, PPIA, PGK1, B2M, RPL13A,
HRPT1 and TBP, were estimated using three statistical
algorithm software packages: geNorm, NormFinder and BestKeeper.
Evaluation with geNorm as well as NormFinder indicated that B2M,
PPIA and RPL13A were the most stably expressed RGs. In
addition, the two programs rendered ACTB, 18S and TBP
the least stably expressed RGs and the rank orders were also
coincident. However, the results of the analysis using BestKeeper
significantly differed from those obtained using geNorm and
NormFinder. BestKeeper indicated that ACTB, TBP and
RPL13A were the three most stable RGs, while 18S,
HPRT and PPIA were least stable. Of note, ACTB
was ranked as most stable by BestKeeper, but least stable by geNorm
and NormFinder. Rank orders obtained were expected to differ
between the three software programs given their distinct
statistical algorithms. For example, unlike geNorm and BestKeeper,
NormFinder used a mathematical model based on assessment of intra-
and intergroup variations to determine the optimal RGs and
therefore showed decreased sensitivity toward co-regulation of RGs.
There was no consensus regarding which was the best method. In the
present study, the most stable RGs (RPL13A, PPIA and
B2M) determined by geNorm and NormFinder were identical, as
were the least stable genes (ACTB, 18S and TBP).
ACTB is widely used as an RG to evaluate
target gene expression levels in MSCs (42–46).
However, in the present study, ACTB was found to have the
lowest stability among the selected RGs in BM-MSCs and FT-MSCs.
GAPDH was previously considered as the gold standard RG,
despite a lack of experimental evidence (37,47–49).
However, GAPDH was ranked sixth by geNorm and fifth by
NormFinder and BestKeeper in the present study. It was therefore
concluded that GAPDH and ACTB were not reliable RGs
for the normalization of qPCR data in BM- and FT-MSC research and
they are not recommended for use throughout this field of
research.
In conclusion, the present study identified
RPL13A, PPIA and B2M as suitable genes for the
normalization of qPCR data and indicated that 18S, ACTB, and
TBP were unsuitable for normalization of mRNA expression
levels of BM- and FT-MSCs.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of Jilin Province of China (no.
20130101167JC).
References
|
1
|
Friedenstein AJ, Petrakova KV, Kurolesova
AI and Frolova GP: Heterotopic of bone marrow. Analysis of
precursor cells for osteogenic and hematopoietic tissues.
Transplantation. 6:230–247. 1968. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Friedenstein AJ, Chailakhyan RK and
Gerasimov UV: Bone marrow osteogenic stem cells: in vitro
cultivation and transplantation in diffusion chambers. Cell Tissue
Kinet. 20:263–272. 1987.PubMed/NCBI
|
|
3
|
Pittenger MF, Mackay AM, Beck SC, Jaiswal
RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S and
Marshak DR: Multilineage potential of adult human mesenchymal stem
cells. Science. 284:143–147. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Keating A: Mesenchymal stromal cells. Curr
Opin Hematol. 13:419–425. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Arthur A, Zannettino A and Gronthos S: The
therapeutic applications of multipotential mesenchymal/stromal stem
cells in skeletal tissue repair. J Cell Physiol. 218:237–245. 2009.
View Article : Google Scholar
|
|
6
|
Young RG, Butler DL, Weber W, Caplan AI,
Gordon SL and Fink DJ: Use of mesenchymal stem cells in a collagen
matrix for Achilles tendon repair. J Orthop Res. 16:406–413. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wakitani S, Saito T and Caplan AI:
Myogenic cells derived from rat bone marrow mesenchymal stem cells
exposed to 5-azacytidine. Muscle Nerve. 18:1417–1426. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dennis JE and Caplan AI: Differentiation
potential of conditionally immortalized mesenchymal progenitor
cells from adult marrow of a H-2Kb-tsA58 transgenic mouse. J Cell
Physiol. 167:523–538. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rubinstein P, Rosenfield RE, Adamson JW
and Stevens CE: Stored placental blood for unrelated bone marrow
reconstitution. Blood. 81:1679–1690. 1993.PubMed/NCBI
|
|
10
|
Gullo F and De Bari C: Prospective
purification of a subpopulation of human synovial mesenchymal stem
cells with enhanced chondro-osteogenic potency. Rheumatology
(Oxford). 52:1758–1768. 2013. View Article : Google Scholar
|
|
11
|
Joshi M, B Patil P, He Z, Holgersson J,
Olausson M and Sumitran-Holgersson S: Fetal liver-derived
mesenchymal stromal cells augment engraftment of transplanted
hepatocytes. Cytotherapy. 14:657–669. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
in ‘t Anker PS1, Noort WA, Scherjon SA, et
al: Mesenchymal stem cells in human second-trimester bone marrow,
liver, lung, and spleen exhibit a similar immunophenotype but a
heterogeneous multilineage differentiation potential.
Haematologica. 88:845–852. 2003.
|
|
13
|
O’Donoghue K and Chan J: Human fetal
mesenchymal stem cells. Curr Stem Cell Res Ther. 1:371–386. 2006.
View Article : Google Scholar
|
|
14
|
Jo CH, Yoon PW, Kim H, Kang KS and Yoon
KS: Comparative evaluation of in vivo osteogenic differentiation of
fetal and adult mesenchymal stem cell in rat critical-sized femoral
defect model. Cell Tissue Res. 353:41–52. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dheda K, Huggett JF, Bustin SA, Johnson
MA, Rook G and Zumla A: Validation of housekeeping genes for
normalizing RNA expression in real-time PCR. Biotechniques.
37:112–114. 116118–119. 2004.PubMed/NCBI
|
|
16
|
Radonić A, Thulke S, Mackay IM, Landt O,
Siegert W and Nitsche A: Guideline to reference gene selection for
quantitative real-time PCR. Biochem Biophys Res Commun.
313:856–862. 2004. View Article : Google Scholar
|
|
17
|
Bustin SA: Absolute quantification of mRNA
using real-time reverse transcription polymerase chain reaction
assays. J Mol Endocrinol. 25:169–193. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Suzuki T, Higgins PJ and Crawford DR:
Control selection for RNA quantitation. Biotechniques. 29:332–337.
2000.PubMed/NCBI
|
|
19
|
Cordoba EM, Die JV, González-Verdejo CI,
Nadal S and Román B: Selection of reference genes in Hedysarum
coronarium under various stresses and stages of development. Anal
Biochem. 409:236–243. 2011. View Article : Google Scholar
|
|
20
|
Schmittgen TD and Zakrajsek BA: Effect of
experimental treatment on housekeeping gene expression: validation
by real-time, quantitative RT-PCR. J Biochem Biophys Methods.
46:69–81. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vandesompele J, De Preter K, Pattyn F,
Poppe B, Van Roy N, De Paepe A and Speleman F: Accurate
normalization of real-time quantitative RT-PCR data by geometric
averaging of multiple internal control genes. Genome Biol.
3:RESEARCH00342002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Andersen CL, Jensen JL and Ørntoft TF:
Normalization of real-time quantitative reverse transcription-PCR
data: a model-based variance estimation approach to identify genes
suited for normalization, applied to bladder and colon cancer data
sets. Cancer Res. 64:5245–5250. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pfaffl MW, Tichopad A, Prgomet C and
Neuvians TP: Determination of stable housekeeping genes,
differentially regulated target genes and sample integrity:
BestKeeper - Excel-based tool using pair-wise correlations.
Biotechnol Lett. 26:509–515. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Riekstina U, Cakstina I, Parfejevs V,
Hoogduijn M, Jankovskis G, Muiznieks I, Muceniece R and Ancans J:
Embryonic stem cell marker expression pattern in human mesenchymal
stem cells derived from bone marrow, adipose tissue, heart and
dermis. Stem Cell Rev. 5:378–386. 2009. View Article : Google Scholar
|
|
25
|
Gang EJ, Bosnakovski D, Figueiredo CA,
Visser JW and Perlingeiro RC: SSEA-4 identifies mesenchymal stem
cells from bone marrow. Blood. 109:1743–1751. 2007. View Article : Google Scholar
|
|
26
|
Martinez C, Hofmann TJ, Marino R, Dominici
M and Horwitz EM: Human bone marrow mesenchymal stromal cells
express the neural ganglioside GD2: a novel surface marker for the
identification of MSCs. Blood. 109:4245–4248. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pozzobon M, Piccoli M, Ditadi A, Bollini
S, Destro R, André-Schmutz I, Masiero L, Lenzini E, Zanesco L,
Petrelli L, et al: Mesenchymal stromal cells can be derived from
bone marrow CD133+ cells: implications for therapy. Stem Cells Dev.
18:497–510. 2009. View Article : Google Scholar
|
|
28
|
Drost AC, Weng S, Feil G, Schäfer J,
Baumann S, Kanz L, Sievert KD, Stenzl A and Möhle R: In vitro
myogenic differentiation of human bone marrow-derived mesenchymal
stem cells as a potential treatment for urethral sphincter muscle
repair. Ann NY Acad Sci. 1176:135–143. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tang KC, Trzaska KA, Smirnov SV, Kotenko
SV, Schwander SK, Ellner JJ and Rameshwar P: Down-regulation of MHC
II in mesenchymal stem cells at high IFN-gamma can be partly
explained by cytoplasmic retention of CIITA. J Immunol.
180:1826–1833. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Greco SJ, Zhou C, Ye JH and Rameshwar P: A
method to generate human mesenchymal stem cell-derived neurons
which express and are excited by multiple neurotransmitters. Biol
Proced Online. 10:90–101. 2008.PubMed/NCBI
|
|
31
|
Trzaska KA, Reddy BY, Munoz JL, Li KY, Ye
JH and Rameshwar P: Loss of RE-1 silencing factor in mesenchymal
stem cell-derived dopamine progenitors induces functional maturity.
Mol Cell Neurosci. 39:285–290. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Trzaska KA, Kuzhikandathil EV and
Rameshwar P: Specification of a dopaminergic phenotype from adult
human mesenchymal stem cells. Stem Cells. 25:2797–2808. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Muguruma Y, Reyes M, Nakamura Y, Sato T,
Matsuzawa H, Miyatake H, Akatsuka A, Itoh J, Yahata T, Ando K, et
al: In vivo and in vitro differentiation of myocytes from human
bone marrow-derived multipotent progenitor cells. Exp Hematol.
31:1323–1330. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Greco SJ, Zhou C, Ye JH and Rameshwar P:
An interdisciplinary approach and characterization of neuronal
cells transdifferentiated from human mesenchymal stem cells. Stem
Cells Dev. 16:811–826. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gresner SM, Golanska E, Kulczycka-Wojdala
D, Jaskolski DJ, Papierz W and Liberski PP: Selection of reference
genes for gene expression studies in astrocytomas. Anal Biochem.
408:163–165. 2011. View Article : Google Scholar
|
|
36
|
Zampieri M, Ciccarone F, Guastafierro T,
Bacalini MG, Calabrese R, Moreno-Villanueva M, Reale A, Chevanne M,
Bürkle A and Caiafa P: Validation of suitable internal control
genes for expression studies in aging. Mech Ageing Dev. 131:89–95.
2010. View Article : Google Scholar
|
|
37
|
Raicevic G, Najar M, Stamatopoulos B, De
Bruyn C, Meuleman N, Bron D, Toungouz M and Lagneaux L: The source
of human mesenchymal stromal cells influences their TLR profile as
well as their functional properties. Cell Immunol. 270:207–216.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jung KH, Yi T, Son MK, Song SU and Hong
SS: Therapeutic effect of human clonal bone marrow-derived
mesenchymal stem cells in severe acute pancreatitis. Arch Pharm
Res. August 21–2014.(Epub ahead of print).
|
|
39
|
Deng Y, Zhou H, Gu P and Fan X: Repair of
canine medial orbital bone defects with miR-31-modified bone marrow
mesenchymal stem cells. Invest Ophthalmol Vis Sci. 55:6016–6023.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wei L, Lei GH, Yi HW and Sheng PY: Bone
formation in rabbit’s leg muscle after autologous transplantation
of bone marrow-derived mesenchymal stem cells expressing human bone
morphogenic protein-2. Indian J Orthop. 48:347–353. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Jones GN, Moschidou D, Abdulrazzak H, et
al: Potential of human fetal chorionic stem cells for the treatment
of osteogenesis imperfecta. Stem Cells Dev. 23:262–276. 2014.
View Article : Google Scholar
|
|
42
|
Najar M, Raicevic G, Boufker HI, Fayyad
Kazan H, De Bruyn C, Meuleman N, Bron D, Toungouz M and Lagneaux L:
Mesenchymal stromal cells use PGE2 to modulate activation and
proliferation of lymphocyte subsets: Combined comparison of adipose
tissue, Wharton’s Jelly and bone marrow sources. Cell Immunol.
264:171–179. 2010. View Article : Google Scholar
|
|
43
|
Raicevic G, Rouas R, Najar M, Stordeur P,
Boufker HI, Bron D, Martiat P, Goldman M, Nevessignsky MT and
Lagneaux L: Inflammation modifies the pattern and the function of
Toll-like receptors expressed by human mesenchymal stromal cells.
Hum Immunol. 71:235–244. 2010. View Article : Google Scholar
|
|
44
|
Yañez R, Oviedo A, Aldea M, Bueren JA and
Lamana ML: Prostaglandin E2 plays a key role in the
immunosuppressive properties of adipose and bone marrow
tissue-derived mesenchymal stromal cells. Exp Cell Res.
316:3109–3123. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen MY, Lie PC, Li ZL and Wei X:
Endothelial differentiation of Wharton’s jelly-derived mesenchymal
stem cells in comparison with bone marrow-derived mesenchymal stem
cells. Exp Hematol. 37:629–640. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yoo KH, Jang IK, Lee MW, Kim HE, Yang MS,
Eom Y, Lee JE, Kim YJ, Yang SK, Jung HL, et al: Comparison of
immunomodulatory properties of mesenchymal stem cells derived from
adult human tissues. Cell Immunol. 259:150–156. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Noël D, Caton D, Roche S, Bony C, Lehmann
S, Casteilla L, Jorgensen C and Cousin B: Cell specific differences
between human adipose-derived and mesenchymal-stromal cells despite
similar differentiation potentials. Exp Cell Res. 314:1575–1584.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Uccelli A, Moretta L and Pistoia V:
Mesenchymal stem cells in health and disease. Nat Rev Immunol.
8:726–736. 2008. View Article : Google Scholar
|
|
49
|
Pozzobon M, Ghionzoli M and De Coppi P:
ES, iPS, MSC, and AFS cells. Stem cells exploitation for Pediatric
Surgery: current research and perspective. Pediatr Surg Int.
26:3–10. 2010. View Article : Google Scholar
|