Introduction
MicroRNAs (miRNAs) are a class of small,
endogenously expressed, well-conserved noncoding RNAs with 18–25
nucleotides. These RNA molecules suppress protein expression
predominantly by base pairing with the 3′ untranslated region
(3′UTR) of their target mRNA (1).
miRNAs are considered as powerful post-transcriptional regulators
of various biological processes, including cell proliferation,
migration, differentiation and apoptosis (2–4).
Previous studies have demonstrated that miRNA dysregulation is
closely associated with the development and progression of various
types of human cancer (5,6).
Hepatocellular carcinoma (HCC) is the fifth most
common cancer type worldwide and the third leading cause of
cancer-associated mortality, resulting in ~700,000 mortalities each
year (7,8). Although its mortality was reduced due
to the advancements of liver transplantation and surgical
resection, the long-term prognosis remains unsatisfactory due to
late-stage diagnosis and the high recurrence rate (9). It is widely accepted that
environmental factors and epigengetic/genetic alterations cooperate
in the initiation and progression of HCC (10). miRNA expression profiles have
demonstrated that a subset of miRNAs were aberrantly expressed in
HCC (10,11). Furthermore, several deregulated
miRNAs have been validated to regulate HCC cell proliferation,
migration and apoptosis (12–16).
These observations indicated that the dysregulation of miRNAs may
be implicated in the generation and progression of HCC.
Previous studies have demonstrated that miRNA-218
(miR-218) and miR-520a are associated with tumor pathogenesis and
the development of various types of human cancer. The expression of
miR-218 is downregulated and may serve as a potential tumor
suppressor in glioblastoma, clear cell renal cell carcinoma and
gastric, nasopharyngeal, cervical, breast, oral and non-small cell
lung cancer (17–24). In addition, Li et al
(25) reported that miR-218 is
downregulated in HCC tissues and may inhibit cell proliferation and
promote cell apoptosis. Regarding miR-520a, fewer studies have been
performed; however, it has been reported to inhibit cell
proliferation and invasion by directly targeting ErbB4 in
esophageal squamous cell carcinoma (26). However, the role of miR-218 and
miR-520a in HCC and the molecular mechanisms by which the miRNAs
exert their functions have remained elusive.
In the present study, it was hypothesized that
miR-218 and miR-520a are downregulated in HCC cells compared with
normal hepatic cells. In addition, the restoration of miR-218 and
miR-520a was suggested to inhibit cell proliferation by inducing
cell cycle arrest at the G0/G1 phase
checkpoint. The present study aimed to provide evidence that
miR-218 directly targets E2F2 to regulate its expression in HCC.
Additionally, miR-520a was hypothesized to affect E2F2
expression.
Materials and methods
Cell culture and transfection
The human HCC cell lines HepG2, Huh7, MHCC-97H,
BEL-7402 and the normal hepatic cell line L02 were obtained from
the Type Culture Collection of the Chinese Academy of Sciences
(Shanghai, China). The cells were maintained at 37°C under 5%
CO2 in Dulbecco’s modified Eagle’s medium (Gibco-BRL,
Invitrogen Life Technologies, Carlsbad, CA, USA) supplemented with
10% fetal bovine serum (Gibco-BRL). Mimics of miR-218 and miR-520a
and the negative control (NC) were purchased from Shanghai
GenePharma, Co., Ltd. (Shanghai, China). miRNA transient
transfection was conducted with Lipofectamine 2000 (Invitrogen Life
Technologies) according to the manufacturer’s instructions.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA from cultured cells was extracted using
TRIzol reagent (Invitrogen Life Technologies). In order to measure
miR-218 and miR-520a expression levels, cDNA was synthesized using
the TaqMan miRNA Reverse Transcription kit (Applied Biosystems,
Life Technologies, Thermo Fisher Scientific, Waltham, MA, USA). The
expression levels of miR-218, miR-520a and the endogenous control
U6 were quantified using the TaqMan MicroRNA Assay kit (Applied
Biosystems). To estimate the mRNA levels of E2F2, a total of 500 ng
total RNA was reverse-transcribed using the PrimeScript RT reagent
kit (Takara Biotechnology Co., Ltd., Dalian, China). RT-qPCR was
conducted using the 7500 Real-Time PCR system (Applied Biosystems)
using SYBR Premix Ex Taq (Takara Biotechnology Co., Ltd.) and
β-actin was used as an internal control. The primers used in the
present study were designed and synthesized by GeneCore
BioTechnologies Co., Ltd. (Shanghai, China); the sequences were as
follows: E2F2 forward, 5′-CGT CCC TGA GTT CCC AAC C-3′ and reverse,
5′-GCG AAG TGT CAT ACC GAG TCT T-3′; and β-actin forward, 5′-AGG
CAC CAG GGC GTG AT-3′ and reverse, 5′-TGC TCC CAG TTG GTG ACG
AT-3′. Each sample was run in triplicate.
Western blot analysis
Total proteins were extracted from cells using
radioimmunoprecipitation assay lysis buffer (Sigma-Aldrich, St.
Louis, MO, USA) and quantified by the Bradford assay (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). Equal amounts of protein
were separated using 8% SDS-PAGE (Affymetrix, Inc., Santa Clara,
CA, USA) prior to being transferred to polyvinylidene difluoride
membranes (Bio-Rad Laboratories, Inc.). Following blocking with 5%
skimmed milk, the membranes were incubated with rabbit anti-human
E2F2 polyclonal antibody (1:200; sc-632; Santa Cruz Biotechnology,
Inc., Santa Cruz, CA, USA) overnight at 4°C. Subsequent to washing
with Tris-buffered saline (Affymetrix, Inc.) containing Tween 20
(TBST; Sigma-Aldrich), horseradish peroxidase-conjugated secondary
goat anti-rabbit immunoglobulin G antibodies (1:1,000; Santa Cruz
Biotechnology, Inc.) were incubated with membranes for 1 h at room
temperature. Following washing again using TBST, the protein bands
were detected by chemiluminescence (Amersham ECL Plus Western
Blotting Detection system; GE Healthcare Life Sciences, Pittsburgh,
PA, USA). Digital images were captured using a chemiluminescent
imaging system (FluorChem SP; Alpha Innotech, San Leandro, CA,
USA). β-Actin was used as a protein-loading control. The intensity
of the protein fragments was quantified using Quantity One
software, version 4.5.0 (Bio-Rad Laboratories, Inc.).
Cell proliferation and colony formation
assays
Cell proliferation was determined using MTT assays.
Following transfection with NC, miR-218 or miR-520a, respectively,
for 48 h, Huh7 and MHCC-97H cells were plated in 96-well plates at
a density of 3,000 cells/well. Following the plating of the cells
for 12, 24, 36 and 48 h, 20 μl MTT (Sigma-Aldrich) was added
to each well. Following incubation with MTT for an additional 4 h
at 37°C, the cells were lysed in 150 μl dimethyl sulfoxide
(Sigma-Aldrich) and cell proliferation was evaluated by measuring
the absorbance at 490 nm (SpectraMax M5; Molecular Devices,
Sunnyvale, CA, USA). For the colony formation assays, 400 cells
were plated onto six-well plates and incubated at 37°C until the
cells grew to visible colonies. The colonies were washed with
phosphate-buffered saline (PBS) twice, fixed with methanol
(Sigma-Aldrich) and stained with 0.1% crystal violet
(Sigma-Aldrich), then the numbers of colonies per well were
counted. All assays were performed in triplicate.
Cell cycle analysis
Huh7 and MHCC-97H cells were plated in 60-mm dishes
and transfected with NC, miR-218 or miR-520a, respectively. At 48 h
post-transfection, the cells were harvested and washed with PBS,
then fixed in 70% ethanol overnight at 4°C. Subsequent to washing
in cold PBS three times, the cells were incubated with 100
μl RNase (Nanjing KeyGen Biotech. Co. Ltd., Nanjing, China)
for 30 min at 37°C and were stained with 400 μl propidium
iodide (Nanjing KeyGen Biotech. Co. Ltd.) for an additional 30 min.
Samples were then analyzed for the cell-cycle distribution using a
BD FACSCalibur Cell Analyzer (BD Biosciences, Franklin Lakes, NJ,
USA).
Dual-luciferase reporter assay
Target genes of miR-218 and miR-520a were assessed
using the miRNA target prediction tool miRanda (http://www.microrna.org). To investigate whether E2F2
is a direct downstream target gene of miR-218 and miR-520a,
dual-luciferase reporter assays were performed. The wild-type (WT)
3′-UTR of E2F2 containing the potential binding site of miR-218 or
miR-520a and the corresponding mutational 3′-UTR of E2F2 were
cloned into the psiCheck2 dual luciferase reporter vector (Promega
Corporation, Madison, WI, USA). Huh7 cells were transiently
co-transfected with the reporter vector containing the respective
3′-UTR and either miR-218 mimics, miR-520a mimics or the NC.
Luciferase activity was assayed at 48 h post-transfection using a
Pikkagene Dual Luciferase Reporter Assay system (TOYO B-Net Co.,
Ltd., Tokyo, Japan).
Statistical analysis
All experiments were conducted in triplicate. Values
are expressed as the mean ± standard deviation. Differences between
two groups and in more than two groups were analyzed using
Student’s t-test and one-way analysis of variance, respectively.
Statistical analyses were performed using SPSS version 16.0
software (SPSS Inc., Chicago, IL, USA). All statistical analyses
were two-tailed and P<0.05 was considered to indicate a
statistically significant difference between values.
Results
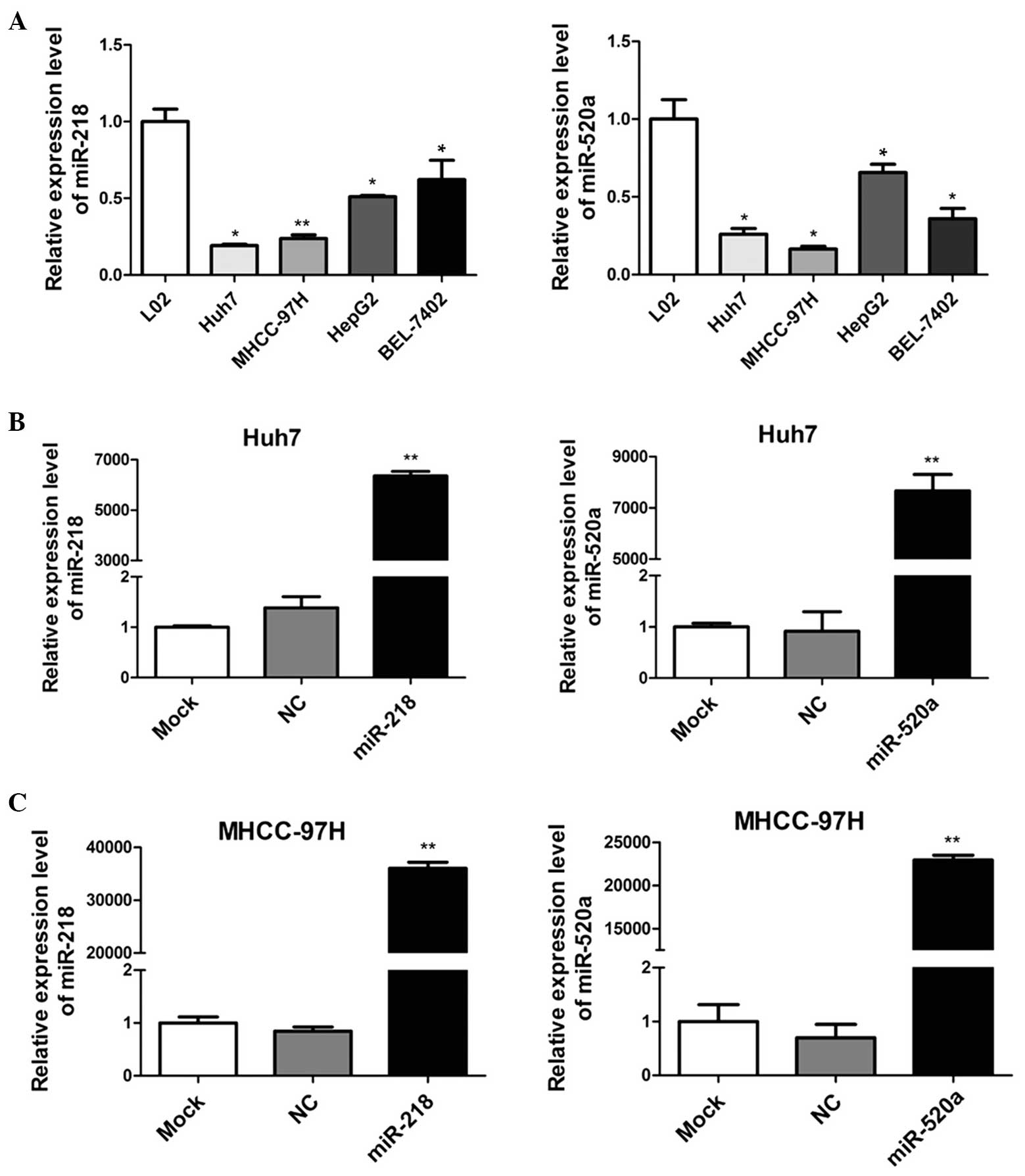
miR-218 and miR-520a expression are
downregulated in human HCC cell lines
To detect the expression of miR-218 and miR-520a in
human HCC cells, RT-qPCR was applied to determine the miRNA
expression in four HCC cell lines (Huh7, MHCC-97H, HepG2 and
BEL-7402), compared with a human normal hepatic cell line (L02). As
demonstrated in Fig. 1A, miR-218
and miR-520a expression levels were significantly downregulated in
human HCC cells. In addition, Huh7 and MHCC-97H cells appeared to
express lower levels of miR-218 and miR-520a than HepG2 and
BEL-7402 cells. These results suggested that miR-218 and miR-520a
may serve as suppressors in the development of HCC.
miR-218 and miR-520a inhibit the
proliferation of HCC cells
To investigate the roles of miR-218 and miR-520a in
HCC, mimics of miR-218 and miR-520a were transfected into Huh7 and
MHCC-97H cells. Fig. 1B
demonstrates that the expression levels of miR-218 and miR-520a
were significantly upregulated (P<0.01) following transfection,
indicating that Huh7 and MHCC-97H cells were effective and
adjustable models for the functional study of miR-218 and miR-520a
expression.
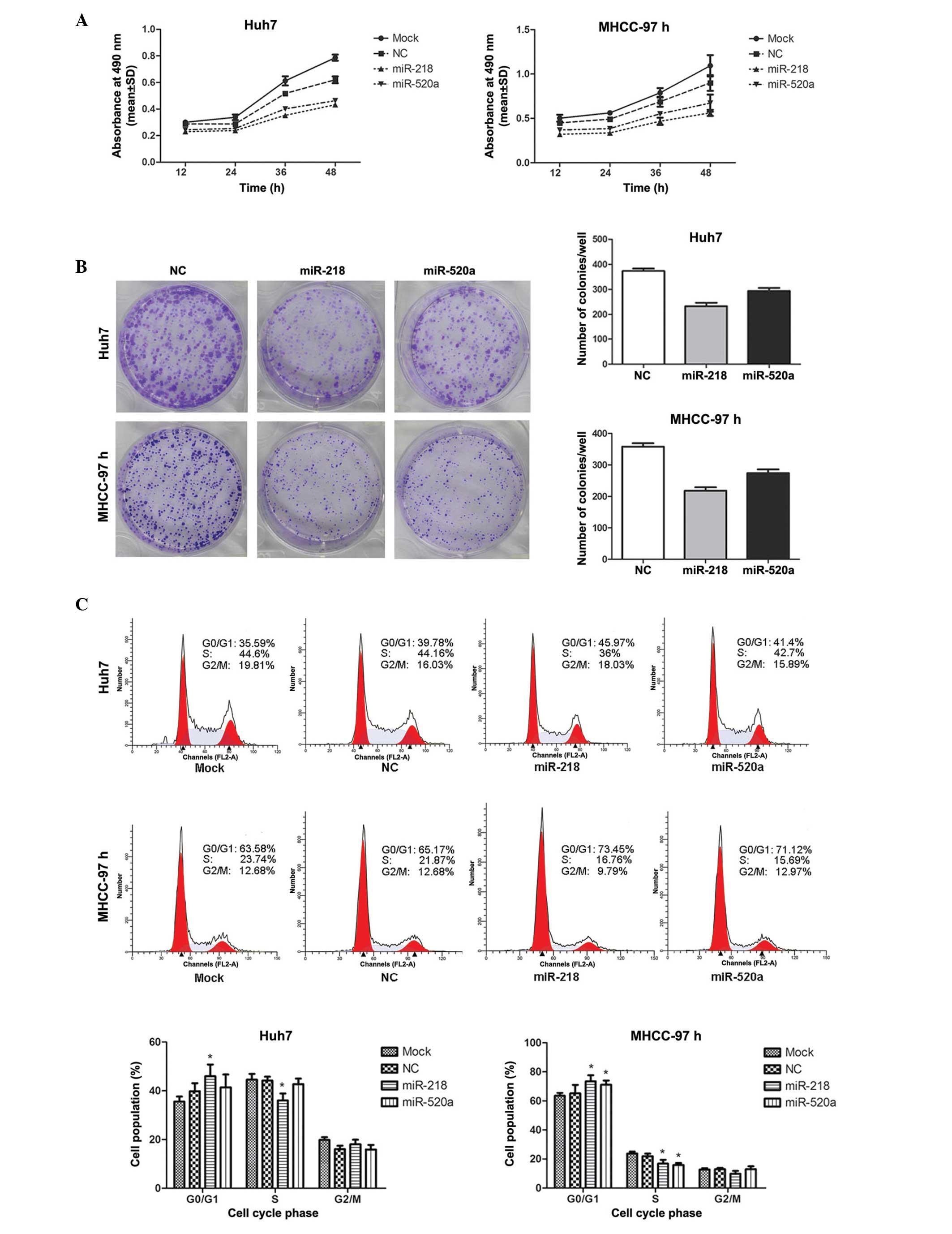
As presented in Fig.
2A, cell viability was measured by MTT assay. MTT growth curves
indicated that the cell proliferative abilities were markedly
reduced when miR-218 and miR-520a were overexpressed (P=0.001 for
miR-218 in Huh7; P= 0.007 for miR-520a in Huh7; P= 0.002 for
miR-218 in MHCC-97; P=0.013 for miR-520a in MHCC-97). Furthermore,
the colony formation assay demonstrated that the colony number in
cells transfected with miR-218 or miR-520a mimics was significantly
lower than that in the NC (P<0.001 for miR-218 in Huh7; P=0.001
for miR-520a in Huh7; P<0.001 for miR-218 in MHCC-97; P=0.001
for miR-520a in MHCC-97), suggesting that the colony-forming
abilities of Huh7 and MHCC-97H cells were suppressed by the
upregulation of miR-218 or miR-520a (Fig. 2B). These results suggested that
miR-218 and miR-520a inhibited the proliferation of the HCC cell
lines Huh7 and MHCC-97H.
miR-218 and miR-520a induce cell cycle
arrest in G0–G1 phase
Flow cytometric analyses demonstrated that the
percentages of miR-218-transfected Huh7 and MHCC-97H cells in
G0-G1 phase were 16% (Huh7) and 13%
(MHCC-97H) greater than those in the NC group, which is consistent
with the reductions of 18% (Huh7) and 23% (MHCC-97H) in the
percentage sof cells in S phase. The percentage of
miR-520a-transfected MHCC-97H cells in G0-G1
phase was 9% greater than that in the NC group, which was in
parallel with a 27% reduction in S phase, whereas the upregulation
of miR-520a exerted no significant effect on the cell cycle
distribution of Huh7 cells (Fig.
2C). These data indicated that miR-218 and miR-520a reduced
cell proliferation via the induction of cell cycle arrest in
G0–G1 phase.
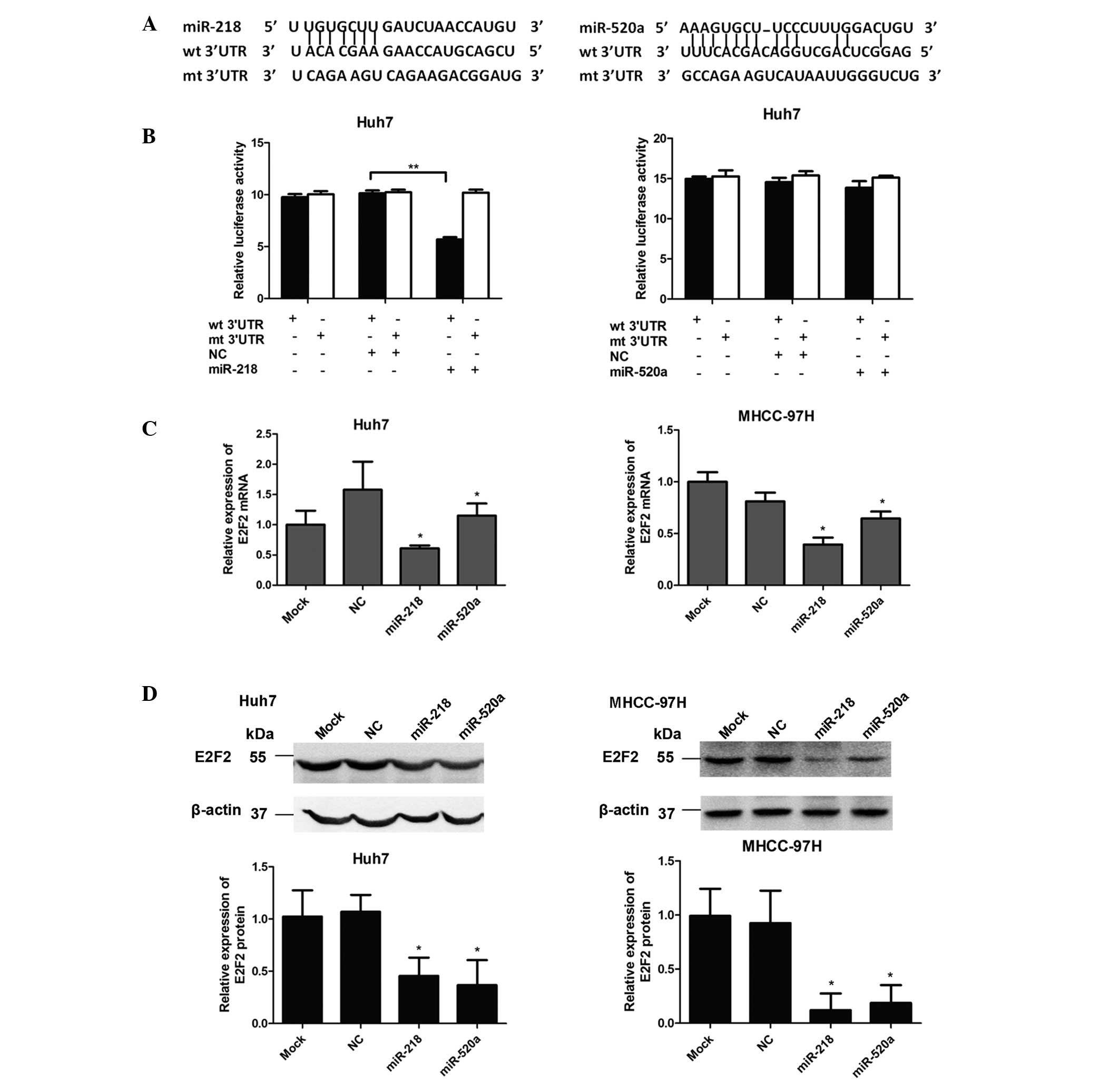
miR-218 directly targets the 3′-UTR of
E2F2
To further investigate the mechanism of miR-218- and
miR-520a-mediated inhibition of cell proliferation, the candidate
target genes were assessed using the miRNA target prediction tool
miRanda (http://www.microrna.org). Among these
genes, including BMI1, MDGA2 and CDK6, E2F2 was predicted as a
potential target of miR-218 and miR-520a.
Dual-luciferase reporter assays were conducted in
Huh7 cells to investigate whether E2F2 was a direct target of
miR-218 and miR-520a. Fig. 3A
presents the potential binding sites of miR-218 and miR-520a in the
3′-UTR of E2F2, with the corresponding sequences of the mutated
3′-UTRs of E2F2 also illustrated. The dual-luciferase reporter
assay demonstrated that the luciferase activity was significantly
reduced (P<0.01) following co-transfection with miR-218 mimics
and the reporter vector bearing the WT 3′-UTR of E2F2. However, no
significant difference in luciferase activity was observed when
miR-520a mimics were co-transfected with the WT 3′-UTR of E2F2. In
addition, the activity in the reporter vector with the mutant
3′-UTR of E2F2 was affected by neither miR-218 nor miR-520a
(Fig. 3B). These results suggested
that E2F2 is a direct target of miR-218 but not miR-520a.
miR-218 and miR-520a suppress E2F2
expression
The effects of miR-218 and miR-520a on E2F2
expression were next investigated. As presented in Fig. 3C, compared with the NC group,
relative mRNA expression of E2F2 was significantly downregulated
(P<0.05) in Huh7 and MHCC-97 cells trans-fected with either
miR-218 or miR-520a mimics. Consistent with the results of RT-qPCR,
western blot analysis indicated that transfection of Huh7 and
MHCC-97 cells with either miR-218 or miR-520a resulted in a
significant reduction in E2F2 protein expression (P<0.05;
Fig. 3D). Collectively, these
results indicated that miR-218 negatively regulated the expression
of E2F2 via directly binding to its 3′-UTR, while miR-520a affected
E2F2 expression indirectly.
Discussion
It is widely accepted that miRNAs regulate diverse
biological processes, including tumorigenesis. The tumor suppressor
miR-218 was reported to be frequently downregulated in various
types of cancer (17–25). A previous study has indicated that
miR-520a acts as a tumor suppressor in esophageal squamous cell
carcinoma (26). In the present
study, the expression of miR-218 and miR-520a in HCC cells and the
molecular mechanisms by which the miRNAs exert their functions were
investigated. First, RT-qPCR was conducted in order to examine the
expression levels of miR-218 and miR-520a in four human HCC cell
lines and a normal hepatic cell line. The results indicated that
the levels of miR-218 and miR-520a were downregulated in cancer
cells, which is consistent with the results of previous studies
(17–26). In addition, Huh7 and MHCC-97H cells
were observed to exhibit low levels of miR-218 and miR-520a
expression amongst the four HCC cell lines examined. In accordance
with this, the Huh7 and MHCC-97H cell lines were selected for the
experiments in the present study. It was hypothesized that miR-218
and miR-520a may serve as tumor suppressors in HCC. The MTT and
colony formation assay demonstrated that HCC cells transfected with
miR-218 or miR-520a mimics exhibited reduced proliferative
abilities compared with those of the control cells. Furthermore,
overexpression of miR-218 or miR-520a was observed to induce cell
cycle arrest at the G0/G1 phase checkpoint.
Therefore, it was inferred that miR-218 and miR-520a function as
tumor suppressors in HCC.
To understand the molecular mechanisms by which
miR-218 and miR-520a inhibit cell proliferation in HCC,
bioinformatics-based predictions and a dual-luciferase reporter
assay were utilized. E2F2 was identified to be a direct target of
miR-218 but not miR-520a in HCC. In addition, RT-qPCR and western
blot analyses were conducted in order to investigate whether
miR-218 and miR-520a may regulate E2F2 expression. The results
demonstrated that upregulation of miR-218 or miR-520a downregulated
E2F2 mRNA and protein levels. Collectively, it was concluded that
miR-218 negatively regulates E2F2 expression via directly binding
to its 3′-UTR, whereas miR-520a affects E2F2 expression
indirectly.
As critical cell cycle regulators, E2F proteins
function downstream of cell cycle signaling cascades and are vital
in cell proliferation and growth via the modulation of genes
involved in cell cycle progression (27–29).
E2F2, a member of the E2F family, activates the transcription of
E2F target genes and regulates the G1/S-phase transition
(28). Previous studies have
demonstrated that E2F2 has a strong oncogenic capacity and is able
to promote cell-cycle progression (30). Additional previous studies have
provided evidence that E2F2 is upregulated in HCC and may be
crucial in the promotion of cell proliferation (31,32).
In normal cell cycle control, the phosphorylation of
cyclin-dependent kinases (CDKs) is essential, which results in
phosphorylation of Rb family proteins. Rb subsequently activates
E2Fs and allows G1/S-phase transition (33). miR-218 has been reported to inhibit
CDK4 expression in colon cancer (34). Based on these factors, it is
hypothesized that miR-218 regulates E2F2 expression; this occurs
not only by directly targeting E2F2 but additionally via the
suppression of CDK4; however, further studies are required to
validate this hypothesis. Regarding miR-520a, even though E2F2 is
not the direct target, the molecular mechanism may be associated
with E2F2.
In conclusion, the present study provided evidence
that miR-218 and miR-520a are downregulated in HCC, and that these
miRNAs act as tumor suppressors to inhibit cell proliferation,
partly by regulating E2F2 expression. This suggested that miR-218
and miR-520a may be promising candidates for therapeutic
intervention in HCC.
Acknowledgments
The authors would like to thank Dr Xiaohuan Chen
(Southern Medical University, Guangzhou, China) for his technical
assistance.
References
|
1
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Baker AH and van Rooij E: miRNA
overexpression induces cardiomyocyte proliferation in vivo. Mol
Ther. 21:497–498. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Heinzelmann J, Henning B, Sanjmyatav J, et
al: Specific miRNA signatures are associated with metastasis and
poor prognosis in clear cell renal cell carcinoma. World J Urol.
29:367–373. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang JF, Fu WM, He ML, et al: MiRNA-20a
promotes osteogenic differentiation of human mesenchymal stem cells
by co-regulating BMP signaling. RNA Biol. 8:829–838. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ventura A and Jacks T: MicroRNAs and
cancer: short RNAs go a long way. Cell. 136:586–591. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bosch FX, Ribes J, Díaz M and Cléries R:
Primary liver cancer: worldwide incidence and trends.
Gastroenterology. 127:S5–S16. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
El-Serag HB: Hepatocellular carcinoma. N
Engl J Med. 365:1118–1127. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Giordano S and Columbano A: MicroRNAs: new
tools for diagnosis, prognosis, and therapy in hepatocellular
carcinoma? Hepatology. 57:840–847. 2013. View Article : Google Scholar
|
|
11
|
Augello C, Vaira V, Caruso L, et al:
MicroRNA profiling of hepatocarcinogenesis identifies C19MC cluster
as a novel prognostic biomarker in hepatocellular carcinoma. Liver
Int. 32:772–782. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Garofalo M, Di Leva G, Romano G, et al:
miR-221&222 regulate TRAIL resistance and enhance
tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell.
16:498–509. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gramantieri L, Ferracin M, Fornari F, et
al: Cyclin G1 is a target of miR-122a, a microRNA frequently
down-regulated in human hepatocellular carcinoma. Cancer Res.
67:6092–6099. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kota J, Chivukula RR, O’Donnell KA, et al:
Therapeutic microRNA delivery suppresses tumorigenesis in a murine
liver cancer model. Cell. 137:1005–1017. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhu Y, Lu Y, Zhang Q, et al:
MicroRNA-26a/b and their host genes cooperate to inhibit the G1/S
transition by activating the pRb protein. Nucleic Acids Res.
40:4615–4625. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Alajez NM, Lenarduzzi M, Ito E, et al:
MiR-218 suppresses nasopharyngeal cancer progression through
downregulation of survivin and the SLIT2-ROBO1 pathway. Cancer Res.
71:2381–2391. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gao CP, Zhang Z, Liu W, Xiao S, Gu W and
Lu H: Reduced microRNA-218 expression is associated with high
nuclear factor kappa B activation in gastric cancer. Cancer.
116:41–49. 2010.
|
|
19
|
Li Q, Zhu F and Chen P: miR-7 and miR-218
epigenetically control tumor suppressor genes RASSF1A and Claudin-6
by targeting HoxB3 in breast cancer. Biochem Biophys Res Commun.
424:28–33. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu Y, Yan W, Zhang W, et al: MiR-218
reverses high invasiveness of glioblastoma cells by targeting the
oncogenic transcription factor LEF1. Oncol Rep. 28:1013–1021.
2012.PubMed/NCBI
|
|
21
|
Martinez I, Gardiner AS, Board KF, Monzon
FA, Edwards RP and Khan SA: Human papillomavirus type 16 reduces
the expression of microRNA-218 in cervical carcinoma cells.
Oncogene. 27:2575–2582. 2008. View Article : Google Scholar :
|
|
22
|
Uesugi A, Kozaki K, Tsuruta T, et al: The
tumor suppressive microRNA miR-218 targets the mTOR component
rictor and inhibits AKT phosphorylation in oral cancer. Cancer
Research. 71:5765–5778. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
White NMA, Bao TT, Grigull J, et al: miRNA
profiling for clear cell renal cell carcinoma: biomarker discovery
and identification of potential controls and consequences of miRNA
dysregulation. J Urol. 186:1077–1083. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu DW, Cheng YW, Wang J, Chen CY and Lee
H: Paxillin predicts survival and relapse in non-small cell lung
cancer by microRNA-218 targeting. Cancer Res. 70:10392–10401. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li C, Tu K, Zheng X, et al: MicroRNA-218
expression and its role in hepatocellular carcinoma. Nan Fang Yi Ke
Da Xue Xue Bao. 33:1127–1131. 2013.In Chinese. PubMed/NCBI
|
|
26
|
Ye W, Yao Q, Zhang M, Wen Q and Wang J:
miR-520a regulates ErbB4 expression and suppresses proliferation
and invasion of esophageal squamous cell carcinoma. Nan Fang Yi Ke
Da Xue Xue Bao. 34:164–168. 2014.In Chinese. PubMed/NCBI
|
|
27
|
Attwooll C, Lazzerini Denchi E and Helin
K: The E2F family: specific functions and overlapping interests.
EMBO J. 23:4709–4716. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dimova DK and Dyson NJ: The E2F
transcriptional network: old acquaintances with new faces.
Oncogene. 24:2810–2826. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ren B, Cam H, Takahashi Y, et al: E2F
integrates cell cycle progression with DNA repair, replication, and
G(2)/M checkpoints. Genes Dev. 16:245–256. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen C and Wells AD: Comparative analysis
of E2F family member oncogenic activity. PLoS ONE. 2:e9122007.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Santoni-Rugiu E, Jensen MR and
Thorgeirsson SS: Disruption of the pRb/E2F pathway and inhibition
of apoptosis are major oncogenic events in liver constitutively
expressing c-myc and transforming growth factor alpha. Cancer Res.
58:123–134. 1998.PubMed/NCBI
|
|
32
|
Zhan L, Huang C, Meng XM, et al: Promising
roles of mammalian E2Fs in hepatocellular carcinoma. Cell Signal.
26:1075–1081. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dong Q, Meng P, Wang T, et al: MicroRNA
let-7a inhibits proliferation of human prostate cancer cells in
vitro and in vivo by targeting E2F2 and CCND2. PLoS ONE.
5:e101472010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
He XQ, Dong YJ, Wu CW, et al: MicroRNA-218
inhibits cell cycle progression and promotes apoptosis in colon
cancer by downregulating BMI1 polycomb ring finger oncogene. Mol
Med. 18:1491–1498. 2012.PubMed/NCBI
|