Introduction
Distal-less homeobox gene 4 (DLX4) is located on
chromosome 17q21-22 and is a member of the DLX family of homeobox
genes (1). Although absent in the
majority of normal adult tissues, DLX4 has been reported to be
widely expressed in leukemia and lung, breast, ovarian and prostate
cancers (2–5). The expression of DLX4 mRNA has been
shown to be significantly increased in tumors with lymph node
metastasis and a high histological grade (6). The expression levels and potential
roles of DLX4 in HCC, however, remain to be determined.
Hepatocellular carcinoma (HCC), has a high incidence of tumor
recurrence and metastasis, and is considered to be a major
worldwide health problem (7). The
oncogenic and tumor suppressive functions of numerous genes have
been characterized; however, the molecular mechanisms of HCC are
complex and remain to be fully elucidated. As a member of the
homeobox family, DLX4 may be involved in HCC progression.
MicroRNAs (miRNAs) are a broad class of small,
non-coding endogenous single RNA molecules that function in gene
expression through directly binding to the 3′-untranslated region
(3′UTR) of the target gene mRNA, resulting in mRNA cleavage or
translational repression (8).
miRNAs are differentially expressed in human cancers and have
essential roles in carcinogenesis, including the development of
HCC. Microarray analyses have started to identify the numbers of
miRNAs that are dysregulated in HCC tissues as compared with normal
tissues, including miR-122. It has been reported that microRNA-122
(miR-122), which accounts for 70% of the total miRNA in the liver,
has a central function in the liver (9). However, there are few reports
investigating the association between miR-122 and DLX4 in HCC.
In the present study, the expression levels of DLX4
in HCC tissues were investigated and compared with those in the
adjacent normal tissues. The association between DLX4 and miR-122
in HCC cancer development was next investigated. The effect of DLX4
knockdown, deletion of the 3′UTR of DLX4 or its inhibition by
miR-122 on DLXR levels, cell proliferation and colony formation
were investigated. This study may provide a novel insight into the
mechanism of miR-122/DLX4 axis in hepatocellular carcinoma.
Materials and methods
Clinical HCC specimens and RNA
isolation
Paired samples of primary HCCs and corresponding
adjacent liver tissues from patients were obtained from the First
Affiliated Hospital of Zhengzhou University (Henan, China) with the
informed consent of the patients. Ethics approval was granted by
the Ethics Committee of Zhengzhou University. Total RNA was
extracted using the TRIzol™ Reagent (Invitrogen Life Technologies,
Carlsbad, CA), according to the manufacturer’s instructions. The
average age of the patients, including four males and three
females, was 55.0±5.6 years. Six patients were identified as
hepatitis B positive. Tumors with moderate differentiation were
identified in five patients and tumors with poor differentiation in
two patients. According to the tumor, nodes and metastasis (TNM)
classification of malignant tumors, there was one patient in stage
I, three in stage II, and three in stage III or IV.
Cell culture and transfection
Hep3B cells (HB-8064; ATCC Manassas, VA, USA) were
cultured in Dulbecco’s modified Eagle’s medium supplemented with
10% fetal bovine serum, 100 U/ml penicillin and 100 μg/ml
streptomycin (Gibco-BRL, Grand Island, NY, USA). Hep3B cells were
incubated at 37°C in a humidified chamber supplemented with 5%
CO2. The miRNA-122 mimic, negative control (NC) and DLX4
siRNA were all purchased from RiboBio (Guangzhou, China). Cells
were plated at 30–50% confluency and transfected with 100 nM
miR-122 mimics or negative control using Lipofectamine™ 2000 in
Opti-MEM (Invitrogen Life Sciences), according to the
manufacturer’s instructions.
Cell growth assay
Cells were seeded in 96-well plates at 8,000 cells
per well and transfected on the following day. MTT assay was used
to determine the relative cell growth at 12, 24, 36 and 48 h after
transfection. A total of 20 μl MTT solution was added to 100
μl culture media and the cells were incubated for a further
4 h at 37°C. The optical density was then measured at 570 nm (OD
570).
Colony formation assay
Following transfection, the cells were seeded into
12-well plates at a density of 200 cells/well and the media was
changed every three days. After ~10 days, most of the cell clones
contained >50 cells. The clones were washed once with phosphate
buffered saline and stained with crystal violet for ~5 min.
Finally, images were taken of the clones, and the colonies were
counted. The colony formation rate = (number of clones)/(number of
seeded cells) x100%.
Quantitative polymerase chain reaction
(qPCR)
The stem-loop qPCR method was used to detect the
miR-122 levels in Hep3B cells. The detection of expression levels
of DLX4 mRNA was performed as previously described (10). The primer sequence for DLX4 was
forward, 5′-CAAAGCTGTCTTCCCAGACC-3′; and reverse,
5′-GTTGTAGGGGACAAGCCAAG-3′. The SYBR® Green Mix Taq™ kit
(Takara, Shiga, Japan) was used to trace the amplified DNA.
Western blot analysis
The Hep3B cells were seeded into six-well plates at
a density of 3×105 cells/well. The cells were
transfected once the density reached ~80% confluency on the second
day. The cells were lysed in radioimmunoprecipitation assay buffer
48 h after transfection, for 30 min at 4°C. The protein
concentration was measured by bicinchoninic acid assay and then 20
μg protein was separated by SDS-PAGE for further analysis.
The primary antibodies used were rabbit polyclonal anti-human DLX4
(1:1,000; Abcam, Cambridge, MA, USA) and rabbit monoclonal
anti-human GAPDH (1:1,000; Abcam). The secondary antibody was goat
anti-rabbit immunoglobulin G conjugated with horseradish peroxidase
(1:1,000; Abcam). The bound antibodies were detected with the use
of Enhanced Chemiluminescence Plus Western Blotting Detection
system (GE Healthcare, Buckinghamshire, UK) and the
chemiluminiscent signals were detected with the use of
high-performance chemiluminescence film (GE Healthcare).
Luciferase reporter assay and vector
construction
The 3′-UTR sequence of DLX4 was predicted by
TargetScan (www.targetscan.org/) to interact with miR-122.
Plasmids containing the DLX4-3′UTR, DLX4-3′UTR with a mutated
sequence of the 3′-UTR sequence and DLX4 open reading frame without
the 3′UTR were constructed with technical support from Guangzhou
Zhiyou Biotech Co. Ltd. (Guangdong, China) and inserted into pGL3
vectors (Promega Corporation, Madison, WI, USA). Following
transfection of miR-122 for 24 h, Hep3B cells were transfected with
pGL3/DLX4-3′UTR and pGL3/DLX4-3′UTR mutant plasmids. After 48 h of
transfection, the luciferase activity of Hep3B cells was measured
using the Dual-Luciferase reporter assay system (Promega
Corporation). Construction of the DLX4 siRNA was designed according
to a previous study (10).
Statistical analysis
All data are presented as the mean ± standard
deviation, from three independent experiments. Statistical analyses
were performed using SPSS 16.0 software (SPSS Inc., Chicago, IL,
USA) and statistical significance between treatment and control
groups was assessed by analysis of variance or the Student’s
t-test. A P<0.05 was considered to indicate a statistically
significant difference.
Results
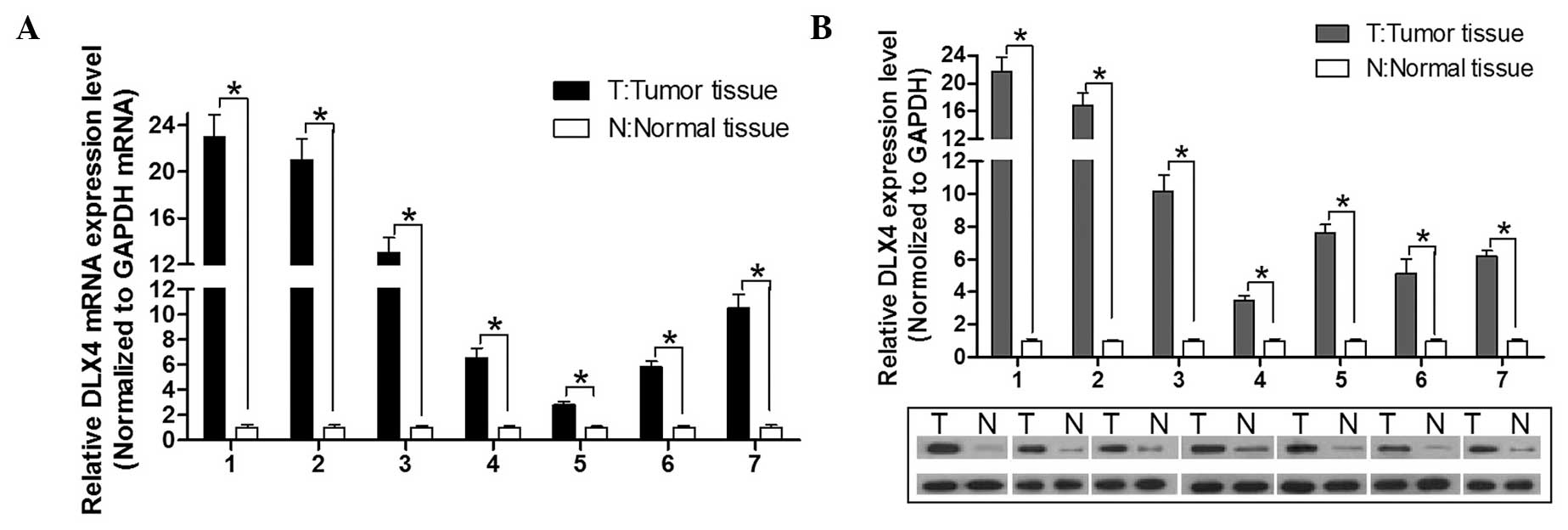
DLX4 is downregulated in HCC tissues as
compared with the adjacent normal tissues
Previous studies have shown that DLX4 is upregulated
in leukemia and lung, breast, ovarian and prostate cancers
(2–5). In the present study, seven paired HCC
tissues were analyzed by qPCR and western blot to detect the
expression status of DLX4. It was identified that both the mRNA and
protein expression of DLX4 was increased as compared with the
adjacent non-tumor tissues (Fig.
1). These data suggested that DLX4 was upregulated in HCC,
implying that DLX4 may act as a tumor oncogene during HCC
development. These data demonstrated that the expression of DLX4 in
HCC was consistent with expression data obtained from other solid
tumors.
DLX4 isoform 1 is targeted by miR-122,
which downregulates its expression
TargetScan was used predict miRNAs that may be
involved in the regulation of DLX4 expression. As shown in Fig. 2A, 14 candidate miRNAs were
identified. Subsequently, a luciferase assay was used to determine
which miRNA was the most effective in regulating DLX4. Hep3B cells
were transfected with a reporter vector along with the 14 miRNA
mimics. The results indicated that miR-122 significantly
downregulated the luciferase intensity of
pGL3/Luciferase-DLX4-3′-UTR, with miR-138 and miR-216b to a lesser
extent (Fig. 2B). Subsequent
studies therefore focused on miR-122, a liver specific and the most
abundant miRNA in the liver. A second luciferase assay was
performed to validate whether the DLX4-3′UTR was the target site
through which miR-122 directly regulates DLX4 expression (Fig. 2C). Hep3B cells were transfected
with the reporter vector along with miR-122 mimics or NC mimics.
The miR-122 mimics significantly decreased the luciferase intensity
of Hep3B cells transfected with the DLX4-3′UTR reporter vector,
whereas it did not affect the luciferase intensity of Hep3B cells
transfected with the DLX4-3′UTR mutated vector (Fig. 2C). The data indicated that DLX4 was
a direct target of miR-122. The effect of miR-122 on the endogenous
expression of DLX4 protein was assayed by western blotting. The
DLX4 protein levels increased 2.95-fold in Hep3B cells transfected
with miR-122 antisense mimics as compared with the control group
(Fig. 2D). The DLX4 protein levels
were decreased by 58% in Hep3B cells transfected with miR-120
mimics as compared with the control group (Fig. 2D). These results indicated that
miR-122 targeted and repressed the expression of DLX4, which may
explain, in part, the upregulation of DLX4 in HCC.
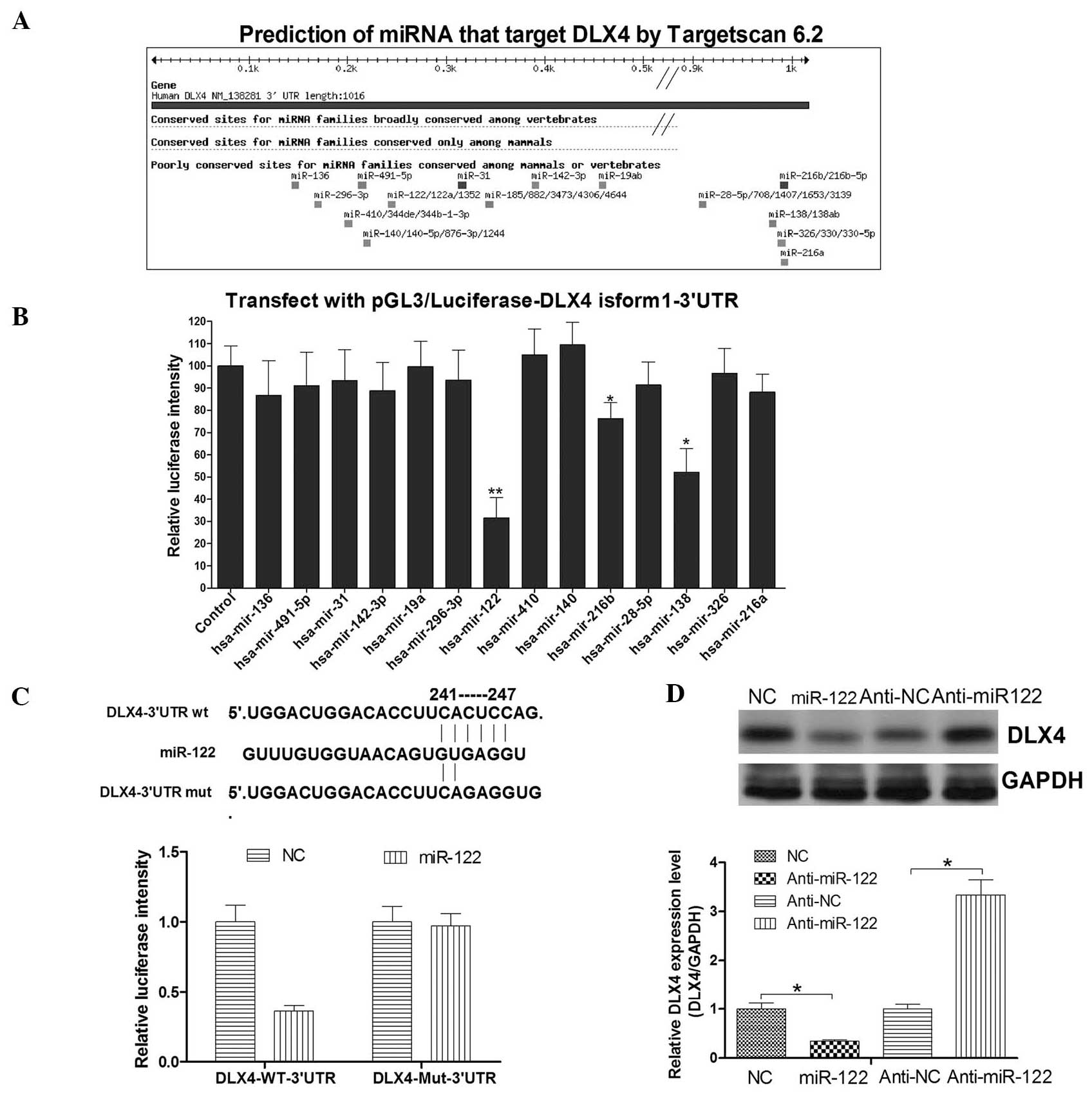 | Figure 2miR-122 directly targets DLX4 and
represses DLX4 in Hep3B cells. (A) A schematic representation
showing the miRNA target prediction algorithms, generated by
Targetscan 6.2, and screen for potential miRNAs that target the
DLX4 mRNA 3′UTR. (B) Hep3B cells were co-transfected with
luciferase constructs expressing the DLX4-3′UTR (pGL3/luciferase
DLX4-3′UTR) and 15 miRNA mimics, including one control mimic.
Luciferase activity was determined 48 h after transfection and
normalized to the control. (C) Luciferase constructs were
transfected into cells transduced with miR-122 mimics and a
negative control. Luciferase activity was determined 48 h after
transfection. The ratio of normalized sensor to control luciferase
activity is shown. (D) miR-122 and antisense miR-122 were
transfected into Hep3B cells, and then the DLX4 protein levels were
measured by western blot analysis. Labwork 4.0 software was used to
quantify the intensity of the DLX4 and GAPDH bands. GAPDH was used
as a loading control. The level of DLX4 in the NC and Anti-NC group
were set as 1.0. All data are expressed as the mean ± standard
deviation of three independent experiments. *P<0.05
as compared with the control group. NC, negative control; WT,
wild-type; Mut, mutant; 3′UTR, 3′-untranslated region; DLX4,
distal-less homeobox 4; Anti, antisense; miR-122, microRNA-122;
hsa, Homo sapiens. |
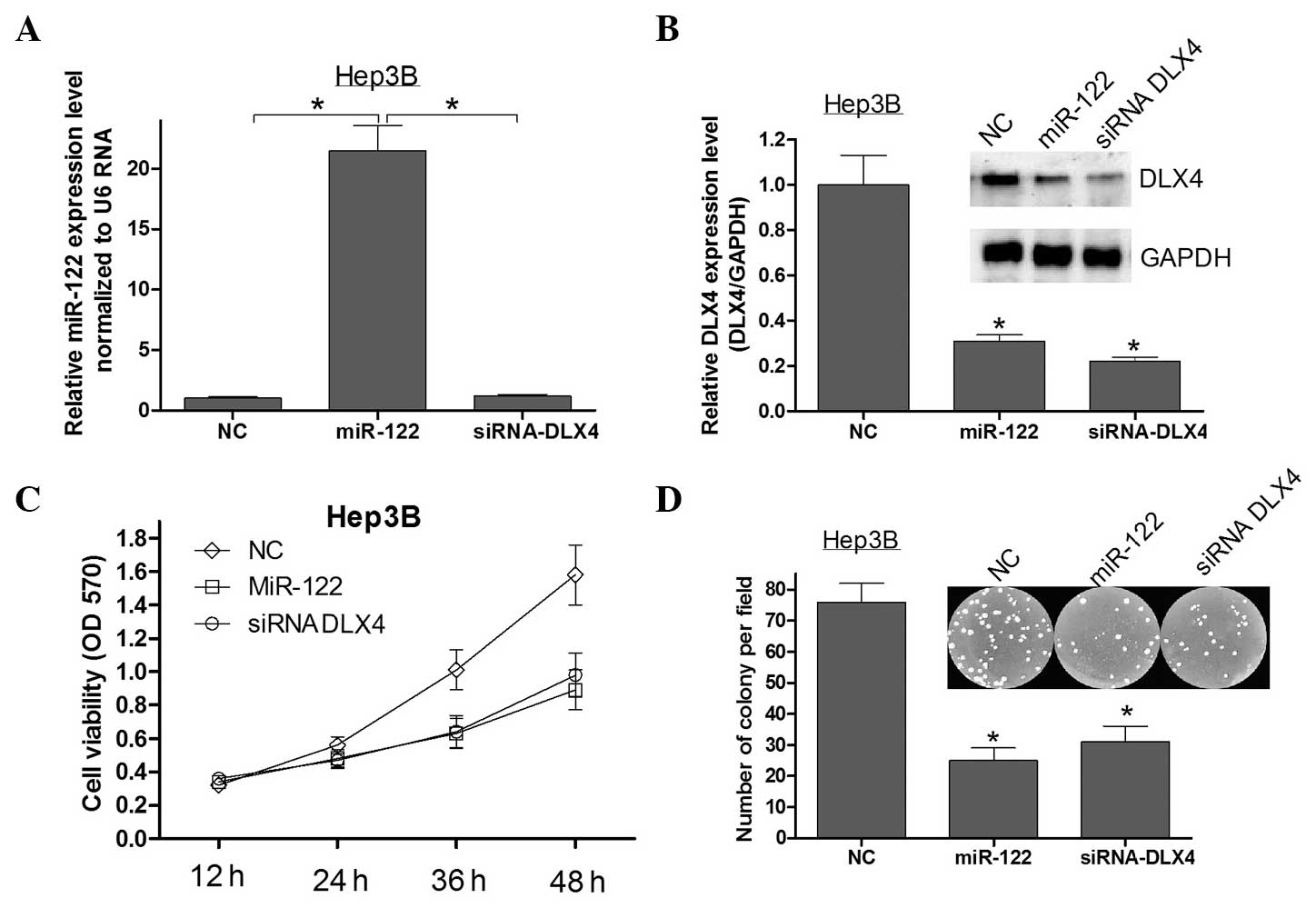
DLX4 is a critical mediator of the
anti-proliferative effects of miR-122
A previous study reported that the overexpression of
miR-122 can inhibit HCC cell growth and promote apoptosis (11). Since it was identified that DLX4
was directly targeted by miR-122, it was further explored whether
DLX4 may be the critical mediator of the role of miR-122 in
cellular proliferation in HCC. Firstly, qPCR and western blot
analysis were used to assess the expression of miR-122 and DLX4 in
Hep3B cells transfected with miR-122 mimics or siRNA DLX4 (Fig. 3A and B). Silencing the expression
of DLX4 using siRNA in Hep3B cells showed that the repression of
DLX4 recapitualted the anti-proliferative effects of miR-122
(Fig. 3C and D). These results
indicated that DLX4 is a critical mediator of the
miR-122-associated anti-proliferative effects in HCC.
Ectopic expression of DLX4 without the
3′UTR counteracts the effects of miR-122 in Hep3B cells
To further confirm that the effects of miR-122 on
Hep3B cell proliferation are in part mediated by DLX4, a vector was
constructed containing the coding sequence of DLX4 without the
3′UTR to avoid miRNA interference. Firstly, the expression of
miR-122 in each Hep3B cell transfection was confirmed (Fig. 4A). Following this, MTT (Fig. 4C), colony formation (Fig. 4D) and western blot assays (Fig. 4B) were used to show that the
ectopic expression of DLX4 alleviated the effects caused by miR-122
in Hep3B cells. These results further confirmed that miR-122
suppressed cell viability and colony formation by downregulating
DLX4 expression in Hep3B cells.
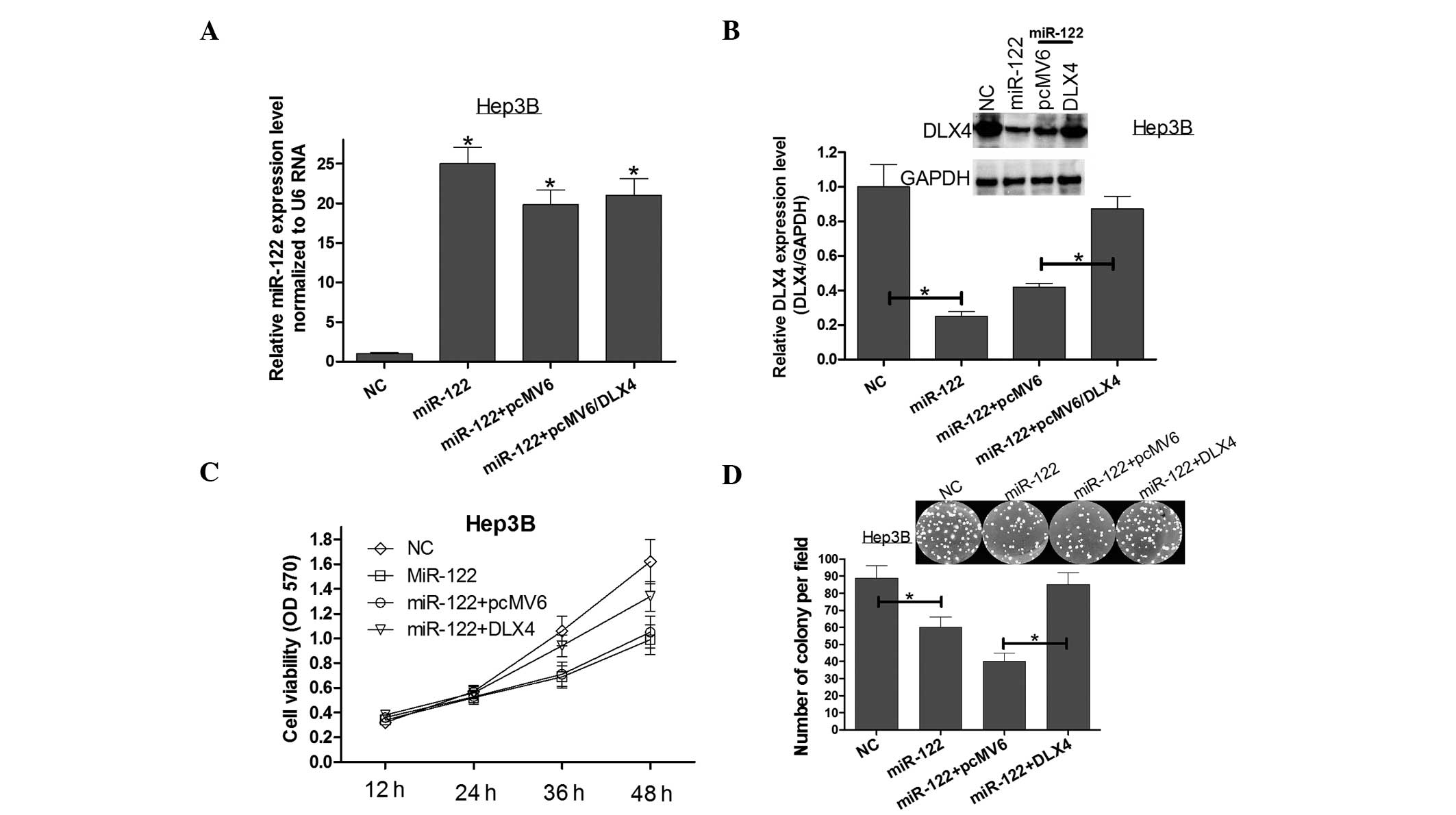 | Figure 4DLX4 alleviates miR-122-induced
cellular phenotypes in Hep3B cells. (A) Hep3B cells were
co-transfected with the pcMV6/DLX4 vector, which did not contain
the 3′-UTR of DLX4, with or without miR-122. Quantitative
polymerase chain reaction was used to validate the expression of
miR-122 in each group. (B) Western blot analysis was used to
validate the expression of DLX4 in each group. (C) Cell viability
was detected by MTT assay at 12, 24, 36 and 48 h after
transfection. (D) A colony formation assay was performed following
transfection. All data are expressed as the mean ± standard
deviation of three independent experiments. *P<0.05
compared with control group. NC, negative control; DLX4,
distal-less homeobox 4; miR-122, microRNA-122; Hep3B, human
hepatocellular carcinoma cells; pcMV6, mammalian expression vector;
3′UTR, 3′-untranslated region. |
Discussion
DLX homeobox genes, originally identified in
Drosophila, are transcription factors that regulate the
transcription of downstream genes. DLX4 belongs to the DLX group of
homeobox genes in humans, and has at least two distinct spliced
variants (12). Loss of function
studies of DLX4 in breast cancer have strongly implicated a role
for this gene in cellular transformation, alterations to the cell
cycle and apoptosis, and progression to a metastatic phenotype
(12). In the present study, it
was demonstrated that DLX4 mRNA and protein were over-expressed in
HCC tissues as compared with the adjacent normal tissues, further
supporting a role in oncogenesis in solid tumors (Fig. 1). Cavalli et al (13) reported that overexpression of DLX4
protein is caused by gene amplification in tumors. However, the
mechanism of DLX4 gene regulation was shown to be complex in
HCC.
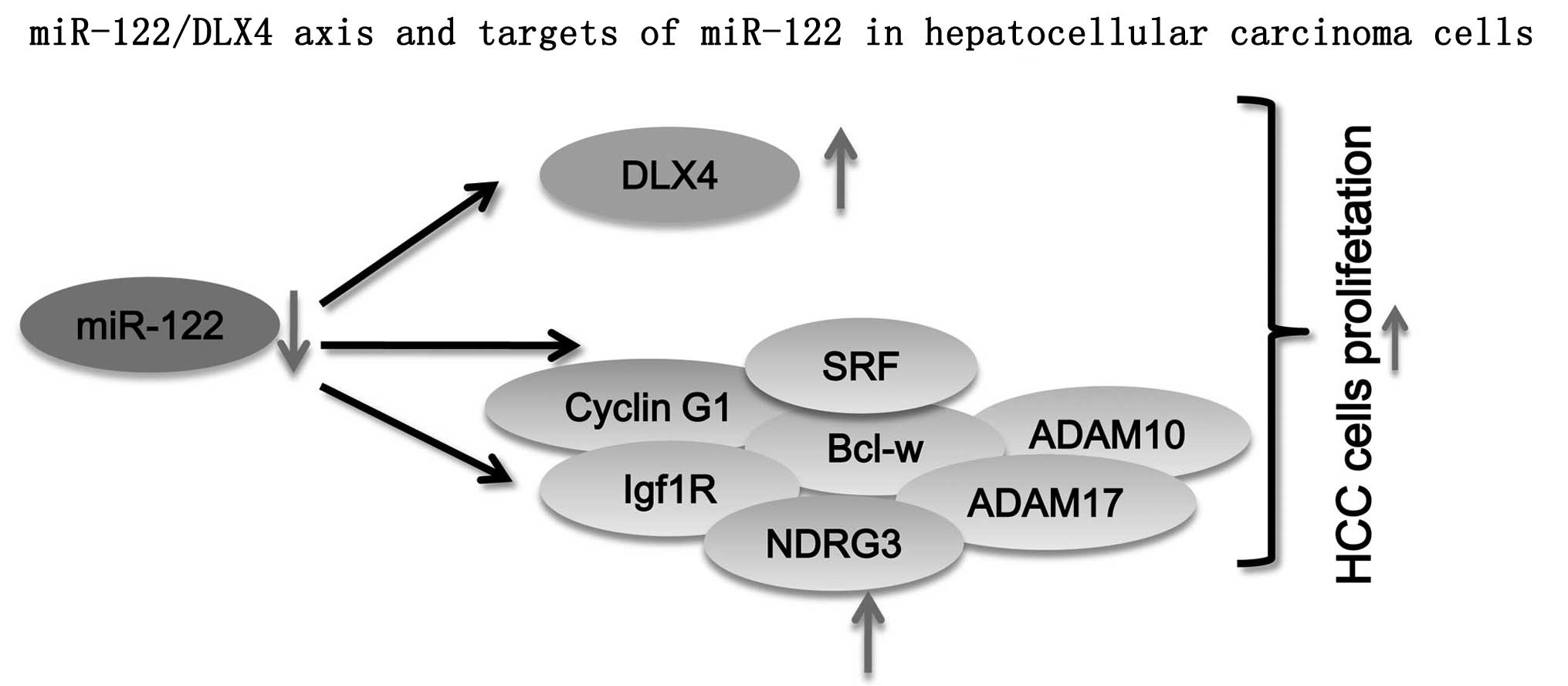
Previous research has estimated that at least 30% of
protein-coding genes in the human genome are regulated by miRNAs,
and that the majority of individual miRNAs target multiple
protein-coding genes (14). MiRNAs
can function as novel types of oncogenes or tumor suppressors and
aberrant regulation of specific miRNAs and their targets is
associated with tumor cell proliferation, apoptosis, angiogenesis,
migration and metastasis (15). In
the present study, a liver specific miRNA, miR-122, was identified
that can directly target DLX4 and regulate its expression in HCC
(Fig. 2). The results presented in
this study, for the first time, to the best of our knowledge,
suggested that miRNA can participate in the regulation of DLX4
expression via mechanisms other than gene amplification (Fig. 5).
MiR-122 is a liver-specific miRNA, and is the most
abundant miRNA in the liver. It acts as a tumor suppressor by
binding to target molecules involved in numerous biological
processes, including cell proliferation, differentiation, apoptosis
and angiogenesis in HCC (16–19).
MiR-122 can modulate cyclin G1 expression in HCC-derived cell
lines, and an inverse correlation between miR-122 and cyclin G1
expression exists in HCCs, indicating that cyclin G1 is a target of
miR-122 (18). MiR-122 also
modulates B-cell lymphoma (Bcl)-w expression by directly targeting
the binding site within the 3′-UTR. The cellular mRNA and protein
levels of Bcl-w were shown to be repressed by elevated levels of
miR-122, which subsequently led to a reduction of cell viability
and activation of caspase-3. This suggested that Bcl-w is a direct
target of miR-122 that functions as an endogenous apoptosis
enhancer in HCC cells (16). Other
miR-122 target genes include SRF, Igf1R, ADAM10, ADAM17 in HCC
(20). In the present study, a new
miR-122/DLX4 axis was identified that inhibited Hep3B cell colony
formation and cell viability. Ectopic expression of DLX4 without
the 3′UTR alleviated the inhibition of colony formation and cell
viability caused by miR-122 in Hep3B cells. Furthermore, DLX4
knockdown inhibited colony formation and cell proliferation in
Hep3B cells. These results were in accordance with the tumor
suppressor function of miR-122. These findings strongly support the
involvement of the signaling pathway of miR-122 and its target
genes in HCC tumorigenesis (Fig.
5).
In conclusion, the present study identified that:
(1) DLX4 was upregulated in HCC
tissues as compared with adjacent normal tissues at both the mRNA
and protein level. (2) A
liver-specific miRNA, miR-122, was identified, which directly
targeted and reduced the expression of DLX4 in HCC Hep3B cells.
This may contribute to the molecular mechanism of deregulation of
DLX4 in HCC. (3) Overexpression of
miR-122 or knockdown of the expression of DLX4 caused a marked
inhibition of proliferation in Hep3B cells. This phenotype could be
rescued by transfection with a DLX4 vector lacking the 3′UTR. These
results provided strong evidence that DLX4 was upregulated in HCC
and functioned as a tumor suppressor. The data supported that DLX4
is regulated by miR-122, which provides novel insight into the
mechanism of the miR-122/DLX4 axis in HCC. Further studies are
required to evaluate the function of DLX4 and the miR-122/DLX4
signaling pathway in other tumors.
References
|
1
|
Haga SB, Fu S, Karp JE, et al: BP1, a new
homeobox gene, is frequently expressed in acute leukemias.
Leukemia. 14:1867–1875. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hara F, Samuel S, Liu J, et al: A homeobox
gene related to Drosophila distal-less promotes ovarian
tumorigenicity by inducing expression of vascular endothelial
growth factor and fibroblast growth factor-2. Am J Pathol.
170:1594–1606. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Man YG, Fu SW, Schwartz A, et al:
Expression of BP1, a novel homeobox gene, correlates with breast
cancer progression and invasion. Breast Cancer Res Treat.
90:241–247. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Man YG, Schwartz A, Levine PH, Teal C and
Berg PE: BP1, a putative signature marker for inflammatory breast
cancer and tumor aggressiveness. Cancer Biomark. 5:9–17.
2009.PubMed/NCBI
|
|
5
|
Schwartz AM, Man YG, Rezaei MK, Simmens SJ
and Berg PE: BP1, a homeoprotein, is significantly expressed in
prostate adenocarcinoma and is concordant with prostatic
intraepithelial neoplasia. Mod Pathol. 22:1–6. 2009. View Article : Google Scholar
|
|
6
|
Yu M, Wan Y and Zou Q: Prognostic
significance of BP1 mRNA expression level in patients with
non-small cell lung cancer. Clin Biochem. 41:824–830. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Parkin DM: The global health burden of
infection-associated cancers in the year 2002. Int J Cancer.
118:3030–3044. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hutvágner G and Zamore PD: A microRNA in a
multiple-turnover RNAi enzyme complex. Science. 297:2056–2060.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tsai WC, Hsu SD, Hsu CS, et al:
MicroRNA-122 plays a critical role in liver homeostasis and
hepatocarcinogenesis. J Clin Invest. 122:2884–2897. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kluk BJ, Fu Y, Formolo TA, et al: BP1, an
isoform of DLX4 homeoprotein, negatively regulates BRCA1 in
sporadic breast cancer. Int J Biol Sci. 6:513–524. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu J, Zhu X, Wu L, et al: MicroRNA-122
suppresses cell proliferation and induces cell apoptosis in
hepatocellular carcinoma by directly targeting Wnt/β-catenin
pathway. Liver Int. 32:752–760. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fu S, Stevenson H, Strovel JW, et al:
Distinct functions of two isoforms of a homeobox gene, BP1 and
DLX7, in the regulation of the beta-globin gene. Gene. 278:131–139.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cavalli LR, Man YG, Schwartz AM, et al:
Amplification of the BP1 homeobox gene in breast cancer. Cancer
Genet Cytogenet. 187:19–24. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lin CJ, Gong HY, Tseng HC, Wang WL and Wu
JL: miR-122 targets an anti-apoptotic gene, Bcl-w, in human
hepatocellular carcinoma cell lines. Biochem Biophys Res Commun.
375:315–320. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ma L, Liu J, Shen J, et al: Expression of
miR-122 mediated by adenoviral vector induces apoptosis and cell
cycle arrest of cancer cells. Cancer Biol Ther. 9:554–561. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gramantieri L, Ferracin M, Fornari F, et
al: Cyclin G1 is a target of miR-122a, a microRNA frequently
down-regulated in human hepatocellular carcinoma. Cancer Res.
67:6092–6099. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu X, Wu S, Tong L, et al: miR-122 affects
the viability and apoptosis of hepatocellular carcinoma cells.
Scand J Gastroenterol. 44:1332–1339. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Saito Y, Suzuki H, Matsuura M, et al:
MicroRNAs in Hepatobiliary and Pancreatic Cancers. Front Genet.
2:662011. View Article : Google Scholar
|