Introduction
Mammalian spermatozoa are produced in the testis and
mature in the epididymis; thus, the testis and epididymis determine
male fertility. The testis generate non-functional sperm from
precursor germ cells (1), which
the epididymis subsequently matures, protects and stores prior to
ejaculation (2,3). Testicular sperm is released into the
seminiferous tubules, and progresses through the epididymal lumen.
During this process, the sperm travels through highly specialized
and localized microenvironments in the seminiferous tubules and
epididymal lumen, which are maintained by the blood-testis and
blood-epididymis barriers, respectively (4,5). The
barriers provide protection for the sperm from toxic substances and
the immune system (6). The
microenvironment may directly contribute to spermatogenesis and
epididymal sperm maturation, and consequentially affects the
quality of the sperm (7). Thus,
the secreted microenvironment proteins in the tubules are targets
for interference, which may result in the disruption of
spermatogenesis or sperm maturation, and provide the basis for the
development of male contraceptive agents (8,9).
Therefore, investigating the interactional association between the
fluid microenvironment and sperm is clinically relevant.
Previous studies of adult human testicular and
epididymal tissue and fluid (sperm-milieu) proteomes have
characterized the testicular and epididymal proteins and their
functions, in addition to suggesting the major pathways in which
these proteins participate (10,11).
Within the sperm-milieu proteome, 524 sperm-milieu proteins have
been identified by two-dimensional gel electrophoresis and
matrix-assisted laser desorption/ionization (MALDI)-time-of-flight
(TOF) or MALDI-TOF/TOF mass spectrometry. Using the corresponding
antibodies, 319 sperm-located domain-specific proteins of
testicular and epididymal origin, were identified, which provided
novel insight into the biology of spermatogenesis and sperm
maturation. However, due to the limited data generated by current
technologies, an overall insight still remains to be gained. The
Human Protein Atlas (HPA; www.proteinatlas.org) has produced a global
immunohistochemistry map of testicular and epididymal protein
expression profiles (12), which
provided a reliable resource for mapping testicular and epididymal
protein profiles. The present study performed a direct comparison
of protein expression levels in testicular and epididymal tissues
to identify the predominantly expressed proteins in the testis and
epididymis. In addition, a secretory protein profile was identified
as a candidate for the creation of the sperm milieu, which
bioinformatically appeared to be important in the maturation and
survival of sperm. The present study aimed to conduct a detailed
comparative mapping of testicular and epididymal protein profiles
in order to extend current knowledge.
Materials and methods
Sample preparation
The testes and epididymides were obtained from five
young fathers (27–33 years old), who had died in car accidents with
no history of pathology that may have affected their reproductive
function, and had consented to donate their bodies to medical
research whilst they were alive. The donation of their organs for
medical research was additionally approved by their immediate
family. All procedures were approved by the Ethics Committee of Yu
Huang Ding Hospital (Yantai, China). The organs from one side were
processed for protein extraction and the other for
immunohistochemistry. Protein extraction was conducted as
previously described (10,11).
Data collection
Staining profiles for proteins in the human testis
and epididymis were downloaded from the Human Protein Atlas
(http://www.proteinatlas.org). The
expression levels of each protein were graded into four levels:
Strong, moderate, weak and negative. The differentially-expressed
proteins referred to proteins with a change of >2 classification
levels between the testis and epididymis, and the resulting
proteins were referred to as predominantly-expressed testicular or
epididymal proteins. A current complet data set of human seminal
fluid proteins was built by integrating the published proteomic
works of seminal fluids (13).
These proteins, combining analysis with the secretory prediction
tool Locate (http://locate.imb.uq.edu.au/), were used as expected
secreted proteins to screen the epididymal fluid proteins. A list
of human sperm proteins was obtained by collecting data from a
previous study (14).
Gene ontology (GO) analysis
The general functions of predominantly expressed
testicular, epididymal and sperm milieu proteins were broadly
classified according to the GO annotation (www.geneontology.org) and protein class annotation in
Panther (http://www.pantherdb.org).
Overrepresentation analysis of
predominantly-expressed proteins
Overrepresentation analysis of the GO terms,
including biological processes and molecular functions, was
conducted using ConsensusPathDB-human (http://cpdb.molgen.mpg.de/CPDB), which is a molecular
functional interaction database. GO level 2 and 3 categories were
selected, and the P-value cutoff was set as 0.01.
Validation experiment
Western blot and immunohistochemical analyses were
performed as previously described (10,11).
The primary antibodies were as follows: Goat polyclonal anti-solute
carrier family 2 (facilitated glucose transporter), member 3
(SLC2A3) (sc-31838; Santa Cruz Biotechnology, Inc., Dallas, TX,
USA), goat polyclonal anti-solute carrier family 25
(carnitine/acylcarnitine translocase), member 20 (SLC25A20)
(sc-103220; Santa Cruz Biotechnology, Inc.), rabbit polyclonal
anti-WAP-type four-disulfide core domain protein 8 (WFDC8)
(sc-86261; Santa Cruz Biotechnology, Inc.) and rabbit polyclonal
anti-prostate and testis expressed 1 (PATE1) (ab173522; Abcam,
Cambridge, MA, USA). Briefly, 50 μg protein was separated by
12.5% (w/v) SDS-PAGE and was then transferred to polyvinylidene
difluoride membranes (Sigma-Aldrich, St. Louis, MO, USA) for
western blotting. The membranes were blocked with 3% (w/v) non-fat
milk (Wondersun Dairy Co., Ltd., Harbin, China) for 1 h and were
incubated with the respective primary antibody at room temperature
for 1 h. Following washing with Tris-buffered saline and
Tween-20® (Sigma-Aldrich) three times, membranes were
incubated with horseradish peroxidase (HRP)-conjugated
anti-immunoglobulin G (OriGene Technologies, Inc., Beijing, China)
for 1 h. A diaminobenzidene (DAB) kit (OriGene Technologies, Inc.)
was used to visualize the immunoreactive complexes and the
membranes were scanned with a Z320 scanner (Founder, Beijing,
China). For immunohistochemistry, the tissues were fixed in Bouin’s
solution (Sigma-Aldrich) for 10 h and embedded with paraffin.
Sections (4 μm) were incubated in a microwave oven for 15
min for antigen retrieval. Incubation of the sections with
H2O2 (3%; v/v) (Science & Technology Co.,
Ltd, Yantai, China) for 10 min was used to to remove endogenous
peroxidases. Following antigen blocking with 3% bovine serum
albumin (Sigma-Aldrich) for 30 min, the appropriate primary
antibody was added to the sections overnight at 4°C. Following
washing of the sections with Tris-buffered saline several times,
HRP-conjugated anti-rabbit IgG (OriGene Technologies, Inc.) was
added for 1 h at 37°C. The DAB kit was used to reveal the binding
sites, and subsequently the sections were counterstained by
hematoxylin (Abcam) and mounted for bright-field microscopy (DM
LB2; Leica Microsystems GmbH, Nussloch, Germany).
Statistical Analysis
Data are presented as the mean ± standard deviation.
Means were compared between two groups using Student’s t-test. A
commercial software package (SPSS 18.0; SPSS, Inc., Chicago, IL,
USA) was used to perform the analysis. P<0.05 was considered to
indicate a statistically significant difference.
Results
Protein profiles of human testis and
epididymis
Proteins with annotations were retrieved from the
Human Protein Atlas database (http://proteinatlas.org). The resulting data included
7,595 proteins expressed in the testicular seminiferous duct cells
and 7,573 proteins in epididymal glandular cells. A total of 5,190
sperm protexins were identified from previous studies; of these,
1,309 and 1,308 proteins were observed to overlap in the testicular
or epididymal samples, respectively.
Differentially expressed testicular and
epididymal proteins
Secretory proteins: The integrated seminal fluid
proteins and secreted proteins identified by the prediction tool
were retrieved to select potential target secretory proteins in
human testis and epididymal expressed proteins. A total of 2,960
proteins were obtained as the background secretory protein dataset.
A total of 1,306 and 1,377 potential secretory proteins were
identified in the testis and epididymis, respectively.
Human testicular and epididymal predominant
expression proteins: Proteins in the HPA were graded into four
expression level categories: High, moderate, low and negative. A
total of 2,451 proteins exhibited high expression levels in the
testis and 2,218 proteins exhibited high expression levels in the
epididymis. By comparing the protein expression levels between
testicular and epididymal samples, a total of 2,783 proteins were
demonstrated to have differential expression levels. By further
screening with a strict criterion of a two-fold difference in
expression, 1,412 proteins were identified to be predominantly
expressed in the testis, including 388 sperm proteins and 173
secretory proteins. A total of 1,371 proteins were observed to be
predominantly expressed in the epididymis, including 363 sperm
proteins and 244 secretory proteins (Table I). These secreted proteins
predominantly expressed in human testis or epididymis were
suggested as promising sperm milieu proteins as previously
described (10,11). Of the 244 secretory proteins
identified in the epididymis, 56 were common with the sperm milieu
identified in a previous study.
 | Table ICharacteristics of testicular and
epididymal expressed proteins. |
Table I
Characteristics of testicular and
epididymal expressed proteins.
| Expression | Weak (n)
| Moderate (n)
| Strong (n)
| Predominant (n)
|
|---|
| Sperm | Secretory | Sperm | Secretory | Sperm | Secretory | Sperm | Secretory | Others |
|---|
| Testicular | 0 | 121 | 849 | 745 | 460 | 440 | 388 | 173 | 969 |
| Epididymal | 0 | 118 | 871 | 815 | 437 | 444 | 363 | 244 | 928 |
Functional analysis
Broad ontological analysis was performed on the
secreted and sperm-located proteins that were predominantly
expressed in the testis and epididymis. All the proteins were
placed into several broad functional categories on the basis of the
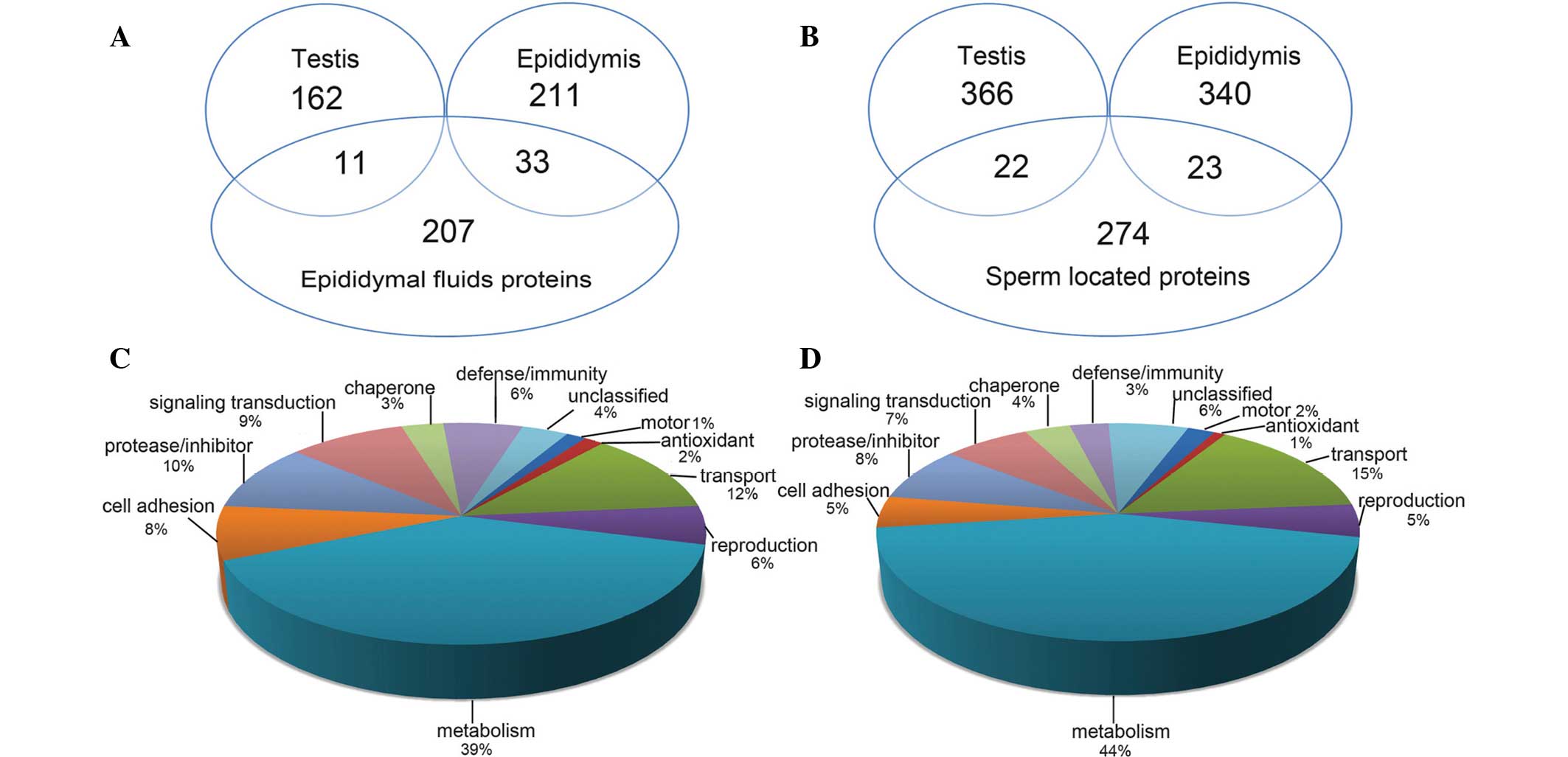
GO and Panther databases. As demonstrated in Fig. 1, the majority of proteins were
involved in metabolic functions, followed by protease/inhibitor
(10%), cell adhesion (8%), defense/immunity (6%) and antioxidant
(2%) functions in the secreted proteins, as well as transport (15%)
and motor (2%) functions in the sperm-located proteins.
Further overrepresentation analysis was conducted on
the predominantly-expressed proteins in the testis, epididymis and
epididymal fluids. The enriched terms reflected their main
functions in spermatogenesis and sperm maturation.
Predominantly-expressed testicular proteins were observed to be
mainly involved in the functions of germ cell development and
differentiation, whereas proteins in the epididymis functioned
predominantly in cell adhesion associated with epithelial cells.
Epididymal fluid proteins were identified to have significant
enzymatic activity (Table
II).
 | Table IIOverrepresentation analysis of
proteins predominantly expressed in the testis and epididymis. |
Table II
Overrepresentation analysis of
proteins predominantly expressed in the testis and epididymis.
A, Predominantly
expressed testicular proteins
|
|---|
| GO term | Candidates contained,
n (%) | P-value |
|---|
| GO:0007276 gamete
generation | 84
(15.8) |
1.89×10−12 |
| GO:0007126
meiosis | 29
(17.7) |
4.04×10−06 |
| GO:0048515 spermatid
differentiation | 19
(22.4) |
5.83×10−06 |
| GO:0051321 meiotic
cell cycle | 29
(17.3) |
6.60×10−06 |
| GO:0009566
fertilization | 21
(19.3) |
2.22×10−05 |
| GO:0007281 germ cell
development | 24
(17.1) |
4.50×10−05 |
| GO:0030317 sperm
motility |
9 (27.3) |
3.53×10−04 |
| GO:0003188 heart
valve formation |
5 (45.5) |
5.67×10−04 |
| GO:0007049 cell
cycle | 130 (9.2) |
1.14×10−03 |
| GO:0007548 gender
differentiation | 31
(12.3) |
1.98×10−03 |
| GO:0007128 meiotic
prophase I |
6 (28.6) |
2.67×10−03 |
| GO:0051301 cell
division | 51
(10.5) |
3.33×10−03 |
| GO:0007530 gender
determination |
6 (25.0) |
5.53×10−03 |
| GO:0048608
reproductive structure development | 30
(11.3) |
7.33×10−03 |
| GO:0035036 sperm-egg
recognition |
5 (26.3) |
8.87×10−03 |
|
B, Predominantly
expressed epididymal proteins
|
| GO term | Candidates contained,
n (%) | P-value |
|
| GO:0007155 cell
adhesion | 91
(9.4) |
3.37×10−03 |
| GO:0042221 response
to chemical stimulus | 241 (8.2) |
5.74×10−03 |
| GO:0030855 epithelial
cell differentiation | 34
(11.1) |
5.88×10−03 |
| GO:0006612 protein
targeting to membrane | 20
(12.8) |
6.95×10−03 |
| GO:0051641 cellular
localization | 178 (8.4) |
7.49×10−03 |
| GO:0008104 protein
localization | 142 (8.6) |
7.50×10−03 |
|
C, Epididymal fluid
proteins
|
| GO term | Candidates contained,
n (%) | P-value |
|
| GO:0006629 lipid
metabolic process | 34
(2.9) |
1.64×10−05 |
| GO:0009056 catabolic
process | 47
(2.3) |
1.09×10−04 |
| GO:2000145
regulation of cell motility | 16
(3.7) |
1.89×10−04 |
| GO:0008233
peptidase activity | 19
(3.2) |
3.37×10−04 |
| GO:0004857 enzyme
inhibitor activity | 13
(4.0) |
4.03×10−04 |
| GO:0006979 response
to oxidative stress | 10
(3.9) |
2.10×10−03 |
| GO:0006508
proteolysis | 24
(2.4) |
3.02×10−03 |
| GO:0009566
fertilization |
6 (5.5) |
3.21×10−03 |
| GO:0051604 protein
maturation |
7 (4.2) |
6.51×10−03 |
| GO:0050776
regulation of immune response | 16
(2.5) |
9.20×10−03 |
Validation of protein expression in the
human testis and epididymis by western blotting and
immunohistochemistry
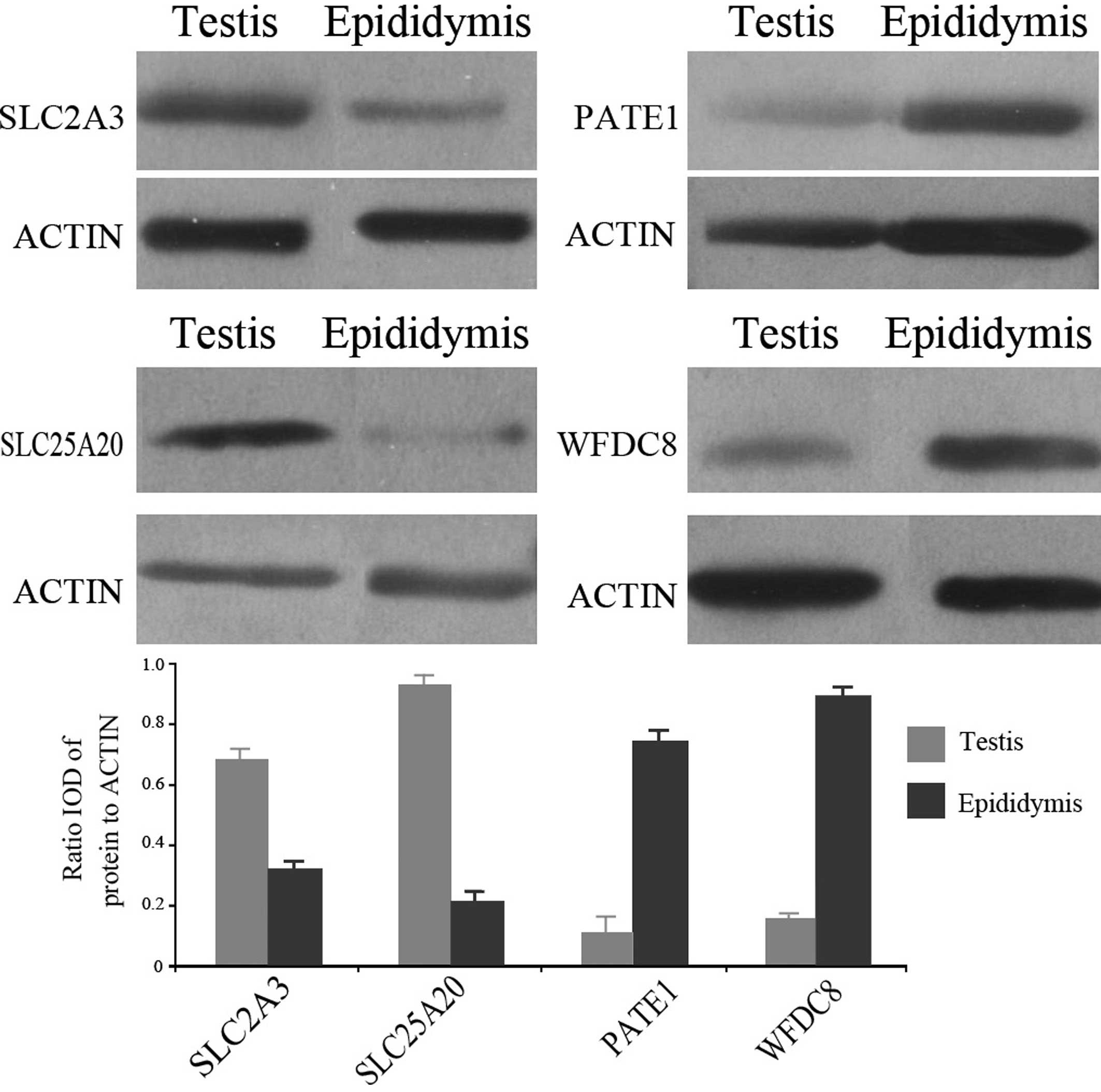
Four proteins (SLC2A3, SLC25A20, WFDC8 and PATE1),
involved in the functions of transport and signal transduction,
were randomly selected for validation by western blotting. The
results indicated higher expression levels of SLC2A3 and SLC25A20
in normal human testis, and higher expression of WFDC8 and PATE1 in
the human epididymis (Fig. 2). In
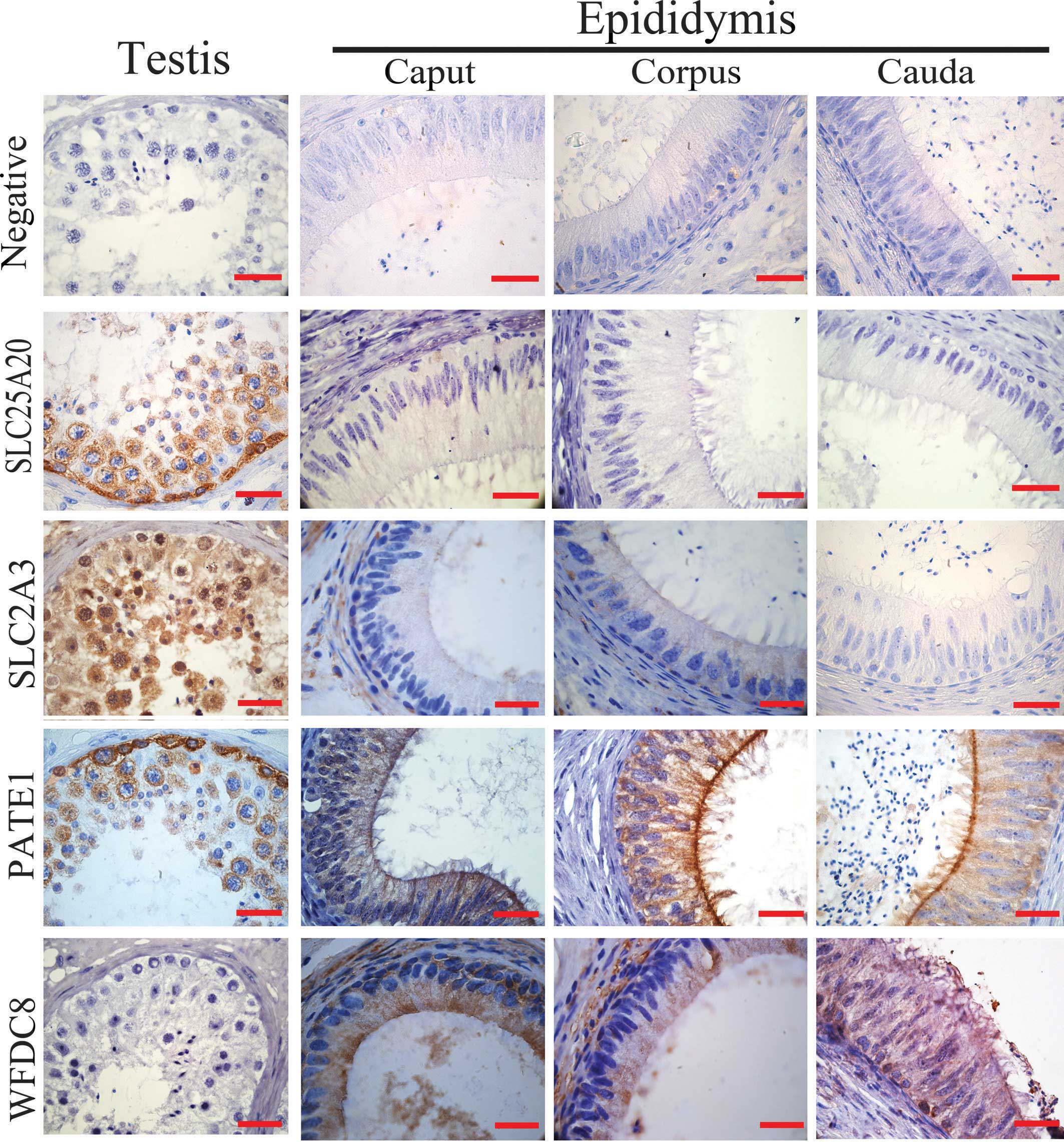
agreement with the results from the western blot analysis, SLC2A3
and SLC25A20 were observed to exhibit strong staining intensity in
the testis with positive staining in the spermatogonium and
spermatocyte, whereas WFDC8 and PATE1 were strongly expressed in
the human epididymis, particularly in the principle cells of corpus
epididymis (Fig. 3).
Discussion
Mammalian testis and epididymis have blood-testis
and blood-epididymis barriers, which create a distinct
microenvironment for spermatogenesis and sperm maturation (15). The fluid environments in the
seminiferous tubules are predominantly produced by the seminiferous
cells, while those in the epididymal lumen are mainly created by
principle cells (4,16). During the progression of sperm
along the testicular and epididymal tubules, they interact with
fluid factors, particularly proteins, in order to complete their
modifications (17). A previous
study mapped the human testicular and epididymal protein profile
and investigated their associations with the sperm proteome by
traditional two-dimensional separation coupled with identification
by MALDI-TOF (10,11). However, the number of identified
proteins by these studies was limited. In order to further
elucidate these interactional associations, data were retrieved
from the HPA in the present study to compare protein expression
levels in the testis and epididymis, and thereby, the identity of
several secretory proteins and their associations with sperm were
determined.
In the present study, a total of 7,595 and 7,573
proteins with immunohistochemistry values in the testis and
epididymis tissues, respectively, were retrieved from the HPA
database. These proteins were used to perform comparative analysis
between the testis and epididymis. The amount of data collected was
markedly greater than the proteomic data currently available
(10,18,19),
and may thus provide an improved understanding of testicular and
epididymal protein profiles. Subsequently, predominantly-expressed
proteins in the testis or epididymis were screened by comparing
differential expression levels between two datasets, and the
results were consistent with the reported transcriptomic elevation
(20). It was hypothesized that
the predominantly expressed proteins in the testis would serve
important roles in spermatogenesis, and those in the epididymis
would have important functions in the maturation of sperm.
Enrichment functional analysis demonstrated that predominantly
expressed proteins in the testis were mainly involved in the
functions of germ cell development and differentiation, and those
in the epididymis mainly functioned in cell adhesion associated
with epithelial cells. The enrichment functions were consistent
with the roles of the corresponding tissues. Thus, in future
studies, further data mining analysis may be performed based on
these data.
Of these proteins, highly expressed secretory
proteins, particularly epididymal secretory proteins, were
investigated due to their direct association with spermatogenesis
and sperm maturation. In the present study, secretory proteins were
further identified by combined retrieval of seminal fluid proteins
and a secretory prediction tool (Locate). These proteins were
suggested to be secreted into the testicular tubules or the
epididymal lumen, thus contributing to the microenvironment (sperm
milieu). Data from the present study suggested that secretory
proteins highly expressed in the testis and epididymis were
predominantly involved in the creation of the testicular and
epidymal microenvironments, respectively. With the exception of
metabolic activity, the secreted proteins were observed to be
predominantly involved in protease/inhibitor, cell adhesion,
defense/immunity and antioxidant functions, which is consistent
with the results of a previous study (21). The proteases and inhibitors were
differentially expressed in testicular and epididymal fluids,
suggesting different roles in spermatogenesis and sperm maturation
(22). Defense/immunity and
antioxidant proteins may serve cooperative functions in sperm
survival (23). Epididymal fluids
were directly involved in sperm maturation, and 251 human
epididymal fluid proteins were identified by 2D-gel separation
combing identification by MALDI-TOF mass spectrometry (11). Secreted proteins in the present
study were identified as potential novel members of the epididymal
sperm milieu. These proteins were involved in various functions
which contribute to a continual and appropriate microenvironment
for sperm maturation, transit and storage.
In conclusion, the present study provided novel
insight into the human testis and epididymis proteomes, and
bioinformatically characterized the predominantly expressed tissue
proteins and secreted proteins. The candidate secreted proteins and
enriched functions require further investigation, which will aid in
a more thorough understanding of the male reproductive proteome,
and additionally facilitate research on sperm maturation.
Acknowledgments
The current study was supported by the National
Natural Science Foundation of China (grant no. 81300533) and the
Shandong Provincial Natural Science Foundation, China (grant no.
ZR2013HQ002).
References
|
1
|
Turner TT: De Graaf’s thread: the human
epididymis. J Androl. 29:237–250. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cornwall GA: New insights into epididymal
biology and function. Hum Reprod Update. 15:213–227. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dacheux JL and Dacheux F: New insights
into epididymal function in relation to sperm maturation.
Reproduction. 147:R27–R42. 2014. View Article : Google Scholar
|
|
4
|
Hinton BT and Palladino MA: Epididymal
epithelium: its contribution to the formation of a luminal fluid
microenvironment. Microsc Res Tech. 30:67–81. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cheng CY, Wong EW, Yan HH and Mruk DD:
Regulation of spermatogenesis in the microenvironment of the
seminiferous epithelium: new insights and advances. Mol Cell
Endocrinol. 315:49–56. 2010. View Article : Google Scholar
|
|
6
|
Mital P, Hinton BT and Dufour JM: The
blood-testis and blood-epididymis barriers are more than just their
tight junctions. Biol Reprod. 84:851–858. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gatti JL, Castella S, Dacheux F, et al:
Post-testicular sperm environment and fertility. Anim Reprod Sci.
82–83:321–339. 2004. View Article : Google Scholar
|
|
8
|
Hinton BT and Cooper TG: The epididymis as
a target for male contraceptive development. Handb Exp Pharmacol.
198:117–137. 2010.PubMed/NCBI
|
|
9
|
Cheng CY and Mruk DD: The blood-testis
barrier and its implications for male contraception. Pharmacol Rev.
64:16–64. 2012. View Article : Google Scholar :
|
|
10
|
Li J, Liu F, Liu X, et al: Mapping of the
human testicular proteome and its relationship with that of the
epididymis and spermatozoa. Mol Cell Proteomics.
10:M110.0046302011. View Article : Google Scholar :
|
|
11
|
Li J, Liu F, Wang H, et al: Systematic
mapping and functional analysis of a family of human epididymal
secretory sperm-located proteins. Mol Cell Proteomics. 9:2517–2528.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Uhlen M, Oksvold P, Fagerberg L, et al:
Towards a knowledge-based Human Protein Atlas. Nat Biotechnol.
28:1248–1250. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rolland AD, Lavigne R, Dauly C, et al:
Identification of genital tract markers in the human seminal plasma
using an integrative genomics approach. Hum Reprod. 28:199–209.
2013. View Article : Google Scholar
|
|
14
|
Amaral A, Castillo J, Ramalho-Santos J and
Oliva R: The combined human sperm proteome: cellular pathways and
implications for basic and clinical science. Hum Reprod Update.
20:40–62. 2014. View Article : Google Scholar
|
|
15
|
Mital P, Hinton BT and Dufour JM: The
blood-testis and blood-edipididymis barriers are more than just
their tight junctions. Biol Reprod. 84:851–858. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rato L, Socorro S, Cavaco JE and Oliveira
PF: Tubular fluid secretion in the seminiferous epithelium: ion
transporters and aquaporins in Sertoli cells. J Membr Biol.
236:215–224. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dacheux JL, Belghazi M, Lanson Y and
Dacheux F: Human epididymal secretome and proteome. Mol Cell
Endocrinol. 250:36–42. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu M, Hu Z, Qi L, et al: Scanning of
novel cancer/testis proteins by human testis proteomic analysis.
Proteomics. 13:1200–1210. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Guo X, Zhang P, Huo R, Zhou Z and Sha J:
Analysis of the human testis proteome by mass spectrometry and
bioinformatics. Proteomics Clin Appl. 2:1651–1657. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li JY, Wang HY, Liu J, et al:
Transcriptome analysis of a cDNA library from adult human
epididymis. DNA Res. 15:115–122. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fu-Jun L and Xiao-Fang S: Comparative
analysis of human reproductive proteomes identifies candidate
proteins of sperm maturation. Mol Biol Rep. 39:10257–10263. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Métayer S, Dacheux F, Dacheux JL and Gatti
JL: Comparison, characterization, and identification of proteases
and protease inhibitors in epididymal fluids of domestic mammals.
Matrix metalloproteinases are major fluid gelatinases. Biol Reprod.
66:1219–1229. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fujii J, Iuchi Y, Matsuki S and Ishii T:
Cooperative function of antioxidant and redox systems against
oxidative stress in male reproductive tissues. Asian J Androl.
5:231–242. 2003.PubMed/NCBI
|