Introduction
Tumor necrosis factor-α (TNF-α) is a pleiotropic
factor, which exhibits antitumor effects, alters endothelial
barrier function, reduces tumor interstitial pressure and mediates
immune responses (1). TNF-α is
associated with a number of cell signalling events, is associated
with cell necrosis and apoptosis, and is important for resistance
to infection and cancer (2,3).
However, due to its potential side effects, including cytotoxicity
to healthy cells as well as cancer cells, for patients with cancer,
it is not seen as a viable antitumor therapy, and its use is
limited to locoregional treatments (4–7).
Matrix metalloproteinases (MMPs) are a class of
endogenous zinc-dependent proteolytic enzymes, which are important
for a number of biological processes (8). MMPs are also involved in cancer cell
invasion and dissemination (9).
MMPs are produced by connective tissue cells, endothelial cells,
macrophages, lymphocytes, thymocytes and tumor cells. Tumor cells
are hypothesized to utilize the extracellular matrix or the basal
lamina degrading capability of these enzymes in order to
metastasize to distant sites. High expression of MMP-1, -2, -7, -9
and -13 in patients with cancer is associated with a poor
prognosis. MMP-12 expression has been shown to increase angiostatin
and decrease vascular endothelial growth factor expression, the
balance of which controls angiogenesis of tumor cells (10,11).
Therefore, MMP activity may be involved in a number of pathological
processes (12,13).
The present study aimed to establish a recombinant
human TNF-α (rhTNF-α) plasmid and to investigate its potential as a
targeted tumor therapy. The pET-42a-c(+) vector was selected as an
expression vector. MMP sequences and a section of the foldon domain
sequence of the T4 bacteriophage fibfitin were inserted into the
receptor binding site in the N-terminal region of mutant hTNF-α.
The foldon domain inhibits the binding of rhTNF-α with TNF
receptors and thus, suppress its bioactivity (14). MMPs present in the tumor cells
hydrolyze fusion proteins, thereby exposing the receptor binding
sites of hTNF-α and consequently inducing tumor cell apoptosis
through the action of rhTNF-α on its receptor.
Materials and methods
Vector, drugs and reagents
The pET-32a-collagen MMP-hTNF-α plasmid was
constructed and stored at −70°C with 20% glycerol. The following
materials were obtained from EMD Millipore, Billerica, MA, USA:
PET-42a-c(+) vector, factor Xa, BugBuster Protein Extraction
Reagent kits and BugBuster GST-Bind Purification kits.
BamHI, SpeI, SacI and T4 DNA ligase were
obtained from Takara Biotechnology Co., Ltd. (Dalian, China).
Isopropyl-β-D-thiogalactoside (IPTG) was obtained from Sheng Gong
Bioscience Technology Limited Company (Qingdao, China). An MTT kit
was purchased from Sigma-Aldrich (St. Louis, MO, USA). The Hoechst
Staining kit was purchased from Beyotime Institute of Biotechnology
(Haimen, China). Anhydrous ethanol, isopropyl alcohol,
hydroxymethanol aminomethane, boric acid, glycerol and ethylene
diamine tetraacetic acid were purchased from Fuyuhuagong (Tianjin,
China).
Primary instruments
Vertical electrophoresis and electrophoresis
apparatus were purchased from Bio-Rad Laboratories (Hercules, CA,
USA). A Thermo Scientific Varioskan Flash was purchased from Thermo
Fisher Scientific (Waltham, MA, USA). A DK-98-II thermostatic water
tank was purchased from Tianjin City Taisite Instrument Co., Ltd.
(Tianjing, China). A microscope and relevant software (TE2000-E;
Nikon Corporation, Tokyo, Japan) were provided by the Guangdong
Medical College (Guangdong, China).
Recombinant plasmid construction
The plasmid pET-42a(+) foldon-MMP-hTNF-α was
constructed by cloning the following sequences into the pET-42a(+)
vector: Foldon sequence, MMP distinguishing sequence and wild
hTNF-α sequence (with 8 amino acids deleted from the N-terminal).
The plasmid pET-42a was used as the template for gene splicing
using overlap extension polymerase chain reaction (PCR). The
following primer was used: Forward:
5′-CGGGATCCTAGGAAGGTGGACAGCAAGACCCATTCACCATCTTTTCGTACGTAGCCCTGGCCATCTCGTGGTGCTTCTGGAATATACCCGGACCCGCGTCCCTCAATACCG-3′
(italics, BamHI endonuclease sites; underlined, a reverse
foldon nucleotide sequence that reversely complements to 16 bp). A
pair of amplification primers was also used. The forward primer
(5′-CGGGATCCTAGGAAGGTG-3′) included the 5′ terminal 18 bp of
the extension primer, which included the restriction enzyme sites
of BamHI, and the reverse (5′-TTCAACTAGTGGTTCTGG-3′)
included restriction enzyme sites of SpeI. The PCR
conditions were as follows: Denaturation at 98°C for 10 sec,
annealing at 55°C for 15 sec and extension at 72°C for 20 sec, for
30 cycles. Subsequently, the pET-42a plasmid and PCR products
containing the foldon sequence were digested using SpeI and
BamHI double enzymes, and connected using T4 DNA ligase. The
recovered PCR product was directionally cloned into the recovered
pET-42a(+) plasmid. Four types of pET-32a-collagen MMP-hTNF-α
plasmids used in a previous study (15) were used as templates for PCR in the
present study, in order to obtain four fragments: MMP-1-hTNF-α,
MMP-2-hTNF-α, MMP-8-hTNF-α and MMP-9-hTNF-α. The four plasmids
shared a common reverse primer, which included the restriction
enzyme sites of SacI (GAGCTC), and is shown, together with
the forward primers, in Table I.
PCR reaction conditions for the double enzyme SacI and
BamHI digestion, and for the DNA connection were the same as
those already described. Positive colonies were screened using
kanamycin (Tiangen Biotech Co., Ltd., Beijing, China), and
identification of recombinants and PCR products was confirmed using
agarose electrophoresis and chain termination method using an ABI
PRISM 310 (Applied Biosystems, Foster City, CA, USA). to analyze
DNA sequences. The recombinant plasmid construction process used in
the present study is summarized in Fig. 1.
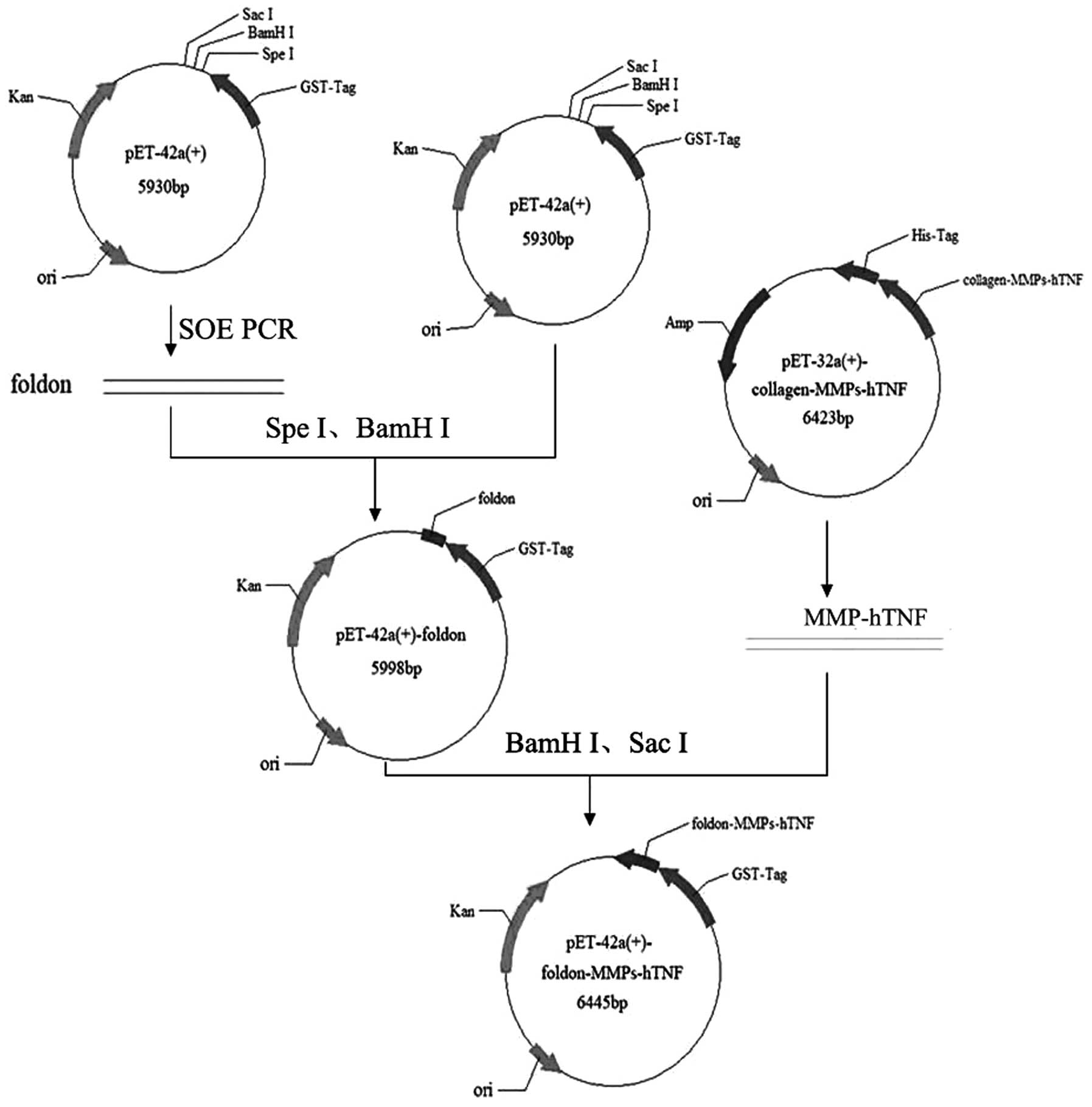 | Figure 1Vector construction of
GST-foldon-MMPs-rhTNF-α. The pET-42a with the foldon signal peptide
was obtained using SOE PCR. The MMP gene was digested with
BamHI and SacI, and ligated with T4 ligase and
eukaryotic expression vector pET-42a-foldon, which was digested
using the same enzymes. rhTNF-α, human recombinant tumor necrosis
factor-α; MMP, matrix metalloproteinase; SOE PCR, splicing by
overlap extension polymerase chain reaction; GST, glutathione
S-transferase; Kan, kanamycin; ori, origin of replication; His-tag,
Histidinetag; Amp, ampicillin. |
 | Table IPrimer sequences used in human
recombinant tumor necrosis factor-α. |
Table I
Primer sequences used in human
recombinant tumor necrosis factor-α.
| Gene | Forward primer
(5′-3′) | Reverse primer
(5′-3′) |
|---|
| MMP-1 |
GCGGATCCCCACTGGCACTGTGGGCACGT
AGTGACAAGCCTGTAGCCCATG |
GCGAGCTCTCCTCACAGGGCAATGATCCC AAAG |
| MMP-2 |
GCGGATCCCCACTGGGTCTGTGGGCACGT
AGTGACAAGCCTGTAGCCCATG |
GCGAGCTCTCCTCACAGGGCAATGATCCC AAAG |
| MMP-8 |
GCGGATCCCCACTGGCATATTGGGCACGT
AGTGACAAGCCTGTAGCCCATG |
GCGAGCTCTCCTCACAGGGCAATGATCCC AAAG |
| MMP-9 |
GCGGATCCCCACTGGGTATGTGGAGCCGT
AGTGACAAGCCTGTAGCCCATG |
GCGAGCTCTCCTCACAGGGCAATGATCCC AAAG |
Expression and purification of the fusion
protein
Four types of rhTNF-α fusion protein plasmids were
inserted into Rosetta 2 (DE3) Escherichia coli (Novagen,
Madison, WI, USA) and selected using kanamycin and chloromycetin
(Tiangen Biotech Co., Ltd.) at appropriate concentrations, in order
to obtain monoclones. Monoclones were added to 4 ml fresh medium LB
and the A600 was measured by UV 2100 spectrophotometry
(Unico, Shanghai, China). The A600 of fresh medium LB
was set as 0, and when the culture reached 0.6, as compared to
fresh LB, IPTG was added to the mixture to obtain a final
concentration of 0.8 mM. Fusion proteins were incubated at 20°C for
12 h, centrifuged at 12,000 × g for 1 min and extracted according
to the manufacturer’s instructions (BugBuster Protein Extraction
Reagent kit; EMD Millipore) and then purified using BugBuster
GST-Bind Purification kit (EMD Millipore). Following affinity
chromatography, proteins were collected into 1.5 ml tubes, and
concentration was determined using the bicinchoninic acid method
(BCA; Thermo Fisher Scientific). Bovine serum albumin (BSA; Thermo
Fisher Scientific) was used as a standard, and concentration
determination was based on the standard curve of BSA. Diluted
thrombin (EMD Millipore) was added to the GST affinity
chromatography column and incubated for 16 h at 20°C. The sepharose
chromatography medium was washed three times with 1X elution buffer
(50 mM Tris-HCl, pH 8.0). The fusion protein was removed from GST
using factor Xa (EMD Millipore) and the target protein was obtained
from the collected elution. Protein concentration was determined
using a BCA method. An uncleaved fusion protein was used as a
control. Fusion proteins were further analyzed using 12% SDS-PAGE.
BandScan 5.0 (Bio-Rad Laboratories) software was used to calculate
the gray values of the protein bands in the gels, and analyze the
results.
Identification of the structure of
rhTNF-α fusion protein
A number of studies have shown that the trimeric
form of TNF-α is required in order to facilitate its biological
activity (16,17). TNF-α consists of high levels of
β-sheets but low levels of α-helix. In its active form, the fusion
protein foldon-MMPs-hTNF-α is a homotrimer. The N-terminal of
foldon may also form trimers independently, promoting the formation
of fusion protein trimer and blocking the hTNF-α receptor site. In
order to confirm the secondary structure of rhTNF-α, the protein
tripolymer was analyzed, using polyacrylamide gel
electrophoresis.
Effects of MMPs on TNF-α activity
The deblocking effect of MMPs 1, 2, 8 and 9 on
rhTNF-α, and the subsequent facilitation of fusion protein activity
and tumor specificity, was investigated. 4-aminophenylmercuric
acetate is involved in the activation of MMP proenzymes.
4-aminophenylmercuric acetate and MMP proenzyme (Novagen) were
mixed at a ratio of 10:1 and incubated for 45 min at 37°C.
Subsequently, the mixtures were prepared, with a ratio of MMP to
protein of 1:20 (µl: µg). They were then incubated
with Tris-HCl (500-mM; pH 8.0) at 37°C for 6 h. The substrate
sequences of MMPs (MMP1, MMP2, MMP8, MMP9) may be digested by
corresponding active MMPs. rhTNF-α products were identified using
12% SDS-PAGE.
Foldon-MMP-1-hTNF-α fusion protein
activity
The bioactivity of purified rhTNF-α was analyzed in
L929 cells using an MTT assay. L929 cells were purchased from the
Type Culture Collection of the Chinese Academy of Sciences
(Shanghai, China). L929 cells are commonly used to study the
cytotoxicity of TNF-α, as TNF-α is not expressed in these cells.
The cells were cultured in RPMI-1640, supplemented with 10% fetal
bovine serum and penicillin-streptomycin (100X; Beyotime Institute
of Biotechnology). The exponentially growing cells were digested
using 0.25% trypsin solution (Life Technologies, Carlsbad, CA, USA)
and the suspended cells were adjusted to 2×105 cell/ml.
Using an RPMI-1640 culture, containing 1 µg/ml of
actinomycin D, cells (100 µl/well) were cultured in a
96-well culture plate and incubated at 37°C with 5% CO2
for 24 h. The following concentrations of fusion protein were used:
1, 10, 100 and 1000 pM. A negative control group was treated with
physiological saline only. Experiments were repeated three times.
Following an incubation period of 24 h at 37°C with 5%
CO2, 10 µl MTT (5 mg/ml) was added to the wells.
The plate was further incubated at 37°C for 4 h. Supernatants were
subsequently aspirated and crystals were dissolved by adding
dimethyl sulfoxide (200 µl) to the wells and incubating for
30 min at 37°C. Absorbance at 570 nm was measured using a Varioskan
Flash 5250030 enzyme-labeled meter (Thermo Fisher Scientific),
following agitation. Cell activity was then calculated and the
dilution at which 50% of cell were inactive, was recorded as the
active unit (U). The value of the median lethal dose was calculated
using SPSS 19.0 (SPSS, Inc., Chicago, IL, USA). Cultures were
incubated for 24 h, subsequently cell morphology was observed using
a fluorescence microscope (TE2000-E; Nikon Corporation).
Hoechst 33258 staining of apoptotic
cells
Asssessment of apoptosis was conducted according to
the methods described in a previous study (18). In the current study, CNE-2Z poorly
differentiated nasopharyngeal carcinoma cells (Department of
Pathology, Affiliated Hospital of Guangdong Medical College) were
used as target cells. They were digested using 0.25% trypsin
solution and the suspended cell concentration was adjusted to
2.5×105 cell/ml. The four treatment groups (identical to
those in the MMT assay), cultured to 50–80 confluence%, were
cultured with the rhTNF-α for 24 h. Results were observed using
fluorescence microscopy.
Statistical analysis
Statistical analysis was conducted using analysis of
variance with SPSS 19.0. Experiments were performed three times.
Data are presented as the mean ± standard deviation. P<0.05 was
considered to indicate a statistically significant difference.
Results
Recombinant plasmid construction
As shown in Fig.
2A, two bands were observed following double SpeI and
BamHI digestion. Sequencing results suggested that the
foldon gene was successfully inserted into the plasmid vector
pET-42a(+), and two bands were observed at 500 and 6000 bp,
following digestion with SacI and BamHI (Fig. 2B). The results of base sequencing
suggested that no base mutation had occurred in the two
fragments.
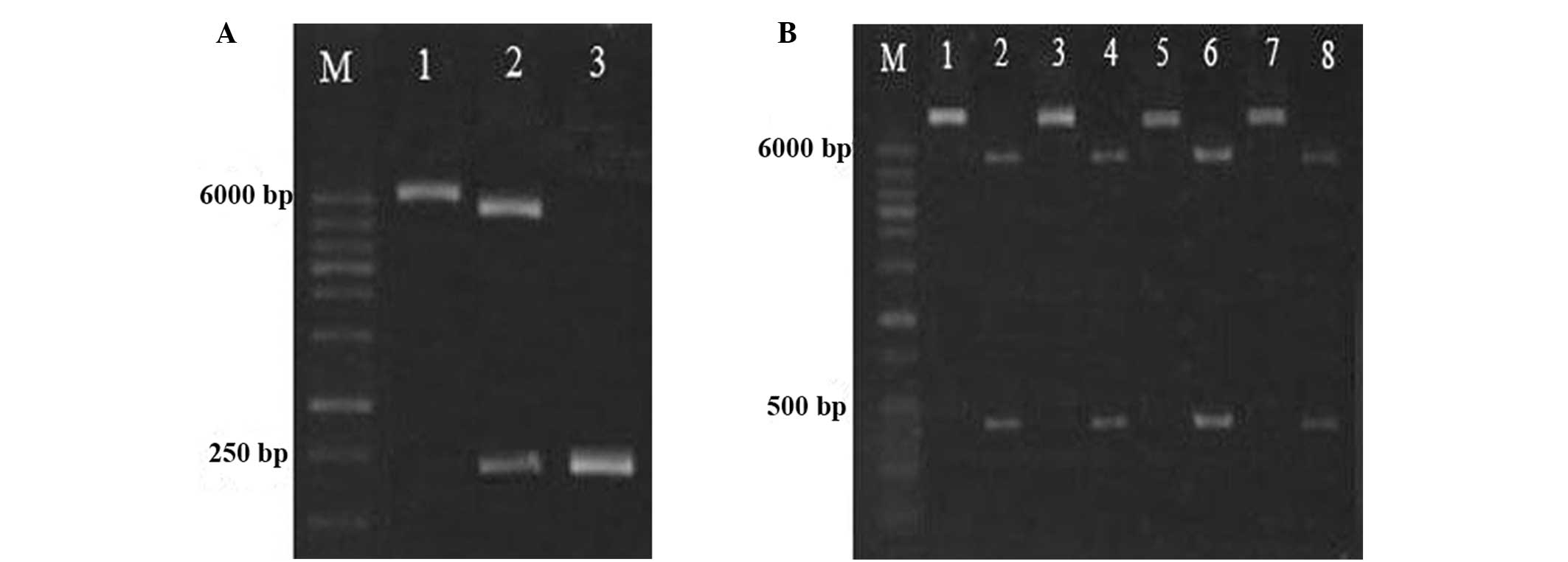 | Figure 2Identification of
GST-foldon-MMPs-hrTNF-α plasmid. (A) Electrophoresis of SpeI
and BamHI digested products and PCR products. Lane 1,
pET-42a(+)-foldon (5,998 bp); lane 2, endonuclease-digested
products (5,950 and 249 bp); lane 3, PCR product (261 bp). (B)
Enzyme-digested products and PCR product of recombinant plasmid.
Lanes 1, 3, 5 and 7 represent recombinant plasmids (5,959 bp), and
lanes 2, 4, 6 and 8 represent enzyme-digested products (484 bp). M,
molecular weight marker (100–6,000 bp); GST, glutathione
S-transferase; rhTNF-α, human recombinant tumor necrosis factor-α;
PCR, polymerase chain reaction. |
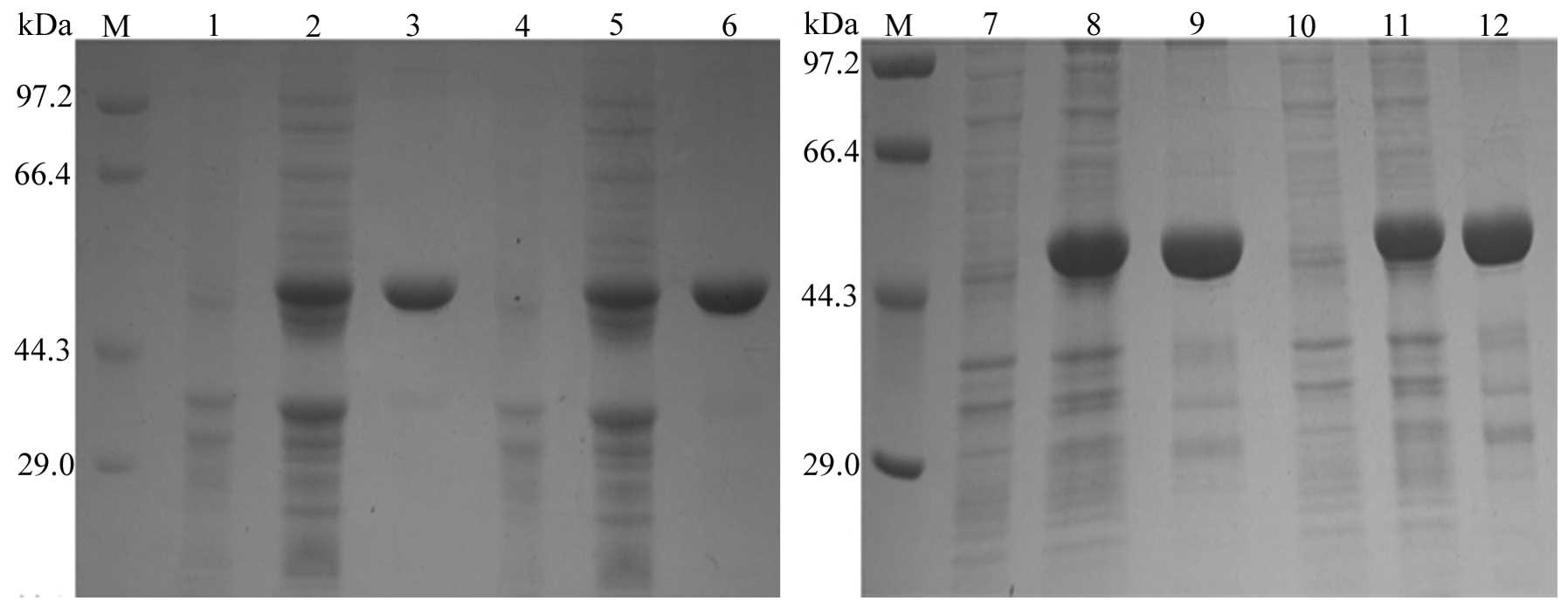
Expression and purification of fusion
proteins
SDS-PAGE analysis suggested that a protein band
>44.3 kDa was amplified, which represents a molecular weight
near to the expected molecular size of 49 kDa. This was in contrast
to the negative control sample, consisting of total bacterial
proteins without the ITPG inducer. According to BandScan software
analysis, the proportions of the four types of purified fusion
proteins in total protein levels were 100, 100, 80.6 and 82.1%,
respectively (Fig. 3).
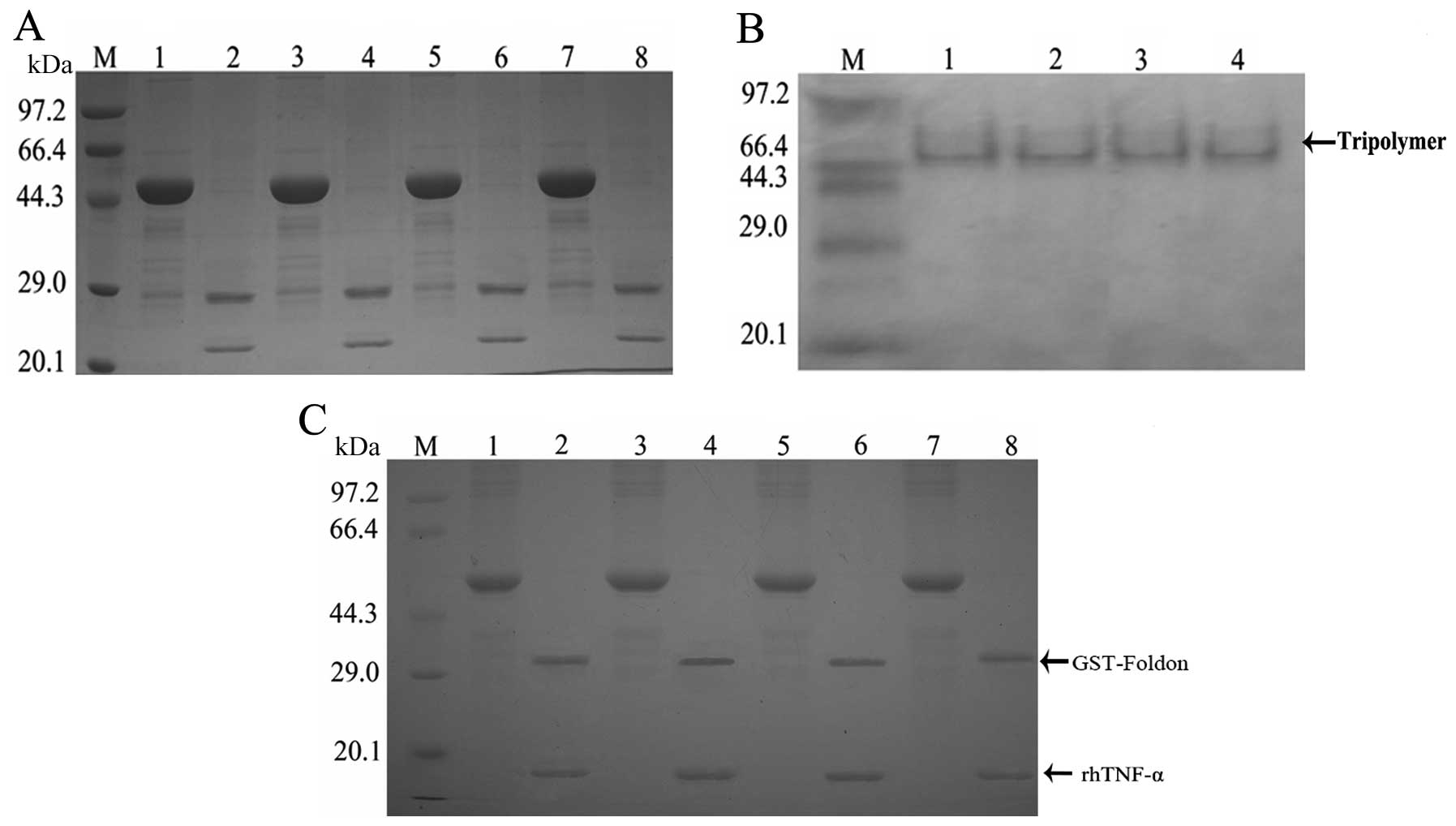
Factor Xa hydrolysis, trimer formation
and MMP influence on the fusion proteins
As predicted the molecular weight of the protein of
interest was ~21 kDa and that of the vector label was ~28 kDa;
there were two protein bands between 20.1 and 29.0 kDa present
following digestion with factor XA (Fig. 4A). The molecular weight of the
trimeric forms of fusion protein was ~63 kDa, and bands appeared at
~66.4 kDa, following gel electrophoresis (Fig. 4B). Two bands at 14.3–20.1 and
29.0–44.3 kDa, were observed in rhTNF-α samples, in the presence of
restriction enzymes. By contrast these bands were not observed in
rhTNF-α samples without restriction enzymes. Furthermore, since the
fusion protein comprises the substrate sequence of MMPs, after MMP
digestion the fusion protein (49 kDa) was cleaved into two parts:
GST-foldon (32 kDa) and rhTNF-α (17 kDa). Therefore, MMP appeared
to successfully deblock the fusion protein (Fig. 4C).
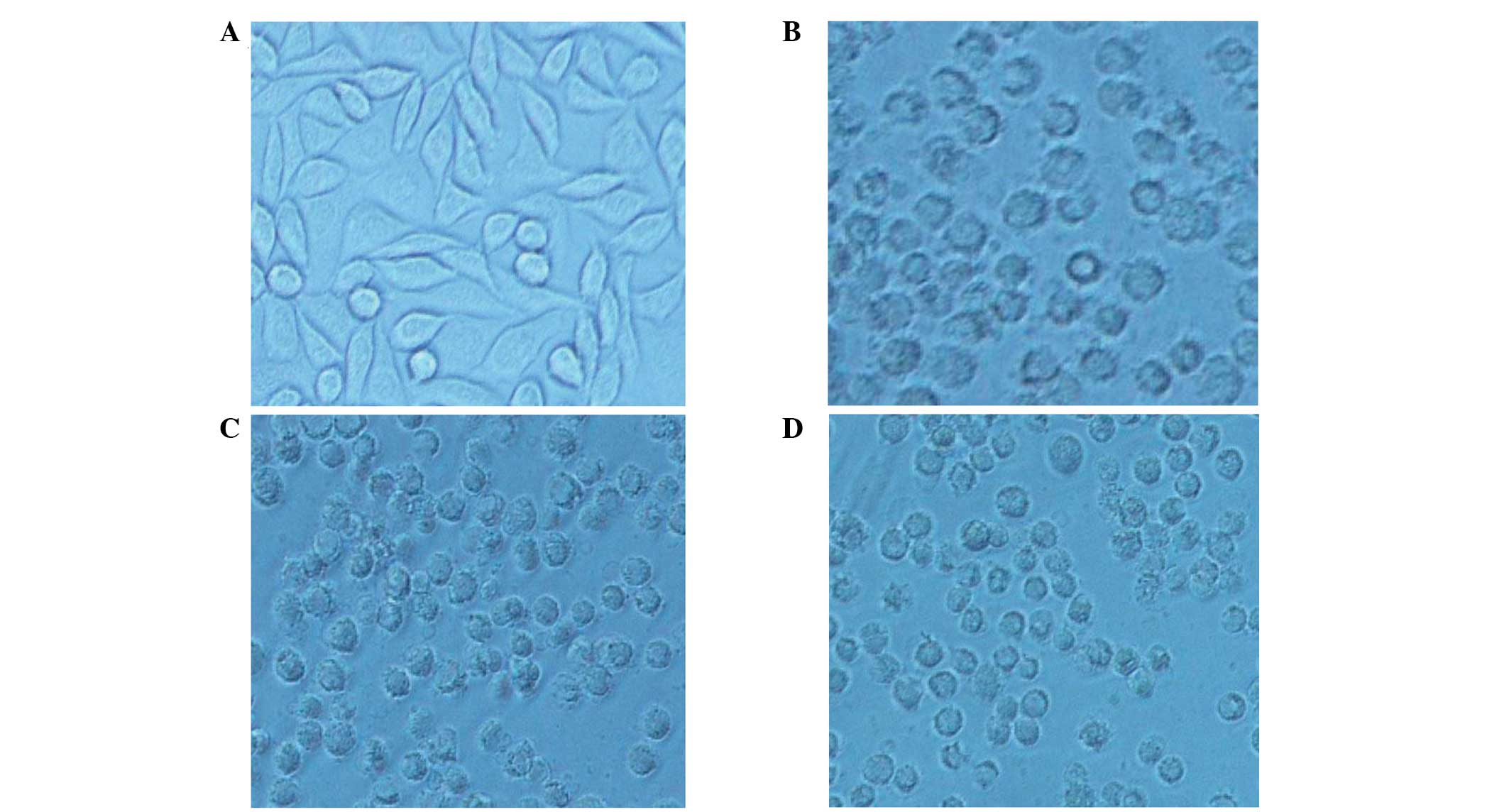
Cytotoxicity assay of rhTNF-α fusion
protein
Compared with the control, the inhibition rate of
MMP-1-hTNF-α was 81.07% in the 1000 pM group (P<0.05) and the
specific activity of the purified MMP-1-hTNF-α was
~6.48×107 U/mg. By contrast, the GST-foldon-MMP-1-hTNF-α
exhibited no significant effects on L929 cells (Table II). Following treatment for 24 h
in the negative control cells, normal cell arrangement was
observed, cells were fusiform in shape, the refractive index was
good and the boundaries of cell membranes were clear. However, the
growth inhibition rates of L929 cells were different in the three
experimental groups (Fig. 5).
 | Table IIResults of MTT assay showing cytotoxic
effects of different proteins on L929 cells. |
Table II
Results of MTT assay showing cytotoxic
effects of different proteins on L929 cells.
| Group | Concentrationa (pM) | A570a | Cytotoxicity
(%)b | LD50 (pM) | Specific
radioactivityc |
|---|
| Negative control | 0 | 0.544±0.04 | | | |
| 1 | 0.435±0.02 | 20.04 | | |
| hTNF-α | 10 | 0.314±0.012 | 42.28 | | |
| 100 | 0.169±0.013 | 68.93 | | |
| 1000 | 0.126±0.011 | 76.84 | 24.95 | 2.35 |
|
GST-foldon-MMP1-rhTNF-α | 1 | 0.543±0.017* | 0.18 | | |
| 10 | 0.401±0.019 | 26.28 | | |
| 100 | 0.338±0.023 | 37.87 | | |
| 1000 | 0.238±0.024 | 56.25 | 401.79 | 0.146 |
| rhTNF-α | 1 | 0.334±0.013 | 38.60 | | |
| 10 | 0.310±0.006 | 43.01 | | |
| 100 | 0.157±0.009 | 71.14 | | |
| 1000 | 0.103±0.008** | 81.07 | 9.045 | 6.48 |
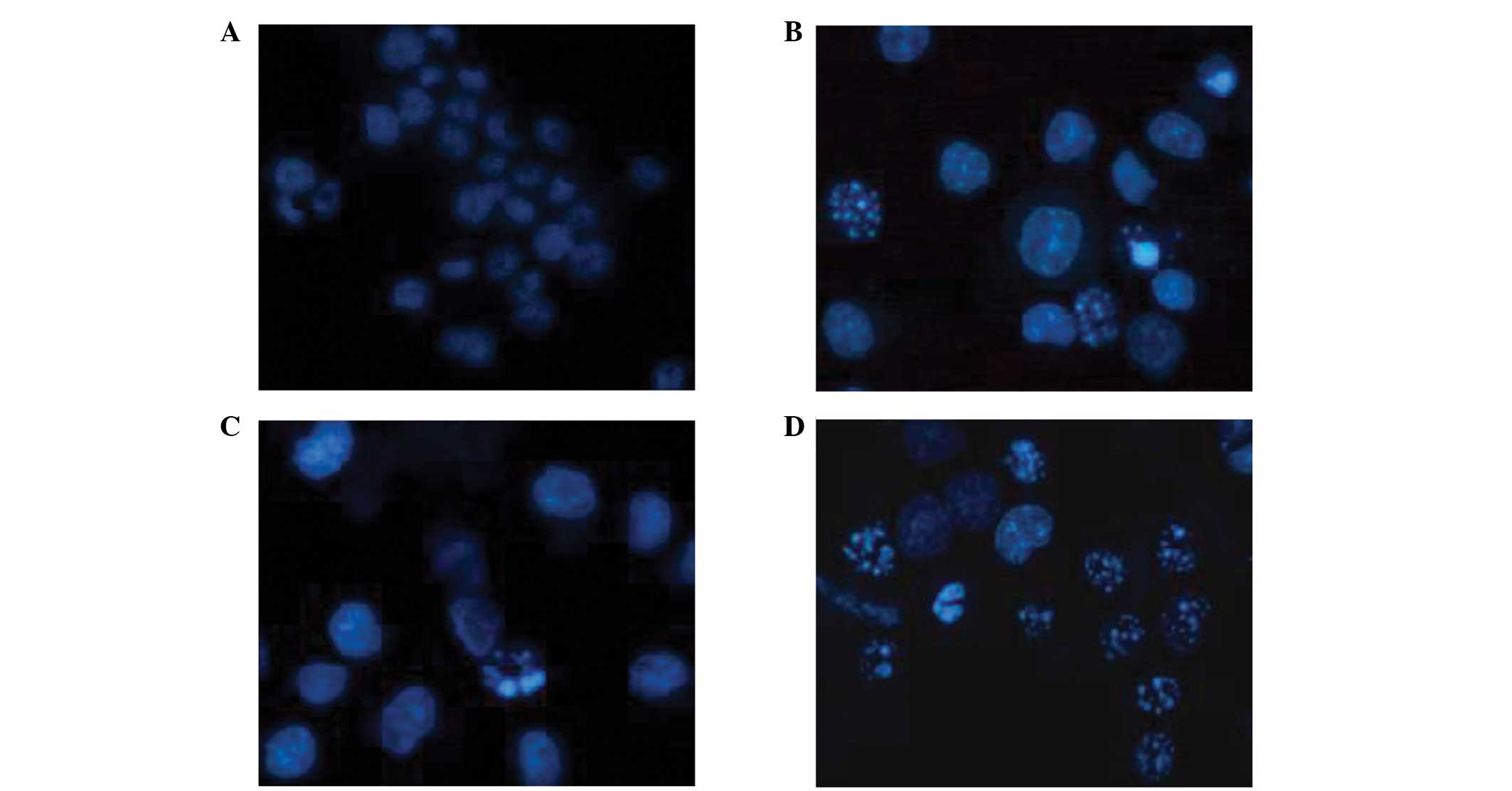
Apoptosis assay
During the terminal phase of cell apoptosis,
chromatin typically condenses and becomes marginalized, the nucleus
breaks down, the nuclear membrane may be dissolved and apoptotic
bodies appear. In the present study, apoptosis was observed in the
rhTNF-α positive control cells (Fig.
6). The influence of rhTNF-α on cell cycle arrest remains to be
investigated.
Discussion
Prodrugs refer to chemical compounds, which are
administered in an inactive form and require metabolic processing
in order to be converted into an active form (19). In tumor cells there anre numerous
types of substances that may be used as prodrug targets, including
overexpressed enzymes, peptides, transport proteins and antigens
(20). TNF-α recombinant fusion
proteins may be constructed using gene engineering technology by
combining certain cytokines or targeting peptides. Prodrugs do not
exhibit bioactivity until the required catalytic reactions occur in
the target tissues. Following MMP hydrolyzation, fusion proteins
are converted to cytotoxic drugs, which result in tumor cell
apoptosis and, therefore, may be an effective antitumor therapy
(21,22).
In the present study, hTNF-α was the active
component, while MMPs were used to specifically target tumor tissue
and form peptide bonds with tumor cells (23). Folden is able to block the TNF-α
receptor binding site, while MMPs hydrolyze the fusion protein and
expose the receptor binding site. The integration of MMPs and
hTNF-α thus allows the hTNF-α fusion protein to retain targeted
antitumor activity, while concomitantly exerting low toxicity on
healthy cells. In the present study, four types of hrTNF-α plasmid
were established using PCR methods. The bioactive form of hrTNF-α
is a non-covalently linked trimer. hrTNF-α monomers consist of two
β-pleated sheets and five antiparallel β-bundles. Extrinsic
β-pleated sheets are enriched with hydrophilic residues, while the
internal β-bundles are hydrophobic. Combined β structures of
hrTNF-α form stable hrTNF-α trimers (24). The molecular weight of fusion
proteins in their trimeric forms is typically ~63 kDa (15). In the present study, bands were
~66.4 kDa according to gel electrophoresis, which is similar to the
known secondary structure of native hrTNF-α (25). This suggests that fusion protein
trimers are formed in vitro.
Furthermore, fusion protein yields may be improved
using affinity tags, such as GST. Certain affinity tags are capable
of promoting fusion protein solubility levels and they may inhibit
the incorporation of fusion proteins into insoluble inclusion
bodies (26,27). GST is a commonly-used tag with
which to express and purify fusion proteins. GST-tags enhance the
ability of fusion proteins to be absorbed into affinity matrices,
containing covalently-bound GSH. The insertion of a GST tag into
the pET-42a(+) vector allowed purification of the extracted fusion
proteins using affinity chromatography. Experiments examining
digestion of the fusion protein using MMP, suggested that MMP
exerted strong and specific activation effects on the fusion
protein.
The present study showed that fusion protein
treatment for 24 h inhibited L929 cell proliferation in a
dose-dependent manner. Theoretically, the rhTNF-α fusion protein
should not exhibit the biological activity of hTNF-α, nor exert
cytotoxic effects on L929 cells. Furthermore, at
~6.48×107 U/mg MMP-1-hTNF-α induced fusion protein
activity. Reduced cytotoxicity was observed following
GST-foldon-MMP-1-hTNF-α treatment, compared with rhTNF-α treatment.
It was hypothesized that L929 cells may produce low levels of MMPs,
which resulted in deblocking of the fusion protein and subsequent
biological activity of hTNF-α biological. A prominent clinical
feature of nasopharyngeal carcinoma is its ability to invade local
tissues and to metastasize (28).
In the present study, rhTNF-α was capable of targeting
nasopharyngeal carcinoma cells, confirming that the fusion protein
activity exhibited antitumor effects in vitro. However, in
order to achieve these effects, the required dose of hTNF-α is
10–50 times the that which the human body is able to tolerate.
Furthermore, tumor cell lines vary in their levels of sensitivity
to TNF-α. In certain cells, TNF-α may promote tumor cell
proliferation. Therefore, further investigation using different
cell lines and dosages is required.
In conclusion, the results of the present study
provide novel insights into MMP-mediated rhTNF-α fusion protein
activity. The fusion protein exhibited tumor-targeting capabilities
and low levels of toxicity in healthy cells. Furthermore, the
present study demonstrated that rhTNF-α is a potential therapeutic
drug for patients with cancer. The development of clinical
applications will facilitate the use of fusion proteins in the
treatment of tumors.
Acknowledgments
The present study was supported by The First Science
and Technology Program of Guangdong province (grant no.
2008B030301023) and the Science and Technology Program of Higher
Learning Institutions in Dongguan (grant nos. 200910815264 and
2012108102016).
References
|
1
|
Locksley RM, Killeen N and Lenardo MJ: The
TNF and TNF receptor superfamilies: Integrating mammalian biology.
Cell. 104:487–501. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Idriss HT and Naismith JH: TNF alpha and
the TNF receptor superfamily: Structure-function relationship(s).
Microsc Res Tech. 50:184–195. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Palladino MA Jr, Patton JS, Figari IS and
Shalaby MR: Possible relationships between in vivo antitumour
activity and toxicity of tumour necrosis factor-alpha. Ciba Found
Symp. 131:21–38. 1987.PubMed/NCBI
|
|
4
|
Asher A, Mulé JJ, Reichert CM, Shiloni E
and Rosenberg SA: Studies on the anti-tumor efficacy of
systemically administered recombinant tumor necrosis factor against
several murine tumors in vivo. J Immunol. 138:963–974.
1987.PubMed/NCBI
|
|
5
|
Rice TW, Wheeler AP, Morris PE, Paz HL,
Russell JA, Edens TR and Bernard GR: Safety and efficacy of
affinity-purified, anti-tumor necrosis factor-alpha, ovine fab for
injection (CytoFab) in severe sepsis. Crit Care Med. 34:2271–2281.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Qiu P, Cui X, Barochia A, Li Y, Natanson C
and Eichacker PQ: The evolving experience with therapeutic TNF
inhibition in sepsis: Considering the potential influence of risk
of death. Expert Opin Investig Drugs. 20:1555–1564. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jiang C, Niu J, Li M, Teng Y, Wang H and
Zhang Y: Tumor vasculature-targeted recombinant mutated human TNF-α
enhanced the antitumor activity of doxorubicin by increasing tumor
vessel permeability in mouse xenograft models. PLoS One.
9:e870362014. View Article : Google Scholar
|
|
8
|
John A and Tuszynski G: The role of matrix
metalloproteinases in tumor angiogenesis and tumor metastasis.
Pathol Oncol Res. 7:14–23. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ryzhakova OS and Solov’eva NI: Matrix
metalloproteinases (MMP) - MMP-1, -2, -9 and its endogenous
activity regulators in transformed by E7 oncogene HPV16 and HPV18
cervical carcinoma cell lines. Biomed Khim. 59:530–540. 2013.In
Russian. View Article : Google Scholar
|
|
10
|
Decock J, Thirkettle S, Wagstaff L and
Edwards DR: Matrix metalloproteinases: Protective roles in cancer.
J Cell Mol Med. 15:1254–1265. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Overall CM and Kleifeld O: Tumour
microenvironment - opinion: Validating matrix metalloproteinases as
drug targets and anti-targets for cancer therapy. Nat Rev Cancer.
6:227–239. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Said AH, Raufman JP and Xie G: The role of
matrix metalloproteinases in colorectal cancer. Cancers (Basel).
6:366–375. 2014. View Article : Google Scholar
|
|
13
|
Spinale FG and Villarreal F: Targeting
matrix metalloproteinases in heart disease: Lessons from endogenous
inhibitors. Biochem Pharmacol. 90:7–15. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Meier S, Güthe S, Kiefhaber T and Grzesiek
S: Foldon, the natural trimerization domain of T4 fibritin,
dissociates into a monomeric A-state form containing a stable
beta-hairpin: Atomic details of trimer dissociation and local
beta-hairpin stability from residual dipolar couplings. J Mol Biol.
344:1051–1069. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao Q, Hou G, Huang D and Chen S:
Construction and expression of hTNF-alpha fusion protein mediated
by MMP1. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi. 28:534–537. 2011.In
Chinese. PubMed/NCBI
|
|
16
|
Soma G, Kitahara N, Tsuji Y, et al:
Improve of cytotoxicity of tumor necrosis factor (TNF) by increase
in basicity of its N-terminal region. Biochem Biophys Res Commun.
148:629–635. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Baeyens KD, De Bondt HL, Raeymaekers A,
Fiers W and De Ranter CJ: The structure of mouse tumournecrosis
factor at 1.4 A resolution: Towards modulation of its selectivity
and trimerization. Acta Crsyrallogr D Biol Crystallogr. 55:772–778.
1999. View Article : Google Scholar
|
|
18
|
Kasibhatla S, Amarante-Mendes GP, Finucane
D, Brunner T, Bossy-Wetzel E and Green DR: Staining of suspension
cells with hoechst 33258 to detect apoptosis. CSH Protoc. 2006:pii:
pdb. prot44922006.
|
|
19
|
Zhang Y, Hong H and Cai W: Tumor-targeted
drug delivery with aptamers. Curr Med Chem. 18:4185–4194. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Graf N and Lippard SJ: Redox activation of
metal-based prodrugs as a strategy for drug delivery. Adv Drug
Deliv Rev. 64:993–1004. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Awad AE, Kandalam V, Chakrabarti S, Wang
X, Penninger JM, Davidge ST, Oudit GY and Kassiri Z: Tumor necrosis
factor induces matrix metalloproteinases in cardiomyocytes and
cardiofibroblasts differentially via superoxide production in a
PI3Kgamma-dependent manner. Am J Physiol Cell Physiol.
298:C679–C692. 2010. View Article : Google Scholar
|
|
22
|
Torbati E, Ghassab RK and Davachi ND:
Recombinant HCV core protein and the secretion of associated
cytokines (IL-6, TNF-α and IFN-γ) in immunized mice. Pak J Biol
Sci. 16:2041–2045. 2013. View Article : Google Scholar
|
|
23
|
Roy R, Yang J and Moses MA: Matrix
metalloproteinases as novel biomarkers and potential therapeutic
targets in human cancer. J Clin Oncol. 27:5287–5297. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Douni E, Rinotas V, Makrinou E, et al: A
RANKL G278R mutation causing osteopetrosis identifies a functional
amino acid essential for trimer assembly in RANKL and TNF. Hum Mol
Genet. 21:784–798. 2012. View Article : Google Scholar
|
|
25
|
Zhang C, Liu Y, Zhao D, Li X, Yu R and Su
Z: Facile purification of Escherichia coli expressed tag-free
recombinant human tumor necrosis factor alpha from supernatant.
Protein Expr Purif. 95:195–203. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Waugh DS: Making the most of affinity
tags. Trends Biotechnol. 23:316–320. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Vinckier NK, Chworos A and Parsons SM:
Improved isolation of proteins tagged with glutathione
S-transferase. Protein Expr Purif. 75:161–164. 2011. View Article : Google Scholar
|
|
28
|
Ben Nasr H, Chahed K, Remadi S, Zakhama A
and Chouchane L: Expression and clinical significance of latent
membrane protein-1, matrix metalloproteinase-1 and Ets-1
transcription factor in tunisian nasopharyngeal carcinoma patients.
Arch Med Res. 40:196–203. 2009. View Article : Google Scholar : PubMed/NCBI
|