Introduction
Mesenchymal stem/stromal cells (MSCs) are able to
extensively self renew and possess a multilineage differentiation
potential (1–3). In addition, MSCs exert a supportive
function through paracrine effects (4–6).
Therefore, these cells have a wide application in cell-based
therapies and tissue engineering. Over the last decade, MSCs
derived from alternative and easily accessible sources have
received increasing attention. MSCs can be isolated from a large
variety of fetal tissues, extraembryonic tissues and organs and a
variety of tissues from children and adults (7).
Human umbilical cord-derived MSCs (HUMSCs) can be
isolated and expanded easily in vitro without ethical
concerns. Therefore, they have received interest as a promising
candidate for several potential clinical applications (8) and several studies have demonstrated
that HUMSCs exhibit this potential (9). HUMSCs may possess a capacity for
autologous and allogenic transplantation due to their
immunoprivileged properties (10).
They are able to effectively suppress mitogen-induced T-cell
proliferation (11) and low levels
of rejection have been observed in animal transplantation studies
(12,13).
HUMSCs and dental pulp-derived stem cells (DPSCs)
offer available sources of autologous cells without ethical
controversy. Human DPSCs were initially reported by Gronthos et
al in 2000 and were observed to share a similar morphology and
in vitro differentiation potential with bone marrow MSCs
(14). DPSCs also possess
immunoregulatory properties and are capable of inducing activated
T-cell apoptosis in vitro and alleviating
inflammatory-related tissue injury (15). Impacted adult wisdom teeth and
naturally-exfoliated deciduous teeth do not ordinarily serve a
function in adults. Therefore, DPSCs can be obtained without
adverse effects on health.
HUMSCs are also amenable to genetic modification.
Kermani et al (16)
transfected HUMSCs with a green fluorescent protein (GFP)-reporter
gene and established a stable cell line. There have been no reports
on the genetic modification of DPSCs to date, to the best of our
knowledge. Survivin (SVV), a 16.5 kDa protein, is a member of the
X-linked inhibitor of apoptosis family and exhibits broadly
cytoprotective properties (17).
In the present study, SVV-modified HUMSCs and DPSCs were
investigated.
The potential clinical application of HUMSCs and
DPSCs remains to be elucidated. In the present study, the
differentiation, SVV-modified effects and molecular basis of the
heterogeneity between HUMSCs and DPSCs were investigated in
vitro and in vivo.
Materials and methods
Preparation of HUMSCs and DPSCs
HUMSCs were isolated, as previously described, with
certain modifications (18). The
human umbilical cords were obtained from babies delivered at
full-term following institutional review board approval. A segment
of umbilical cord, cut to ~7 cm in length was obtained. This
segment was placed immediately into 25 ml complete medium,
comprising Dulbecco’s modified Eagle’s medium (DMEM)/F12 (Gibco
Life Technologies, Darmstadt, Germany) supplemented with 10% fetal
bovine serum (FBS; Gibco Life Technologies, Grand Island, NY, USA),
100 U/ml penicillin and 100 μg/ml streptomycin (Gibco Life
Technologies). The tubes were stored on ice for dissection within 4
h. The umbilical cord segment was washed three times with
phosphate-buffered saline (PBS) without calcium and magnesium
(HyClone, Shanghai, China) and dissected longitudinally using an
aseptic technique. The umbilical vein and the umbilical arteries
were removed and the umbilical cord segment was cut into a 0.5–1
mm3 tissue block and incubated in 3 ml 0.2% collagenase
type II (Sigma-Aldrich, St. Louis, MO, USA) for 1 h at 37°C. A
five-fold volume of complete medium was added and the supernatant
was filtered using a 70-μm filter following free settling for 20
min. The cells were collected following centrifugation at 500 × g
for 5 min and plated in plastic culture flasks at a concentration
of 5×103/cm2. After 3 days, the non-adherent
cells were removed by washing three times with PBS. The medium was
changed every 2–3 days.
The human DPSCs were isolated as follows: Two adult
wisdom teeth from one patient aged between 18 and 30 years old,
which were extracted for clinical purposes at Tonji Hospital, Tonji
University School of Medicine (Shanghai, China), were collected and
the crown and root were separated. The dental pulp was isolated and
digested in 0.3% collagenase type I (Sigma-Aldrich) for 1 h at
37°C. The DPSCs were cultured in α-minimum essential medium (α-MEM;
HyClone) supplemented with 10% FBS at 37°C in 5%
CO2.
The HUMSCs and DPSCs were identified according to
the expression of antigens using flow cytometry with a Human
Mesenchymal Stem Cell Functional Identification kit (BD
Biosciences, San Jose, CA, USA).
The study was approved by the ethics committee of
Tonji Hospital, Tonji University School of Medicine (Shanghai,
China). Written informed consent was obtained from all patients or
their families.
Differentiation characterization
The adipogenic, osteogenic and chondrogenic
differentiation potentials of the HUMSCs and DPSCs were determined
by incubating the cells in differentiation media using a Human
Mesenchymal Stem Cell Functional Identification kit (R&D
Systems, Inc, Minneapolis, MN, USA).
Adipogenic differentiation
The HUMSCs and DPSCs were seeded onto 12 mm cover
slips at a density of 3.7×104 cells/well in the α-MEM
basal medium supplemented with 10% FBS, 100 U/ml penicillin, 100
μg/ml streptomycin and 2 mM L-glutamine. Theells were cultured in
an incubator at 37°C and 5% CO2. At 100% confluence, the
medium was replaced with the adipogenic differentiation medium
containing hydrocortisone, isobutylmethylxanthine, indomethacin in
95% ethanol and α minimum essential medium basal medium. (R&D
Systems, Inc). The medium was replaced every 3 days. After 2 weeks,
the cells on the cover slips were washed with PBS twice and fixed
with 4% paraformaldehyde (Beijing Solarbio Science &
Technology, Beijing, China) for 20 min at room temperature. The
cells were then washed with PBS and stained for 30 min using oil
red O (Sigma-Aldrich). Following washing twice with PBS, images of
cells were captured under a phase contrast microscope (IX71,
Olympus Corp., Tokyo Japan).
Osteogenic differentiation
In order to induce osteogenic differentiation,
7.4×103 HUMSCs and DPSCs per well were cultured for 3
weeks in α-MEM basal medium supplemented with 10% FBS, 100 U/ml
penicillin, 100 μg/ml streptomycin and 2 mM L-glutamine (Gibco Life
Technologies). At 50–70% confluency, the medium was replaced with
the osteogenic differentiation medium (R&D Systems, Inc), which
was replaced every 3 days. The cells were fixed with 4%
paraformaldehyde for 20 min, washed with PBS and stained using
alizarin red S (Sigma-Aldrich). Images of the cells were then
captured under a phase contrast microscope (IX71; Olympus
Corp.).
Chondrogenic differentiation
In order to induce chondrogenic differentiation,
2.5×105 cells were centrifuged at 200 × g for 5 min at
room temperature and resuspended in DMEM/F-12 basal medium
supplemented with 0.5 ml 100X ITS supplement (R&D Systems,
Inc), 100 U/ml penicillin, 100 μg/ml streptomycin and 2 mM
L-glutamine. The cells were then centrifuged at 200 × g for 5 min,
resuspended in chondrogenic differentiation medium (R&D
Systems, Inc) and centrifuged again. The resulting pellets were
incubated at 37°C and 5% CO2 for 3 weeks and the medium
was replaced every 3 days. The pellets were fixed with 4%
paraformaldehyde in PBS for 20 min at room temperature and then
frozen and sectioned using standard cryosectioning methods. The
sections were stained using polyclonal goat anti-human aggrecan
antibody (1:10; #962644) and NorthernLights
557-fluorochrome-conjugated donkey anti-goat secondary antibody
(1:200; #NL001; R&D Systems, Inc) and images were captured
under a fluorescence microscope.
Lentiviral vectors and MSC
transduction
The MSCs were transduced with a pLVX-IRES-ZsGreen1
vector (Clontech, Mountain View, CA, USA) containing an inserted
full length cDNA of SVV. Vectors without this insertion were used
as a control. The volume of lentivirus used for each transduction
was determined by titration as the volume required to generate
90–95% ZsGreen1-positive MSCs after 3 days.
Measurement of the levels of SVV in the
cell culture supernatant
The transduced MSCs were plated in six-well plates
(5,000 cells/cm2) overnight at 37°C in 5%
CO2. The medium was then replaced with 2 ml/well
DMEM/F12 for the HUMSCs and α-MEM for the DPSCs, followed by
incubation for 48 h at 37°C in 5% CO2. The supernatants
were collected in order to confirm overexpression and secretion of
SVV using a human enzyme-linked immunosorbent assay (ELISA) kit
according to the manufacturer’s instructions (Abcam, Cambridge, MA,
USA). The cell number was determined for normalization.
Western blotting
To detect the overexpression of SVV in the HUMSCs
and DPSCs, the proteins in the transduced cells were extracted
using radioimmunoprecipitation assay lysis buffer supplemented with
1 mM phenylmethylsulfonyl fluoride (Beyotime Institute of
Biotechnology, Haimen, China). The proteins were loaded onto 12%
SDS-PAGE gels and transferred to polyvinyline difluoride membranes
(EMD Millipore, Billerica, MA, USA). Following blocking with 5%
defatted milk [defatted milk powder (Quechao, Beijing, China) in
tris-buffered saline supplemented with Tween20] for 1 h, the
membranes were incubated with survivin (FL-142) rabbit polyclonal
antibody raised against amino acids 1-142 of human survivin (1:100;
sc-10811; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA)
overnight at 4°C.
DPSC transplantation
As been previous studies have examined the survival
and differentiation of umbilical cord-derived MSCs in vivo
(19), the current study focused
on the properties of the DPSCs in vivo.
All experimental procedures involving animals were
conducted under the approval of the Animal Care and Use Committee
of Tongji University (Shanghai, China). Female C57BL/6 mice were
housed at 22°C and 65% humidity with a 12 h light/dark cycle and
access to food and water ad libitum.
The mice were randomly assigned to one of the
following three groups: Control mice injected with PBS (n=6); mice
transplanted with DPSCs (n=6) and mice transplanted with
SVV-modified DPSCs (n=6). The adult 8-week-old mice were
anesthetized using 0.5% pentobarbital sodium (Sigma-Aldrich). The
heads of the mice were shaved and cleaned and the mice were then
positioned in the stereotaxic apparatus, with anesthesia maintained
during surgery. A midline incision was made on the scalp and the
skin was retracted to expose the bregma. Burr holes (0.5 mm) were
then placed directly over the striatum (coordinates:
anterior/posterior, +0.5 mm; medial/lateral, +2.0 mm from the
bregma). The control and SVV-modified DPSCs were resuspended at a
density of 100,000 cells/μl in PBS. The cells were transplanted
(−3.5 mm ventral to the skull surface) into the control and
SVV-modified DPSCs at a constant rate of 0.5 μl/min in a total
injection volume of 2 μl/mouse using a 10-μl Hamilton microsyringe
(Dalian Replete Scientific Instrument Co. Ltd, Dalian, Liaoning,
China). The syringe was left in place for 3 mins and then
retracted. The wound was closed and the mice were placed on heat
insulation pads (Kent Scientific Corporation, Torrington, CT, USA)
until conscious.
Immunocytochemistry
Immunocytochemistry was performed in order to
examine the survival and differentiation of the control and
SVV-modified DPSCs in the brains of the mice. Following cell
transplantation, the mice were anesthetized for either 4 or 14 days
using pentobarbital sodium and perfused transcardially with 0.1 M
PBS, followed by 4% paraformaldehyde diluted in 0.1 M PBS (pH 7.4)
to fix the tissues. The brains were removed promptly, post-fixed in
4% paraformaldehyde overnight at 4°C and then transferred to 30%
sucrose in 0.1 M PBS for 2 days at 4°C. Coronal sections (20 μm)
were then cut and mounted on positively-charged microscope slides
(Ruihoge, Nanjing, China). For immunohistochemical analysis, the
tissue was blocked using 10% normal goat serum (CST, Boston, MA,
USA) in PBS containing 0.1% Triton-X (Sigma-Aldrich) for 45 min at
room temperature. The tissues were then cultured with primary
antibodies against rabbit SVV (1:100; sc-10811; Santa Cruz
Biotechnology, Inc.), rabbit anti-human/mouse/rat neuronal nuclei
(NeuN; 1:500; 1:500; Abcam) and rabbit anti-cat/dog/mouse/rat/sheep
glial fibrillary acidic protein (GFAP; 1:500; Nr.z 0334; Dako North
America, Inc. Carpinteria, CA, USA) at 4°C overnight. All
antibodies were polyclonal. The following day, the tissues were
rinsed three times in PBS with 1% bovine serum albumin and
incubated with conjugated secondary antibodies (goat anti-rabbit
AlexaFluor594; 1:500; A-11037; Invitrogen Life Technologies) for 1
h at room temperature. Subsequently, 4′,6-diamidino-2-phenylindole
(1:1,000; Sigma-Aldrich) was used to label the positions of the
cell nuclei. Images of the fluorescent labels were captured using a
Nikon fluorescent microscope (Eclipse Ti-S; Nikon Corporation,
Tokyo, Japan) at ×20 magnification. All the images were analyzed
using ImageJ software (National Institutes of Health; Bethesda, MD,
USA). Briefly, images of the transplanted cells were captured from
five sections of each animal and the average intensity of the
labels in each group was analyzed.
RNA sequencing and analysis
In order to compare the gene expression profile
between the control and the SVV-modified HUMSCs and DPSCs, RNA
isolation was performed on the cells using TRIzol®
reagent (Invitrogen Life Technologies) and the total RNA was then
digested with DNase I (Invitrogen Life Technologies) to ensure that
the samples were not contaminated with genomic DNA. Duplicate
experiments were performed for each sample. Library preparation and
paired end sequencing (101 bp length) were performed using an
Illumina HiSeq 2500 sequencer (Illumina, San Diego, CA, USA). Short
reads were aligned to the reference human genome (Homo
sapiens GRCh37) using TopHat (20). The gene annotations were from the
Ensembl database, packaged using iGenome (Illumina). The sequence
reads were used to calculate the overall gene expression in terms
of the reads per kilobase of exon per million mapped reads (RPKM).
The differentially expressed transcripts were screened using the
cufflinks procedure (21).
Abundant genes
Genes with an RPKM value >100 and levels of gene
expression ≥2-fold higher than the control groups were identified
in the HUMSCs, DPSCs and SVV-modified HUMSCs and DPSCs and
functional enrichment analyses were performed using the Gene
Ontology database (http://www.geneontology.org/GO.downloads.annotations.shtml).
and the Kyoto Encyclopedia of Genes and Genomes pathway database
(http://www.genome.jp/kegg/pathway.html).
Genes associated with stem cell
differentiation
A total of five genes associated with stem cell
differentiation, with RPKM values >5 in the control and
SVV-modified HUMSCs and DPSCs, were selected based on the Gene
Ontology database.
Cytokine-cytokine receptor interaction
genes
A total of eight genes associated with
cytokine-cytokine receptor interactions, with RPKM values >5 in
the control and SVV-modified HUMSCs and DPSCs, were selected based
on the Kyoto Encyclopedia of Genes and Genomes pathway
database.
Data presentation and statistical
analysis
Data are presented as the mean ± standard error of
the mean (error bars). All significant differences were evaluated
using Student’s t-test or analysis of variance. Statistical
analyses were performed using SPSS 15.0 (SPSS, Inc., Chicago, IL,
USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
Phenotypic characterization of isolated
HUMSCs and DPSCs
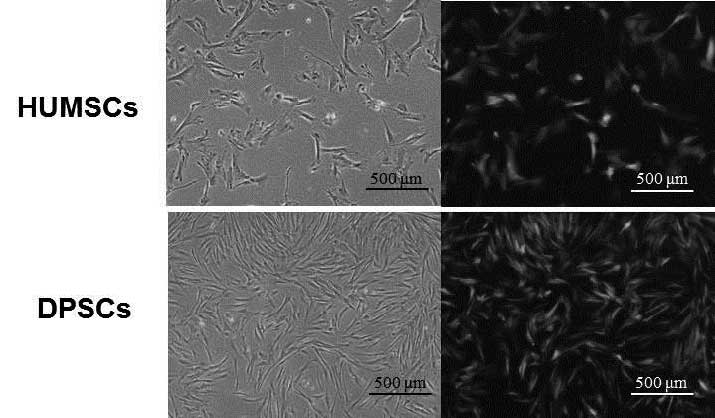
The shapes of the DPSCs were more similar to those
of fibroblasts compared with the HUMSCs (Fig. 1A). Flow cytometric analysis
revealed that the HUMSCs and DPSCs expressed a set of cluster of
differentiation (CD) MSC markers (CD90, CD73 and CD105), however,
they did not express endothelial/hematopoietic markers (CD34, CD45,
CD11b or CD14, CD19 or CD79α or HLA-DR; Fig. 1B and C).
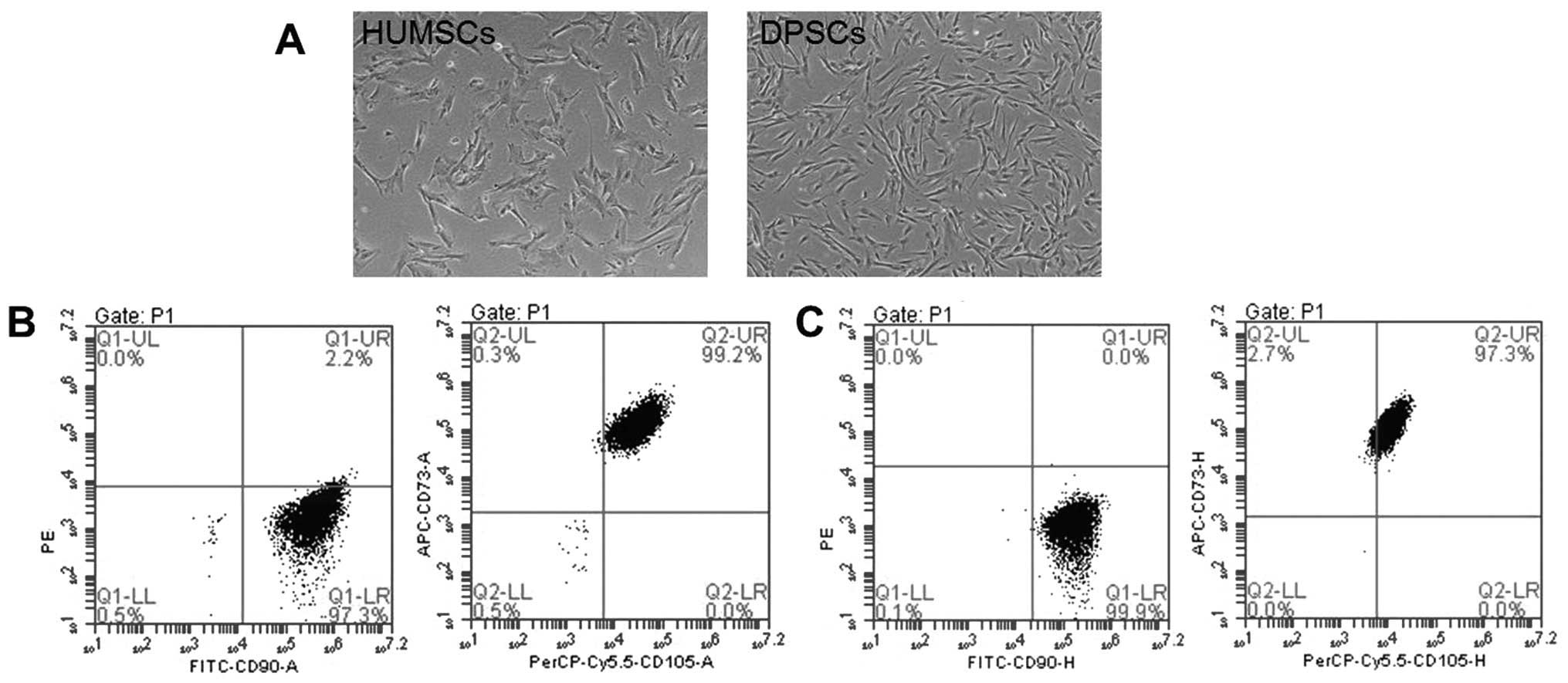 | Figure 1Phenotypic characterization of HUMSCs
and DPSCs. (A) Passage 5 DPSCs were more similar to fibroblasts
than the HUMSCs. Scale bar: 500 μm. (B) 96.5% of the HUMSCs were
positive for CD73, CD90 and CD105, but negative for CD34, CD45,
CD11b or CD14, CD19, CD79α and HLA-DR. (C) 99.2% of the DPSCs were
positive for CD73, CD90 and CD105, but negative for CD34, CD45,
CD11b or CD14, CD19, CD79α and HLA-DR. HUMSCs, human umbilical
cord-derived mesenchymal stem cells; DPSCs, dental pulp-derived
stem cells; FITC, fluorescein isothiocyanate; CD, cluster of
differentiation. |
Identification of differentiation
Following induction with adipogenic differentiation
medium for 2 weeks, lipid droplets, stained using oil red O, were
observed in the HUMSCs (Fig. 2A),
but not the DPSCs (Fig. 2D). For
osteogenic differentiation, the cells were cultured for 3 weeks in
osteogenic media. Calcium precipitation, determined by alizarin red
S staining, was observed in the HUMSCs and the DPSCs (Fig. 2B and E). The chondrogenic
differentiation capacity of the HUMSCs and DPSCs were evaluated by
examining the expression of aggrecan, a chondrogenic marker.
Following culture with the HUMSCs and DPSCs in chondrogenic
differentiation medium for 3 weeks, the cells exhibited increased
expression of aggrecan (Fig. 2C and
F).
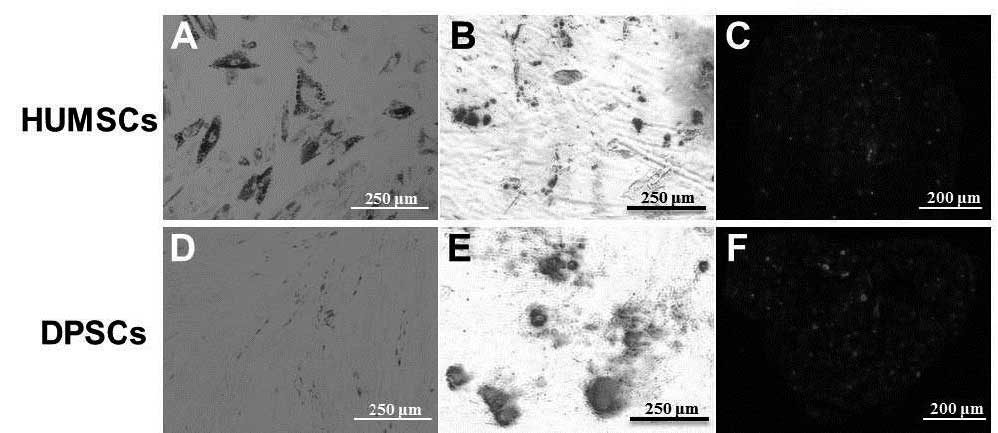 | Figure 2Adipogenic, osteogenic and
chondrogenic differentiation of the (A-C) HUMSCs and (D-F) DPSCs,
respectively. Lipid droplets, stained using oil red O, were present
in the (A) HUMSCs, but not the (D) DPSCs (Scale bar: 250 μm).
Osteogenic differentiation in the (B) HUMSCs and (E) DPSCs was
detected using alizarin red S staining of calcium precipitation
(Scale bar, 250 μm). Aggrecan, a chondrogenic marker, was highly
expressed in the (C) HUMSCs and (F) DPSCs (Scale bar, 200 μm).
HUMSCs, human umbilical cord-derived mesenchymal stem cells; DPSCs,
dental pulp-derived stem cells. |
Efficiency of MSC transduction with
lentiviral vectors containing SVV
Following infection with the SVV recombinant
lentivirus, overexpression of GFP was observed in the HUMSCs and
DPSCs (Fig. 3). The efficiency of
gene transduction was >90%.
Protein expression of SVV
The protein expression levels of SVV were determined
by ELISA and western blotting. The protein levels of SVV were
significantly increased in the cell culture supernatant of the
SVV-modified HUMSCs and DPSCs compared with levels in the control
groups (Fig. 4A). Furthermore, the
secretion of SVV in the modified HUMSCs was significantly higher
compared with that observed in the modified DPSCs. The western
blotting revealed that modified HUMSCs and DPSCs exhibited a
significant increase in the expression of SVV (Fig. 4B and C).
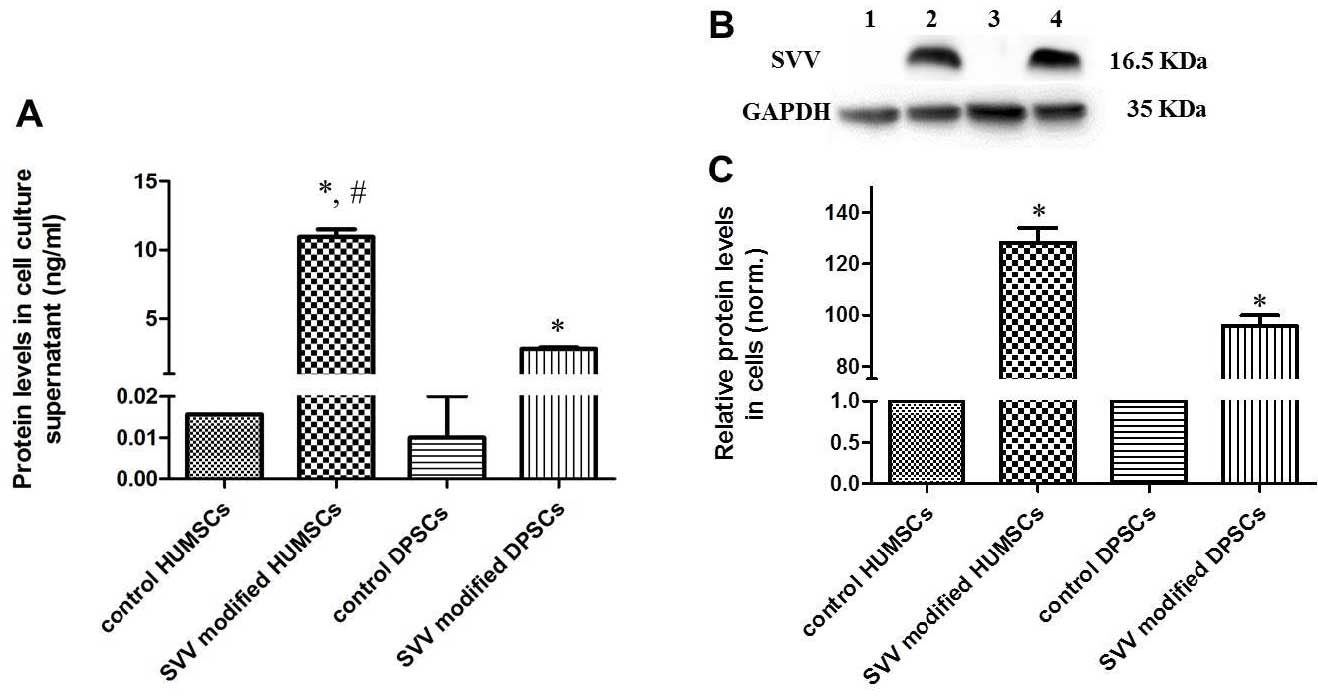 | Figure 4Protein levels of SVV are
significantly increased in the SVV-modified HUMSCs and DPSCs. (A)
Protein levels of SVV were measured in the cell culture supernatant
using an enzyme-linked immunosorbent assay. (B) Representative
western blot analysis of the control, SVV-modified HUMSCs and
DPSCs. The level of GAPDH was used as an internal control. Lanes:
1, HUMSCs; 2, SVV-modified HUMSCs; 3, DPSCs; 4, SVV-modified DPSCs.
(C) Quantitative analysis revealed that the modified HUMSCs and
DPSCs had a significantly higher expression of SVV.
(*P<0.05, compared with the control and
#P<0.05, compared with the SVV-modified DPSCs.
HUMSCs, human umbilical cord-derived mesechymal stem cells; DPSCs,
dental pulp-derived stem cells; SVV, survivin. |
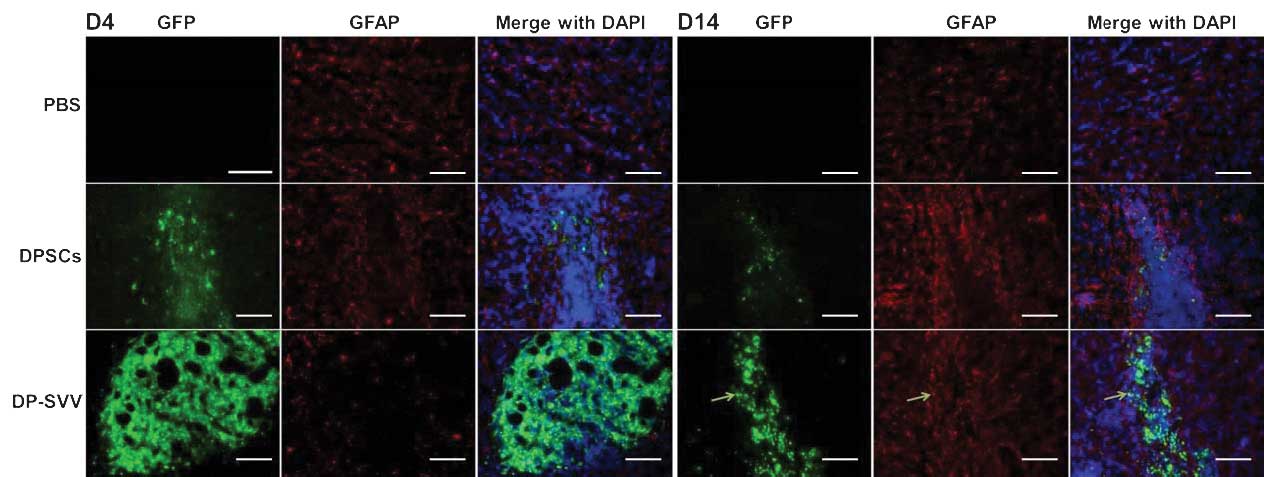
Control and SVV-modified DPSCs survive
following intrastriatal transplantation
In the present study, the survival and the
expression of SVV in the control and SVV-modified DPSCs were
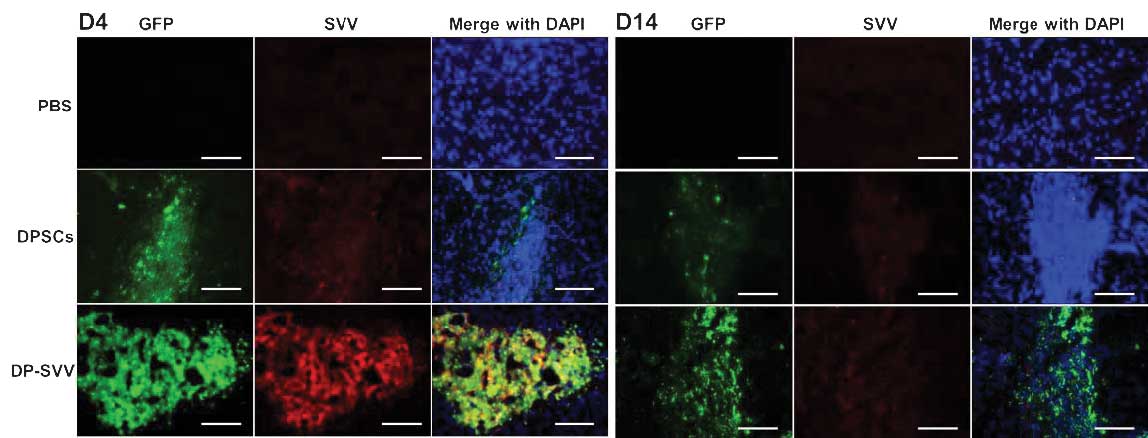
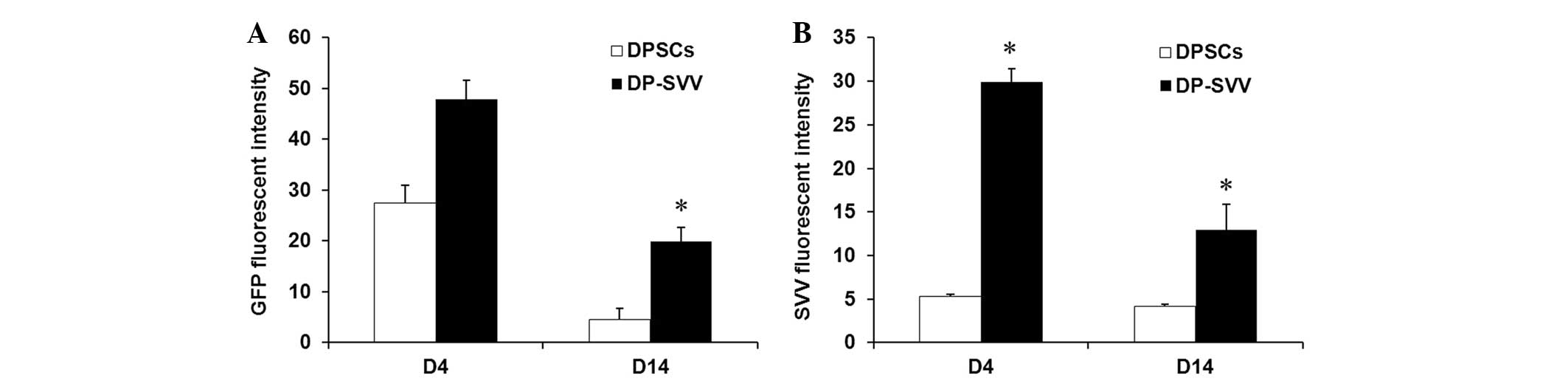
investigated in vivo. The results demonstrated that the GFP
signal, expressed by the inserted pLVX-IRES-ZsGreen1 vector, was
marked in the control and SVV-modified cell lines 4 days after
transplantation, however, it had weakened by day 14 (Fig. 5). In addition, the GFP signal in
SVV-modified DPSCs was more marked compared with that in the
control cells (Fig. 6). These
results suggested that SVV promoted the survival of the DPSCs in
vivo. The expression of SVV in the transplanted SVV-modified
DPSCs was higher than that in the control DPSCs on day 4. However,
the expression of SVV in the transplanted DPSCs decreased 14 days
after transplantation.
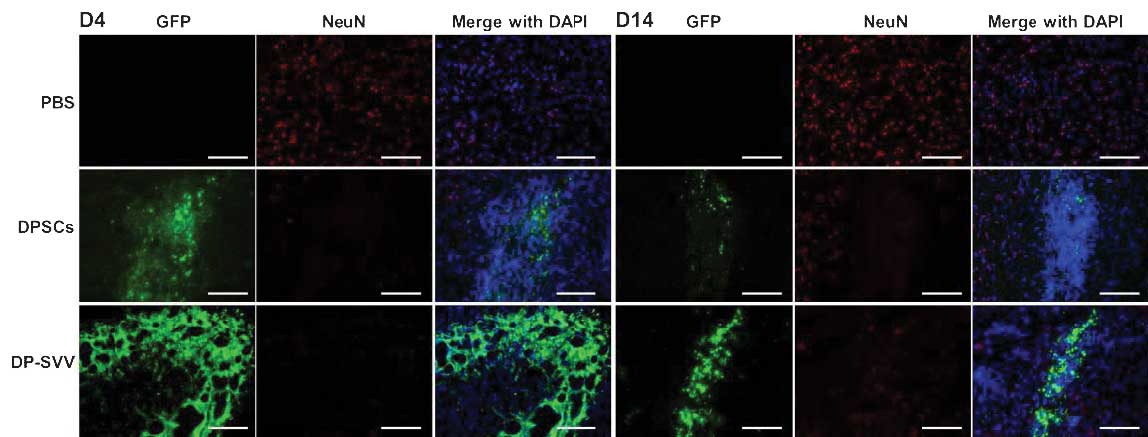
In addition, the present study examined the
differentiation capacity of the control and SVV-modified DPSCs
in vivo. Neither NeuN (a marker for neurons) nor glial
fibrillary acidic protein (GFAP; a marker for astrocytes) were
detected in the control or SVV-modified DPSCs 4 days after
transplantation (Figs. 7 and
8). However, 14 days after
transplantation, a few GFP-labeled cells coexpressed GFAP (Fig. 8), but not NeuN (Fig. 7) in the striatum.
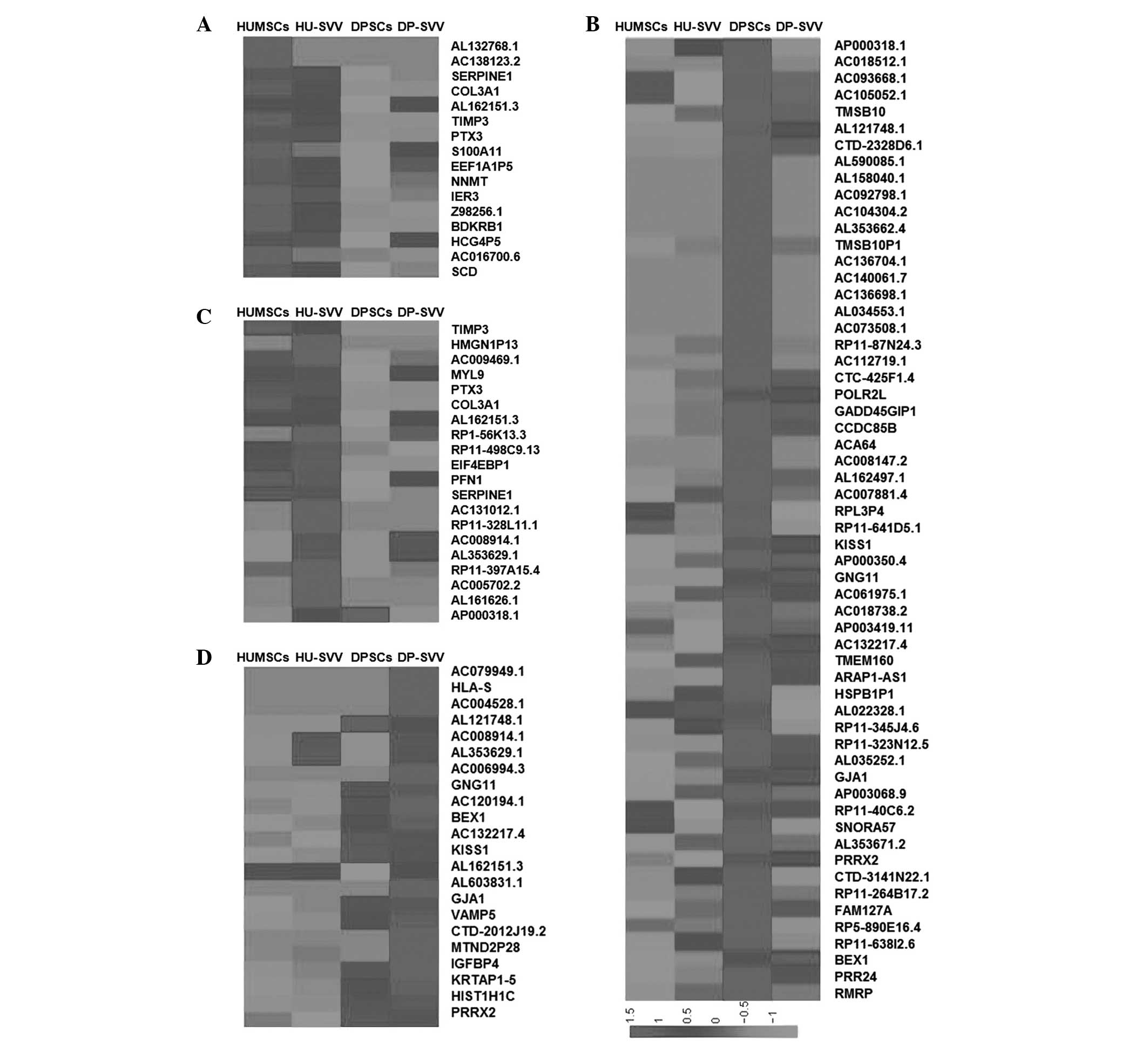
Gene expression data analysis
The heat map of the abundant gene set is shown in
Fig. 9. The genes with RPKM values
>100 were analyzed with respect to each MSC group. A total of
16, 20, 58 and 22 genes were identified with RPKM values >100
and gene expression levels ≥2-fold higher in the HUMSCs,
SVV-modified HUMSCs, DPSCs and SVV-modified DPSCs, respectively,
compared with the control (Fig.
9). The most significant biological processes were
‘inflammatory response’, ‘vascular transport’, ‘platelet
activation’ and ‘vascular transport’ for the HUMSCs, DPSCs,
SVV-modified HUMSCs and SVV-modified DPSCs, respectively. The most
significant pathways were ‘complement and coagulation cascades’,
‘hypoxia-inducible factor-1 signaling pathway’, ‘RNA polymerase’
and ‘SNARE interactions in vesicular transport’ for the HUMSCs,
DPSCs, SVV-modified HUMSCs and SVV-modified DPSCs,
respectively.
The expression of genes associated with biological
activities among the groups was analyzed in order to clarify the
molecular basis of the heterogeneity. Since MSCs possess
multilineage differentiation potential, genes associated with stem
cell differentiation were analyzed in the control and the
SVV-modified HUMSCs and DPSCs. A total of 128 genes were identified
as associated with stem cell differentiation in the four cell lines
and genes with RPKM values >5 were selected. Analysis
demonstrated higher expression levels of the FGF2 and MED10 stem
cell differentiation genes in the SVV-modified HUMSCs, compared
with the HUMSCs, DPSCs and SVV-modified DPSCs (Fig. 10A) and increased expression of
FZD7 in the SVV-modified DPSCs compared with HUMSCs, HU-SVV and
DPSCs. MSCs exhibit anti-inflammatory and immunomodulatory
properties (22). Therefore, a
comparative analysis of genes associated with cytokine-cytokine
receptor interactions in the control and SVV-modified HUMSCs and
DPSCs was also performed. A total of 196 genes associated with
cytokine-cytokine receptor interactions were identified, from which
genes with RPKM values >5 were selected. Clustering highlighted
a group of genes with increased expression in the HUMSCs compared
with HU-SVV, DPSCs and DP-SVV, including tumor necrosis factor
receptor superfamily (TNFRSF)6B, TNFRSF10D, LIF and vascular
endothelial growth factor C (VEGFC; Fig. 10B). Clustering also highlighted a
group of inflammatory-associated genes, the expression levels of
which were highest in the SVV-modified DPSCs, including IL10RB,
PLEKHO2, TNFRSF12A and VEGFC.
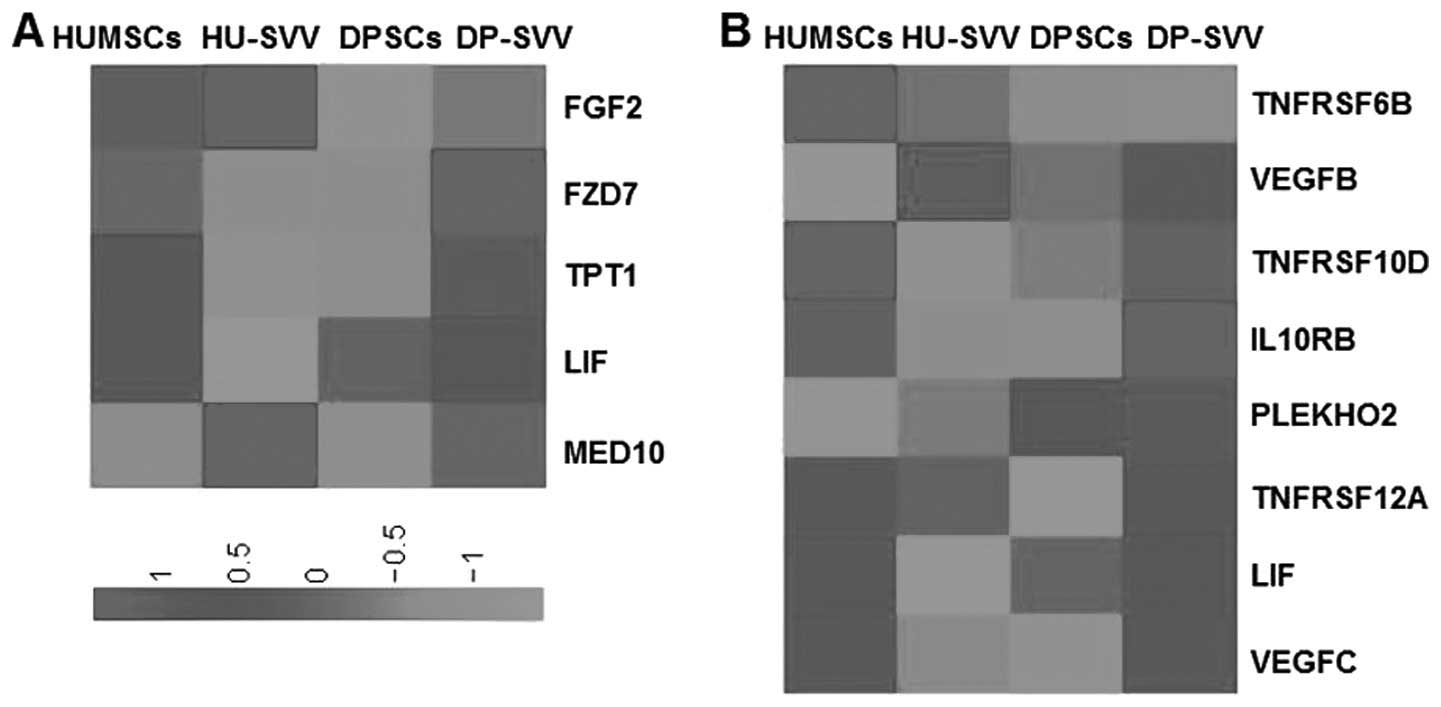 | Figure 10Heat map of genes associated with stem
cell differentiation and cytokine-cytokine receptor interaction.
(A) Genes associated with stem cell differentiation with RPKM
values >5 were compared through RPKM value analysis in the
HUMSCs, HU-SVV, DPSCs and DP-SVV cells. (B) Genes associated wirth
cytokine-cytokine receptor interactions with RPKM values >5 in
the four cell lines. RPKM, reads per kilobase of exon per million
mapped reads; HUMSCs, human umbilical cord-derived mesenchymal stem
cells; DPSCs, dental pulp-derived stem cells; SVV, survivin;
HU-SVV, SVV-modified HUMScs; DP-SVV, SVV-modified DPSCs; TNFRSF,
tumor necrosis factor receptor superfamily; VEGF, vascular
endothelial growth factor. |
Discussion
MSCs may be isolated from a variety of sources,
including bone marrow, umbilical cord, adipose tissue and dental
pulp, and they may be used in several cell-based therapies and
tissue engineering approaches. It is important to select the type
of MSCs that are the most promising candidates for specific
clinical applications. In the present study, the appearance of the
DPSCs were more elongated compared with the HUMSCs. The surface
antigens, CD90, CD73 and CD105, were used for the identification of
HUMSCs and DPSCs, as these markers are indicated by the
International Society for Cellular Therapy as positive markers for
human MSCs (23). These MSCs lack
expression of surface markers of endothelial or hematopoietic
origin, including CD34, CD45, CD11b or CD14, CD19 or CD79α and
HLA-DR.
In the present study, lipid droplets were observed
in the HUMSCs rather than the DPSCs following the induction of
adipogenic differentiation. Osteogenic differentiation was observed
through calcium precipitation in the HUMSCs and DPSCs. The
chondrogenic differentiation capacity of the HUMSCs and DPSCs was
also evaluated by examining the expression level of aggrecan, which
was marked in the two cell lines. These results indicated that the
HUMSCs were more likely to differentiate into adipocytes compared
with the DPSCs and that the two cell types are able to
differentiate into osteoblasts and chondroblasts. These findings
are consistent with those of Mangano et al (14) and Gronthos et al (24), which demonstrated that DPSCs can be
induced to differentiate into osteoblasts, but not adipocytes.
Following transduction of the HUMSCs and DPSCs with
a lentiviral vector, containing the full-length cDNA of survivin
insertion, the expression of GFP indicated that the efficiency of
gene transduction was >90%. The protein expression of SVV in the
HUMSCs and DPSCs was compared using an ELISA assay and western
blotting. The protein levels of SVV significantly increased in the
SVV-modified HUMSCs and DPSCs compared with the controls. However,
the expression of SVV in the cell culture supernatant was
significantly higher in the SVV-modified HUMSCs compared with the
SVV-modified DPSCs. Therefore, modified HUMSCs secreted more SVV
protein compared with the modified DPSCs.
SVV is an important protein for cell death
resistance and has been reported to modulate stem cell
proliferation (25). As MSCs
promote tissue repair by stimulating and modulating tissue-specific
cells (4), SVV may enhance these
stimulating and modulating effects of MSCs. A previous study on the
differentiation of umbilical cord-derived MSCs in vivo
(19), observed no neuronal or
glial differentiation in the transplanted umbilical cord-derived
MSCs. Therefore, the present study focused on the survival and
differentiation of DPSCs in vivo. The results confirmed that
SVV promoted the survival and astrocyte differentiation of the
DPSCs in vivo.
The RNA sequencing analyses provided novel insight
into the variable biological properties among the four groups.
Higher levels of the expression of FGF2, which is involved in the
development of the limb and nervous system, were observed in the
SVV-modified HUMSCs and FZD7, which encodes receptors for Wnt
signaling proteins, in the SVV-modified DPSCs. Genetic analyses
elucidates the MSC-mediated anti-inflammatory and immunomodulatory
properties of the different groups. The expression of IL10RB, an
anti-inflammatory and immunoregulatory gene, was higher in the
SVV-modified DPSCs than in the DPSCs, HUMSCs and SVV-modified
HUMSCs. The expression levels of the TNFRSF6B and TNFRSF10D
anti-inflammatory genes were higher in the HUMSCs than in DPSCs or
SVV-modified DPSCs and HUMSCs. Therefore, HUMSCs express higher
levels of anti-inflammatory, immunoregulatory and stem cell
differentiation-associated genes than DPSCs.
In conclusion, the source of MSCs and their
subsequent modification can be selected according to the difference
in differentiation, the effect of genetic modification and the
molecular basis of the HUMSCs and DPSCs. However, further
investigation is required in order to decide which cell line is
most suitable for a specific clinical application.
Acknowledgements
This study was supported by grants from the Program
of Shanghai Subject Chief Scientist (grant no. 2012XD1404400), the
State Key Program of National Natural Science Foundation of China
(grant no. 81330030), the Excellent Academic Leaders of Shanghai
Health Field (grant no. XBR2013094), the National Science
Foundation for Post-doctoral Scientists of China (grant no.
2012M520933), the Shanghai Municipal Health Bureau Youth Projects
(grant no. 20124y147) and the Fundamental Research Funds for the
Central Universities (grant no. 2012KJ016).
References
|
1
|
Choong PF, Mok PL, Cheong SK, Leong CF and
Then KY: Generating neuron-like cells from BM-derived mesenchymal
stromal cells in vitro. Cytotherapy. 9:170–183. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pittenger MF, Mackay AM, Beck SC, Jaiswal
RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S and
Marshak DR: Multilineage potential of adult human mesenchymal stem
cells. Science. 284:143–147. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xu R, Jiang X, Guo Z, Chen J, Zou Y, Ke Y,
Zhang S, Li Z, Cai Y, Du M, Qin L, Tang Y and Zeng Y: Functional
analysis of neuron-like cells differentiated from neural stem cells
derived from bone marrow stroma cells in vitro. Cell Mol Neurobiol.
28:545–558. 2008. View Article : Google Scholar
|
|
4
|
Cui X, Chen L, Ren Y, Ji Y, Liu W, Liu J,
Yan Q, Cheng L and Sun YE: Genetic modification of mesenchymal stem
cells in spinal cord injury repair strategies. Biosci Trends.
7:202–208. 2013.PubMed/NCBI
|
|
5
|
Kode JA, Mukherjee S, Joglekar MV and
Hardikar AA: Mesenchymal stem cells: immunobiology and role in
immunomodulation and tissue regeneration. Cytotherapy. 11:377–391.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
da Meirelles LS, Fontes AM, Covas DT and
Caplan AI: Mechanisms involved in the therapeutic properties of
mesenchymal stem cells. Cytokine Growth Factor Rev. 20:419–427.
2009. View Article : Google Scholar
|
|
7
|
Davies OG, Smith AJ, Cooper PR, Shelton RM
and Scheven BA: The effects of cryopreservation on cells isolated
from adipose, bone marrow and dental pulp tissues. Cryobiology.
69:342–347. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bakhshi T, Zabriskie RC, Bodie S, Kidd S,
Ramin S, Paganessi LA, Gregory SA, Fung HC and Christopherson KW
II: Mesenchymal stem cells from the Wharton’s jelly of umbilical
cord segments provide stromal support for the maintenance of cord
blood hematopoietic stem cells during long-term ex vivo culture.
Transfusion. 48:2638–2644. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Moretti P, Hatlapatka T, Marten D,
Lavrentieva A, Majore I, Hass R and Kasper C: Mesenchymal stromal
cells derived from human umbilical cord tissues: primitive cells
with potential for clinical and tissue engineering applications.
Adv Biochem Eng Biotechnol. 123:29–54. 2010.
|
|
10
|
Lisianyĭ MI: Mesenchymal stem cells and
their immunological properties. Fiziol Zh. 59:126–134. 2013.(In
Ukranian).
|
|
11
|
Yoo KH, Jang IK, Lee MW, Kim HE, Yang MS,
Eom Y, Lee JE, Kim YJ, Yang SK, Jung HL, et al: Comparison of
immunomodulatory properties of mesenchymal stem cells derived from
adult human tissues. Cell Immunol. 259:150–156. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang CC, Shih YH, Ko MH, Hsu SY, Cheng H
and Fu YS: Transplantation of human umbilical mesenchymal stem
cells from Wharton’s jelly after complete transection of the rat
spinal cord. PLoS One. 3:e33362008. View Article : Google Scholar
|
|
13
|
Ma H, Wu Y, Xu Y, Sun L and Zhang X: Human
umbilical mesenchymal stem cells attenuate the progression of focal
segmental glomerulosclerosis. Am J Med Sci. 346:486–493. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gronthos S, Mankani M, Brahim J, Robey PG
and Shi S: Postnatal human dental pulp stemcells (DPSCs) in vitro
and in vivo. Proc Natl Acadm Sci USA. 97:13625–13630. 2000.
View Article : Google Scholar
|
|
15
|
Zhao Y, Wang L, Jin Y and Shi S: Fas
ligand regulates the immunomodulatory properties of dental pulp
stem cells. J Dent Res. 91:948–954. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kermani AJ, Fathi F and Mowla SJ:
Characterization and genetic manipulation of human umbilical cord
vein mesenchymal stem cells: potential application in cell-based
gene therapy. Rejuvenation Res. 11:379–386. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bhowmick R and Girotti AW: Cytoprotective
signaling associated with nitric oxide upregulation in tumor cells
subjected to photodynamic therapy-like oxidative stress. Free Radic
Biol Med. 57:39–48. 2013. View Article : Google Scholar :
|
|
18
|
Kikuchi-Taura A, Taguchi A, Kanda T, Inoue
T, Kasahara Y, Hirose H, Sato I, Matsuyama T, Nakagomi T, Yamahara
K, et al: Human umbilical cord provides a significant source of
unexpanded mesenchymal stromal cells. Cytotherapy. 14:441–450.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fink KD, Rossignol J, Crane AT, Davis KK,
Bombard MC, Bavar AM, Clerc S, Lowrance SA, Song C, Lescaudron L
and Dunbar GL: Transplantation of umbilical cord-derived
mesenchymal stem cells into the striata of R6/2 mice: behavioral
and neuropathological analysis. Stem Cell Res Ther. 4:1302013.
View Article : Google Scholar :
|
|
20
|
Trapnell C, Pachter L and Salzberg SL:
TopHat: discovering splice junctions with RNA-Seq. Bioinformatics.
25:1105–1111. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Trapnell C, Roberts A, Goff L, Pertea G,
Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL and Pachter L:
Differential geneand transcript expressionanalysis of RNA-seq
experiments with TopHat and Cufflinks. Nat Protoc. 7:562–578. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Baron F and Storb R: Mesenchymal stromal
cells: a new tool against graft-versus-host disease? Biol Blood
Marrow Transplant. 18:822–840. 2012. View Article : Google Scholar :
|
|
23
|
Dominici M, Le Blanc K, Mueller I,
Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A,
Prockop DJ and Horwitz E: Minimal criteria for defining multipotent
mesenchymal stromal cells. The international society for cellular
therapy position statement. Cytotherapy. 8:315–317. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mangano C, De Rosa A, Desiderio V,
d’Aquino R, Piattelli A, De Francesco F, Tirino V, Mangano F and
Papaccio G: The osteoblastic differentiation of dental pulp stem
cells and bone formation on different titanium surface textures.
Biomaterials. 31:3543–3551. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fukuda S, Foster RG, Porter SB and Pelus
LM: The antiapoptosis protein survivin is associated with cell
cycle entry of normal cord blood CD34(+) cells and modulates cell
cycle and proliferation of mouse hematopoietic progenitor cells.
Blood. 100:2463–2471. 2002. View Article : Google Scholar : PubMed/NCBI
|