Introduction
Glioma arises from glial cells and accounts for 80%
of all malignant brain tumors (1).
Although extensive investigations have been performed in order to
understand the mechanisms underlying its pathogenesis, the
molecular pathways involved in the initiation and progression of
glioma remain unclear (1–3).
MicroRNAs (miRs), a class of non-coding RNA
molecules, are involved in the transcriptional and
post-transcriptional regulation of gene expression (4,5).
Recent investigations have demonstrated that glioma growth is
controlled by a number of miRs (6,7).
miR-16 inhibits glioma cell growth and invasion by suppressing the
BCL2 and nuclear factor-κB1/MMP9 signaling pathway (8). Other investigations have demonstrated
that miR-155 overexpression in patients with glioma is an indicator
of poor prognosis (9). Therefore,
an investigation into the dysregulated miRs that are involved in
the development of glioma may help to elucidate the prognostic
value and therapeutic potential of miRs in patients with this
disease.
Previous investigations have indicated that miR-320
is involved in the development of a number of types of tumors. For
example, miR-320 inhibits cell proliferation and induces apoptosis
in breast, prostate and hepatocellular carcinoma (10–13).
However, until now, the molecular mechanisms underlying the
involvement of miR-320 in gliomagenesis remain poorly
understood.
Materials and methods
Cell culture and tissue samples
U251 and SHG-44 glioma cell lines were obtained from
American Type Culture Collection (Rockville, MD, USA). Cells were
cultured in Dulbecco’s Modified Eagle’s Medium (DMEM;
Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% fetal
bovine serum. Cultures were maintained at 37°C in a humidified
atmosphere with 5% CO2. Tumor tissues and adjacent
healthy tissues were collected during elective therapeutic surgery
(tumor resection) at the Department of Neurosurgery, Tongji
Hospital (Shanghai, China) between January and March 2014. All
samples were obtained with informed consent and with the approval
of the institutional review board of Tongji Hospital.
Analysis of miR expression using
TaqMan® reverse transcription-quantitative polymerase
chain reaction (RT-qPCR)
Total RNA from tissue samples and cell lines was
harvested using the miR Isolation kit (Ambion Life Technologies,
Carlsbad, CA, USA). Expression of mature miRs was assayed using a
TaqMan MiR Assay (Applied Biosystems, Beijing, China) specific for
miR-320. Total RNA (5 ng) was reverse transcribed to cDNA using
specific stem-loop RT primers (5′-AAA AGC TGG GTT GAG AGG GCGA-3′
and a reverse complement primer; Ambion Life Technologies). qPCR
was performed using Applied Biosystems 7900 Real-Time PCR System
(Life Technologies, Shanghai, China) and a TaqMan Universal PCR
Master Mix (Life Technologies). Primers were obtained using TaqMan
miR Assays. The primer sequences were as follows: Cyclin B1,
forward 5′-AAT AAG GCG AAG ATC AAC ATGGC-3′ and reverse 5′-TTT GTT
ACC AAT GTC CCC AAGAG-3′; Cyclin E, forward 5′-AAG GAG CGG GAC ACC
ATGA-3′ and reverse 5′-ACG GTC ACG TTT GCC TTCC-3′. Small nuclear
U6 RNA (snRNA; Applied Biosystems) was used as an internal control.
A total of 50 ng/ml cDNA was used in the PCR. PCR conditions
included an initial holding period at 94°C for 3 min, followed by a
two-step PCR program consisting of 95°C for 5 sec and 60°C for 30
sec, for 45 cycles. Relative quantification analysis of gene
expression data was performed according to the 2-ΔΔCt method.
Cell transfection
miR-320 mimics and negative controls (NCs) were
obtained from Ambion Life Technologies. In order to inhibit miR-320
expression, an antisense oligonucleotide against miR-320 (Ambion
Life Technologies) was used. Lipofectamine 2000®
(Invitrogen Life Technologies, Carlsbad, CA, USA) and 25 nM of the
miRs were prepared according to the manufacturer’s instructions.
Once the cells had reached 70–80% confluence they were transfected
(2.5×104 cells/well) with the mimics, antisense
oligonucleotides or NC using Lipofectamine 2000®,
according to the manufacturer’s instructions for 24 or 48 h. The
same NC was used for the mimic and antisense experiments.
Bromodeoxyuridine (BrdU) assays
A cell proliferation enzyme-linked immunosorbent
assay (BrdU kit; Beyotime Biotechnology, Nantong, China) was used
in order to assess the incorporation of BrdU during DNA synthesis,
according to the manufacturer’s instructions. Experiments were
repeated three times. Absorbance was measured at 450 nm using a
Spectra Max 190 ELISA reader (Molecular Devices, Sunnyvale, CA,
USA)
Cell migration assays
Cell migration capability was analyzed using
Transwell® migration chambers (Millipore, Hayward, CA,
USA) and a EL×800 absorbance reader (BioTek, Winooski, VT, USA) was
used to quantify cell migration at 570 nm, according to the
manufacturer’s instructions.
Western blotting
Cells or tissues were harvested and lysed with
ice-cold lysis buffer (50 mM Tris-HCl, pH 6.8; 100 mM 2-ME; 2% w/v
SDS and 10% glycerol; Beyotime Biotechnology). Following
centrifugation at 20,000 × g for 10 min at 4°C, proteins in the
supernatants were quantified, separated using 10% SDS PAGE and
transferred to a nitrocellulose membrane (Amersham Bioscience,
Amersham, UK). Non-fat milk (10%) in phosphate-buffered saline was
used to block the membranes. Subsequently the membranes were
immunoblotted using the following primary antibodies: Rabbit
polyclonal anti-E2F1 (1:1,000; cat. no. 3742; Cell Signaling
Technology, Inc., Beverley, MA, USA) and mouse monoclonal
anti-GAPDH (1:10,000; cat. no. sc-365062; Santa Cruz Biotechnology,
Inc., Dallas, TX, USA) at 4°C for 12 h, followed by
horseradish-linked secondary antibodies (1:2,000; cat. nos. 7054
and 7056; Cell Signaling Technology, Inc.) at 25°C for 2 hr.
Protein expression was detected using a SuperSignal West Pico
Chemiluminescent Substrate kit (Pierce, Rockford, IL, USA)
according to the manufacturer’s instructions. Anti-E2F1 antibodies
were purchased from Cell Signaling Technology, Inc.). Protein
expression levels were standardized against GAPDH, using the mouse
anti-GAPDH antibody.
Luciferase reporter assay
The potential targets of miR-320 were analyzed using
miRWalk software (http://www.umm.
uni-heidelberg.de/apps/zmf/mirwalk/) (14). Total cDNA from U251 cells was used
to amplify the 3′-untranslated region (UTR) of E2F1. The E2F1
3′-UTR was cloned into an miR expression reporter vector system
(pMir-REPORT™ Ambion Life Technologies). Mutations were introduced
into the potential miR-320 binding sites using the QuikChange
site-directed mutagenesis kit (Stratagene, Agilent, San Diego, CA,
USA). SHG-44 and U251 cells were transfected with miR-320 mimics or
NCs using Lipofectamine 2000®. Subsequently, cells were
transfected with the pMir-Report vector or with the mutant
construct for 36 h. A pRL-SV40 vector (Promega Corporation,
Madison, WI, USA) containing the Renilla luciferase gene was
used in order to standardize transfection efficiency. Luciferase
activity was determined using the Dual-Luciferase Reporter Assay
System (Promega Corporation).
Statistical analysis
Data are expressed as the mean ± standard error of
the mean from at least three repeated experiments. Differences
between groups were analyzed using Student’s t-test. All
statistical analyses were conducted using SPPS version 20.0 (IBM,
Armonk, NY, USA) P<0.05 was considered to indicate a
statistically significant difference.
Results
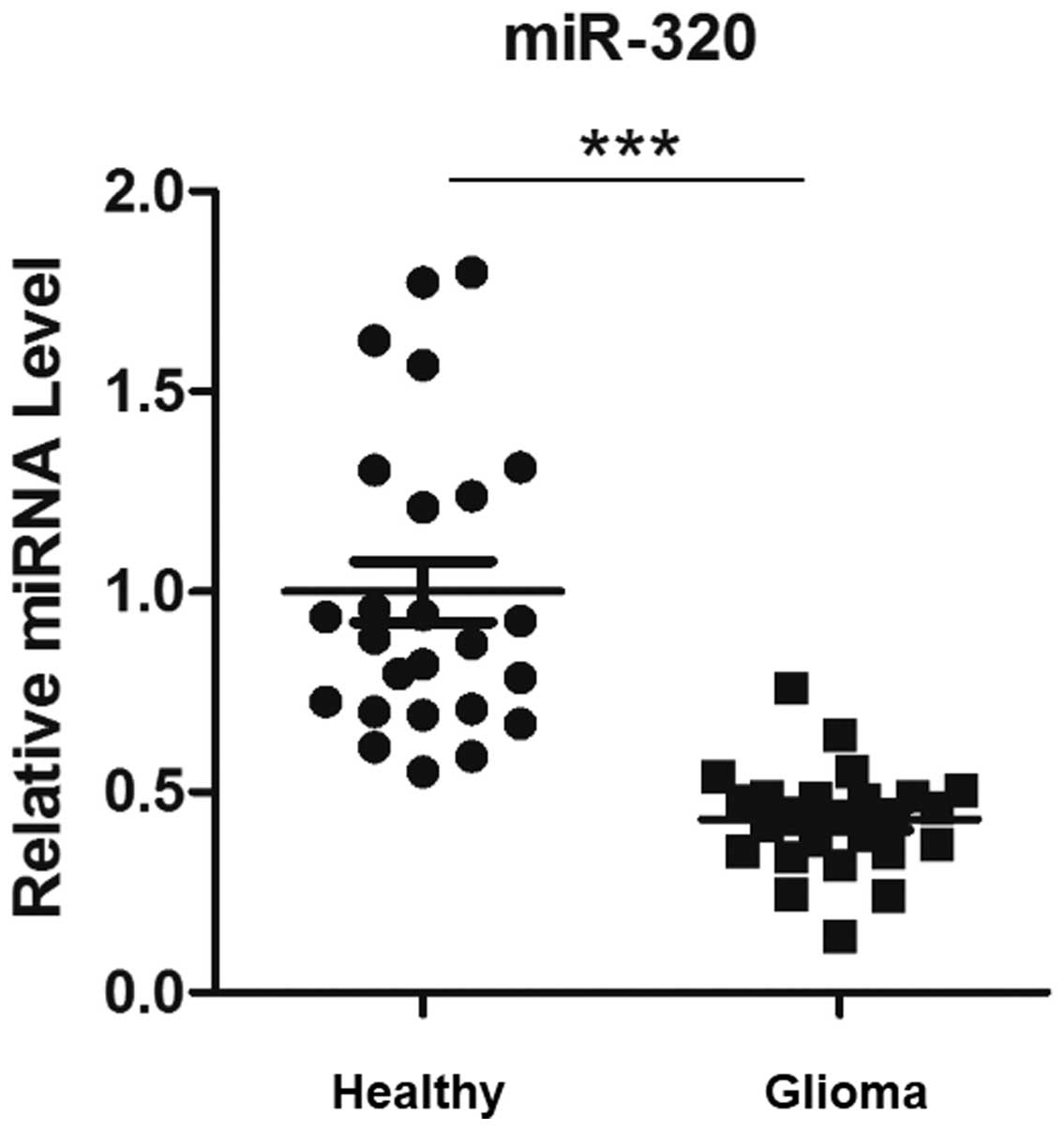
miR-320 expression was downregulated in
patients with glioma
In order to analyze the association between miR-320
expression and glioma, miR-320 expression was determined using
RT-qPCR in 25 pairs of human glioma tissues and adjacent healthy
tissues. Expression of miR-320 was significantly decreased in
glioma tissues, compared with that of adjacent healthy tissues
(Fig. 1).
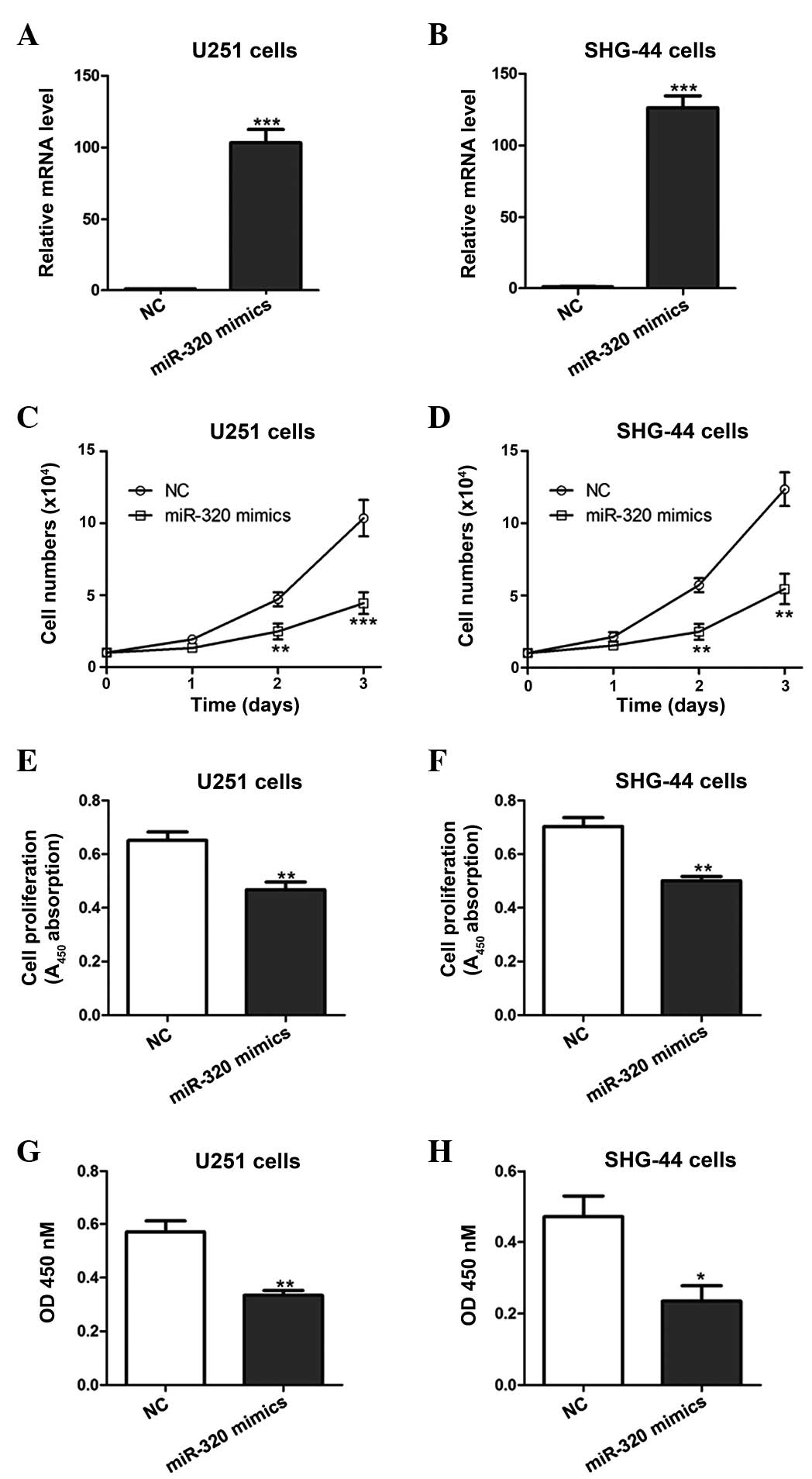
miR-320 overexpression inhibits cell
proliferation
In order to assess the effects of miR-320 on glioma
cell growth, miR-320 mimics or NCs were transfected into U251 and
SHG-44 cells (Fig. 2A and B). In
comparison with NC cells, cell numbers and proliferation were
significantly lower in U251 and SHG-44 cells lines overexpressing
miR-320 mimics (Fig. 2C–F).
Furthermore, miR-320 mimic overexpression significantly inhibited
in vitro migration capabilities of the U251 and SHG-44 cell
lines compared with that of the NC cells (Fig. 2G–H).
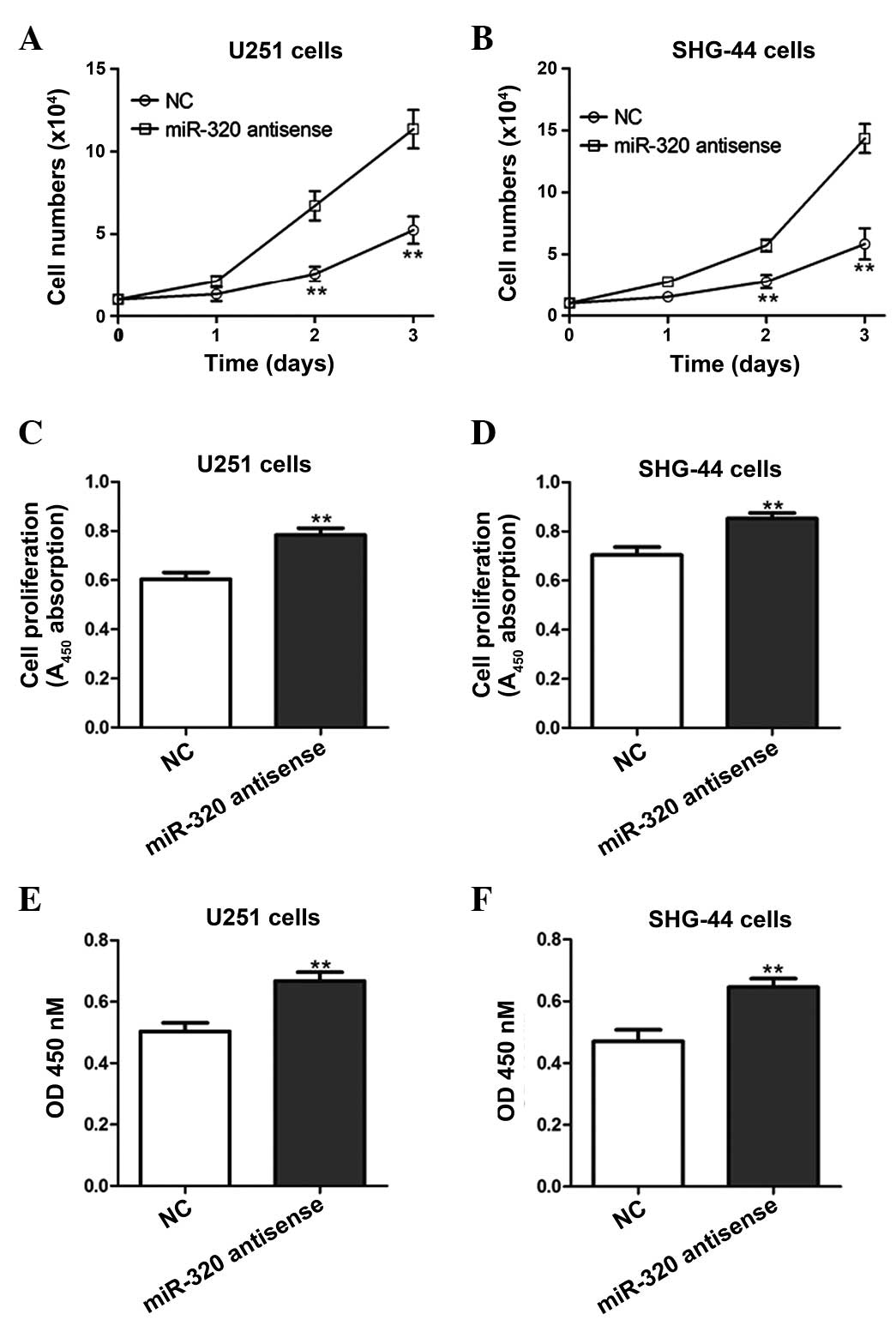
Inhibition of miR-320 promotes the
proliferation of glioma cells
In order to demonstrate the association between
miR-320 expression and glioma cell growth, U251 and SHG-44 cells
were transfected with an antisense oligonucleotide against miR-320,
which inhibited endogenous miR-320 activity. This resulted in
enhanced growth of the antisense-transfected cells compared with
that of NC cells (Fig. 3A–D). In
addition, the in vitro migration capability of
antisense-transfected cells was significantly higher than that of
NC cells (Fig. 3E–F).
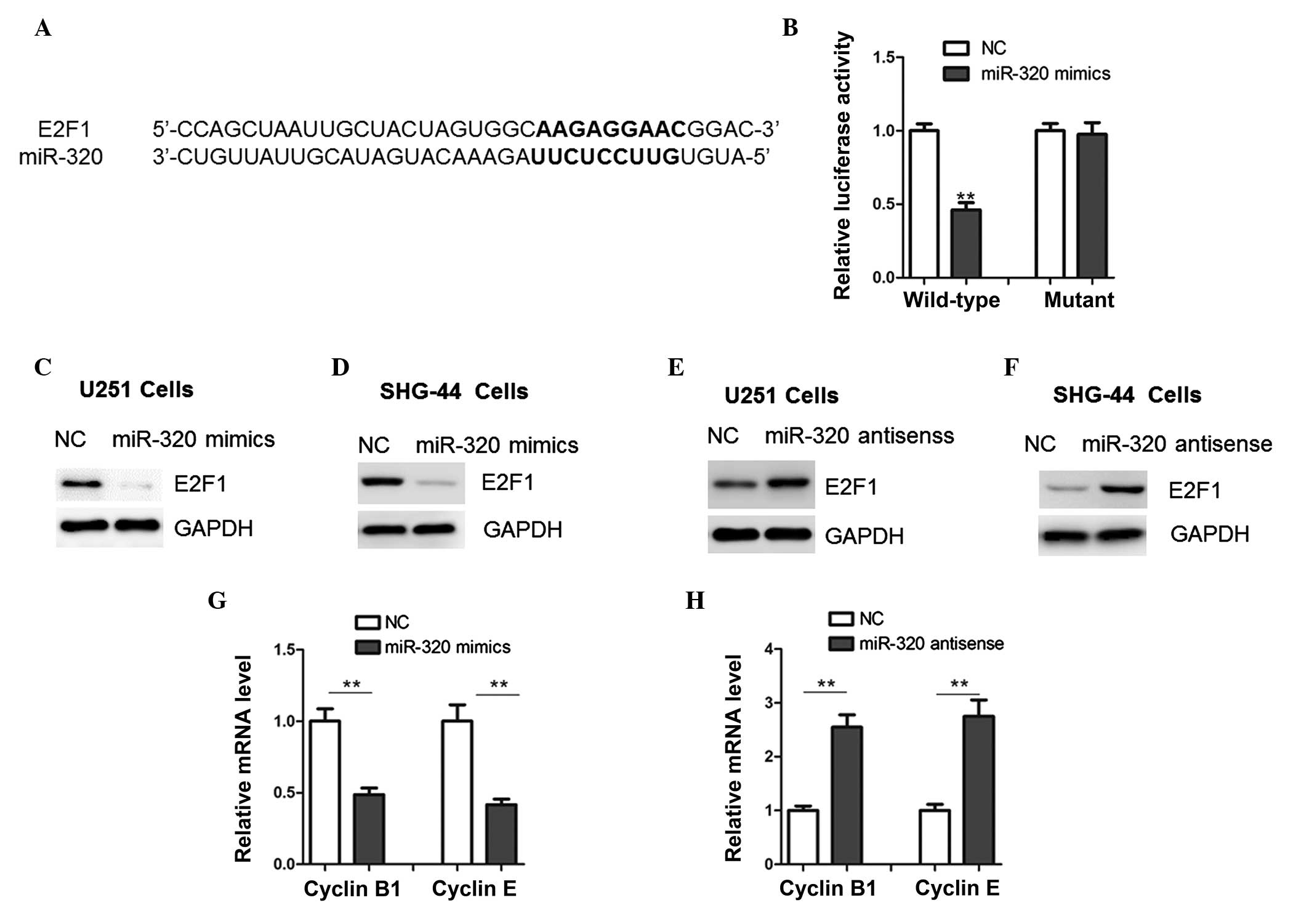
miR-320 directly targets the E2F1 in
glioma cells
Using a bioinformatics approach (miRWalk software),
a number of putative target genes of miR-320 were identified (data
not provided), among which the gene encoding E2F1 harbored a
potential miR-320 binding site (Fig.
4A). Luciferase activity was significantly lower in cells
overexpressing miR-320 compared with NC cells following
transfection with pMir-REPORT vectors (Fig. 4B). No significant difference in
luciferase activity was observed in mutated U251 and SHG-44 cells
(mutation of the conserved miR-320 binding motif) between cells
overexpressing miR-320 and NC cells (Fig. 4B). E2F1 protein expression was
lower in cells overexpressing miR-320 mimics compared with that of
the NC cells (Fig. 4C–D). E2F1
protein expression was significantly higher in anti-sense
miR-320-transfected cells compared with that in the NC cells
(Fig. 4E–F), which indicates that
E2F1 may be a target of miR-320 in glioma cells. Expression levels
of down-stream target genes of E2F1, Cyclin B1 and Cyclin E
(13,15), were significantly higher in cells
overexpressing miR-320 mimics and significantly lower in antisense
miR-320-transfected cells compared with those of the NC cells in
each case (Fig. 4G–H).
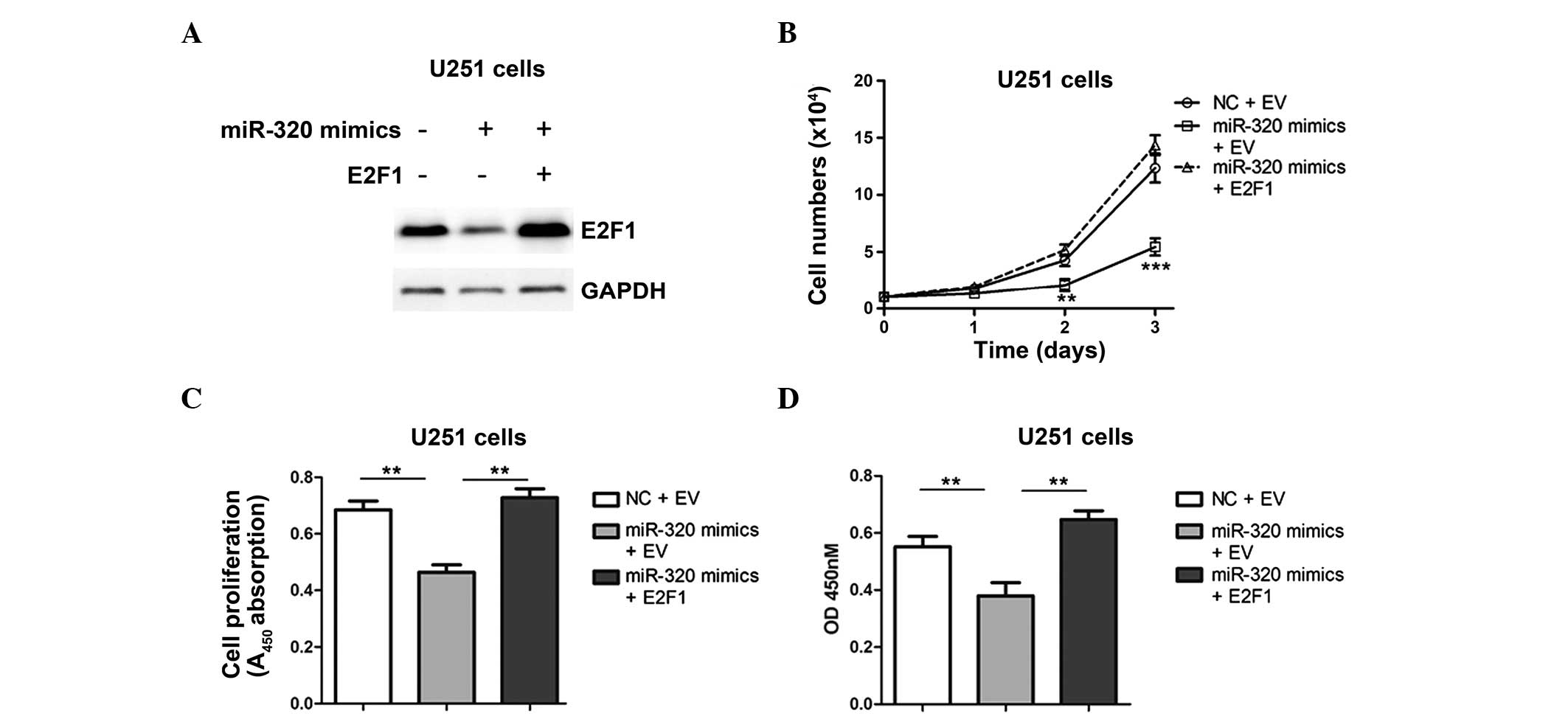
In order to verify the association between miR-320
and E2F1, U251 cells were transfected with lentiviruses containing
an E2F1 gene or an empty vector (EV), following transfection with
miR-320 mimics (Fig. 5A). U251
cell proliferation and migration levels were significantly lower
following transfection with miR-mimic and the EVs compared with
those of cells transfected with miR-320 mimic and E2F1 vectors, and
with NC cells transfected with EVs (Fig. 5B–D). Overall, the results of the
present study suggest that the mechanisms underlying the effect of
miR-320 on glioma development are associated with the regulation of
E2F1.
Discussion
A recent study demonstrated that miR-320 regulates
tumor angiogenesis, which is driven by vascular endothelial cells
in oral cancer by silencing the expression of neuropilin 1
(16). Another study demonstrated
that miR-320 inhibits osteosarcoma cell proliferation by directly
targeting fatty acid synthase (17). In the present study, miR-320
expression was shown to be lower in glioma tissues compared with
that of adjacent healthy tissues. The results suggest that miR-320
overexpression inhibits glioma cell proliferation, metastasis and
invasion, compared with that of NC cells and miR-320
antisense-transfected cells. Therefore, in accordance with previous
reports, the present study suggests that miR-320 may suppress tumor
development and progression.
Computational algorithms have been used to predict
multiple genes as potential targets of miR-320 (16,17).
The results of the present study suggest that miR-320 targets the
3′UTR of E2F1 and is involved in regulating E2F1 expression. E2F1
is a transcription factor involved in the pRb/E2F1 pathway and in
the regulation of the G1/S phase (18). Tumor suppression triggered by pRb
has been associated with its ability to inhibit E2F1-responsive
promoters (18). E2F1-responsive
promoters were more active in glioma cells than in healthy cells in
an investigation by Alonso et al (19), which suggests that there is more
‘free’ E2F1 and fewer pRb/E2F1 repressor complexes in glioma cells
compared with healthy cells. E2F1 activity is capable of
suppressing and promoting tumor development (20). Although the precise mechanisms are
currently unclear, E2F1 activity may be dependent on the nature of
associated oncogenic mutations. A cluster of miRs, including
miR-106a, miR-138 and miR-329, promoted glioma cell proliferation
by targeting E2F1 (21–23), which suggests that E2F1 is involved
in glioma cell proliferation and that E2F1 expression may be
regulated by a number of factors.
In conclusion, the present study demonstrated that
miR-320 was downregulated in glioma tissues and could inhibit cell
proliferation by targeting E2F1. Since E2F1 has a critical role in
cell proliferation, differentiation and apoptosis, further studies
are required to establish the role of miR-320 in glioma.
References
|
1
|
Goodenberger ML and Jenkins RB: Genetics
of adult glioma. Cancer Genet. 205:613–621. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Weller M and Wick W: Neuro-oncology in
2013: improving outcome in newly diagnosed malignant glioma. Nat
Rev Neurol. 10:68–70. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Castro MG and Lowenstein PR:
Neuro-oncology: The long and winding road - gene therapy for
glioma. Nat Rev Neurol. 9:609–610. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sun K and Lai EC: Adult-specific functions
of animal microRNAs. Nat Rev Genet. 14:535–548. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ameres SL and Zamore PD: Diversifying
microRNA sequence and function. Nat Rev Mol Cell Biol. 14:475–488.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Auffinger B, Thaci B, Ahmed A, Ulasov I
and Lesniak MS: MicroRNA targeting as a therapeutic strategy
against glioma. Curr Mol Med. 13:535–542. 2013. View Article : Google Scholar
|
|
7
|
Palumbo S, Miracco C, Pirtoli L and
Comincini S: Emerging roles of microRNA in modulating cell-death
processes in malignant glioma. J Cell Physiol. 229:277–286. 2014.
View Article : Google Scholar
|
|
8
|
Yang TQ, Lu XJ, Wu TF, et al: MicroRNA-16
inhibits glioma cell growth and invasion through suppression of
BCL2 and the nuclear factor-κB1/MMP9 signaling pathway. Cancer Sci.
105:265–271. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun J, Shi H, Lai N, Liao K, Zhang S and
Lu X: Overexpression of microRNA-155 predicts poor prognosis in
glioma patients. Med Oncol. 31:9112014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bronisz A, Godlewski J, Wallace JA, et al:
Reprogramming of the tumour microenvironment by stromal
PTEN-regulated miR-320. Nat Cell Biol. 14:159–167. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hsieh IS, Chang KC, Tsai YT, et al:
MicroRNA-320 suppresses the stem cell-like characteristics of
prostate cancer cells by downregulating the Wnt/beta-catenin
signaling pathway. Carcinogenesis. 34:530–538. 2013. View Article : Google Scholar
|
|
12
|
Yao J, Liang LH, Zhang Y, et al: GNAI1
suppresses tumor cell migration and invasion and is
post-transcriptionally regulated by Mir-320a/c/d in hepatocellular
carcinoma. Cancer Biol Med. 9:234–241. 2012.
|
|
13
|
Engelmann D and Pützer BM: The dark side
of E2F1: in transit beyond apoptosis. Cancer Res. 72:571–575. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dweep H, Sticht C, Pandey P and Gretz N:
miRWalk - database: Prediction of possible miRNA binding sites by
"walking" the genes of three genomes. J Biomed Inform. 44:839–847.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Biswas AK and Johnson DG: Transcriptional
and nontranscriptional functions of E2F1 in response to DNA damage.
Cancer Res. 72:13–17. 2012. View Article : Google Scholar
|
|
16
|
Wu YY, Chen YL, Jao YC, Hsieh IS, Chang KC
and Hong TM: miR-320 regulates tumor angiogenesis driven by
vascular endothelial cells in oral cancer by silencing neuropilin
1. Angiogenesis. 17:247–260. 2014. View Article : Google Scholar
|
|
17
|
Cheng C, Chen ZQ and Shi XT: MicroRNA-320
inhibits osteosarcoma cells proliferation by directly targeting
fatty acid synthase. Tumour Biol. 35:4177–4183. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pützer BM and Engelmann D: E2F1 apoptosis
counterattacked: evil strikes back. Trends Mol Med. 19:89–98. 2013.
View Article : Google Scholar
|
|
19
|
Alonso MM, Alemany R, Fueyo J and
Gomez-Manzano C: E2F1 in gliomas: a paradigm of oncogene addiction.
Cancer Lett. 263:157–163. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fueyo J, Gomez-Manzano C, Liu TJ and Yung
WK: Delivery of cell cycle genes to block astrocytoma growth. J
Neurooncol. 51:277–287. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang G, Zhang R, Chen X, et al: MiR-106a
inhibits glioma cell growth by targeting E2F1 independent of p53
status. J Mol Med (Berl). 89:1037–1050. 2011. View Article : Google Scholar
|
|
22
|
Qiu S, Huang D, Yin D, et al: Suppression
of tumorigenicity by microRNA-138 through inhibition of
EZH2-CDK4/6-pRb-E2F1 signal loop in glioblastoma multiforme.
Biochim Biophys Acta. 1832:1697–1707. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xiao B, Tan L, He B, Liu Z and Xu R:
MiRNA-329 targeting E2F1 inhibits cell proliferation in glioma
cells. J Transl Med. 11:1722013. View Article : Google Scholar : PubMed/NCBI
|