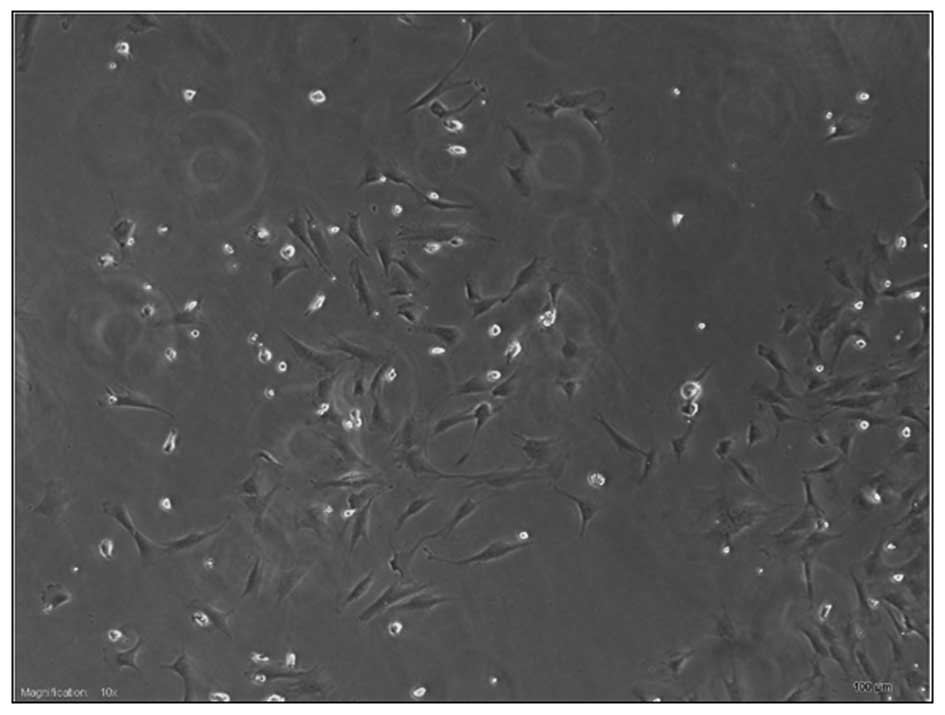
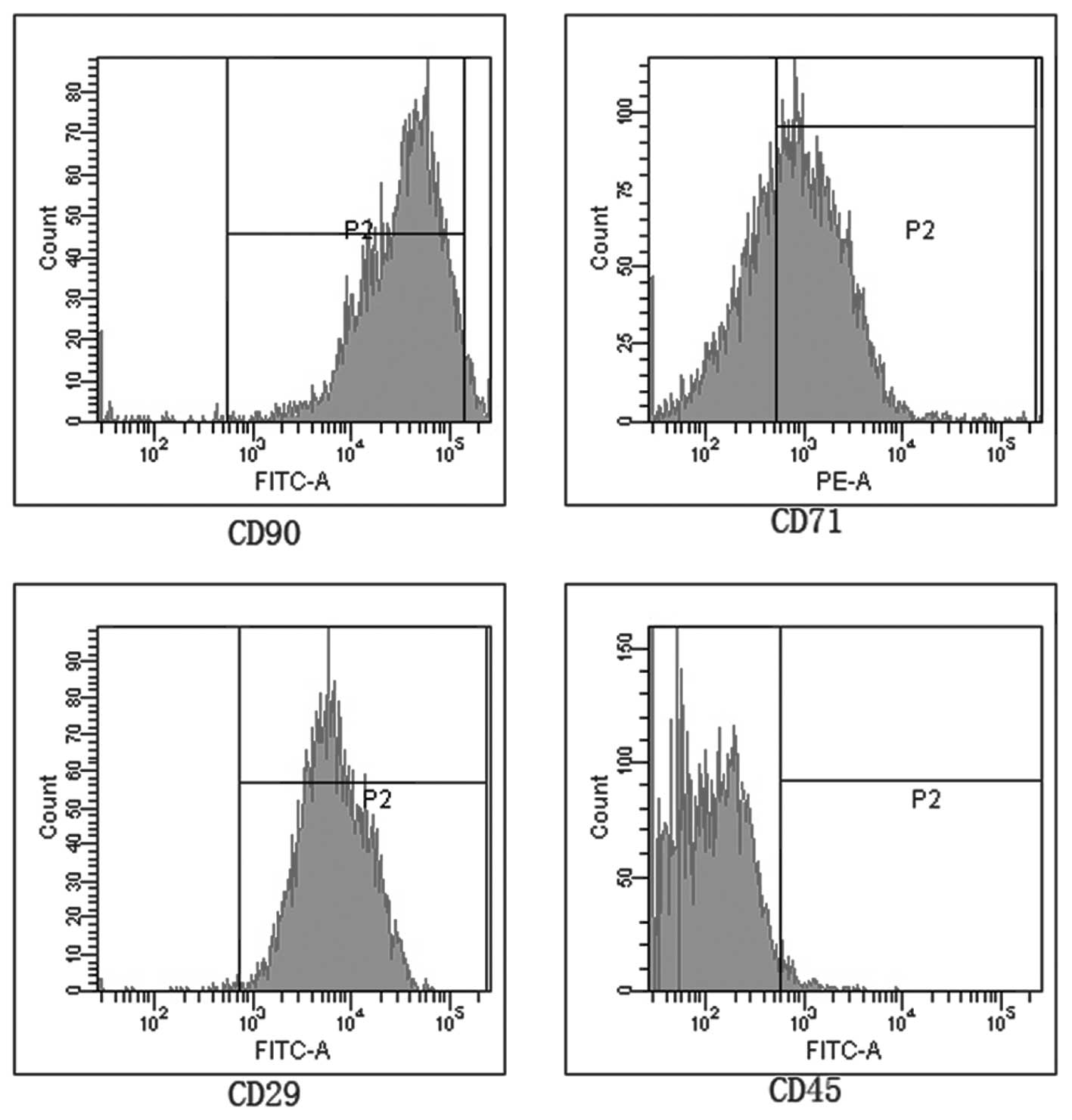
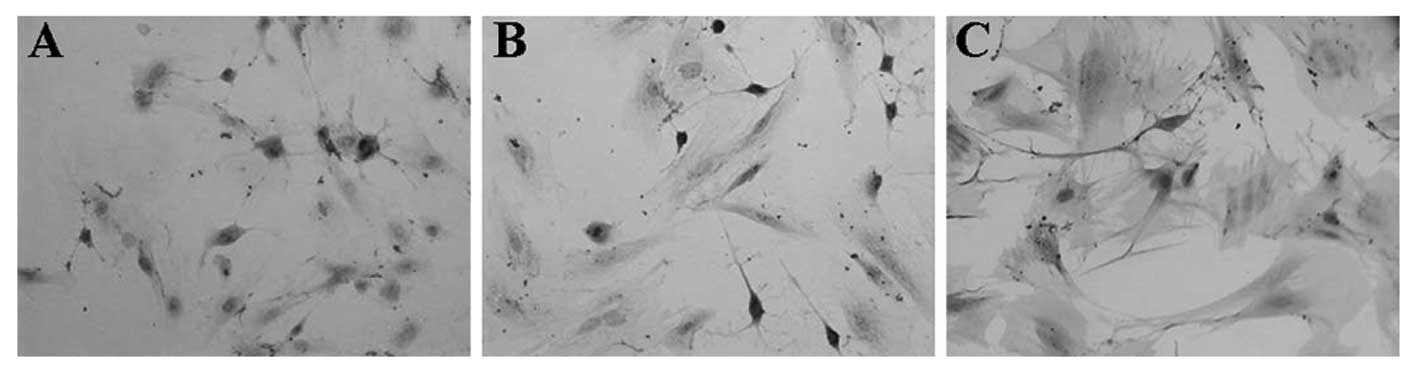
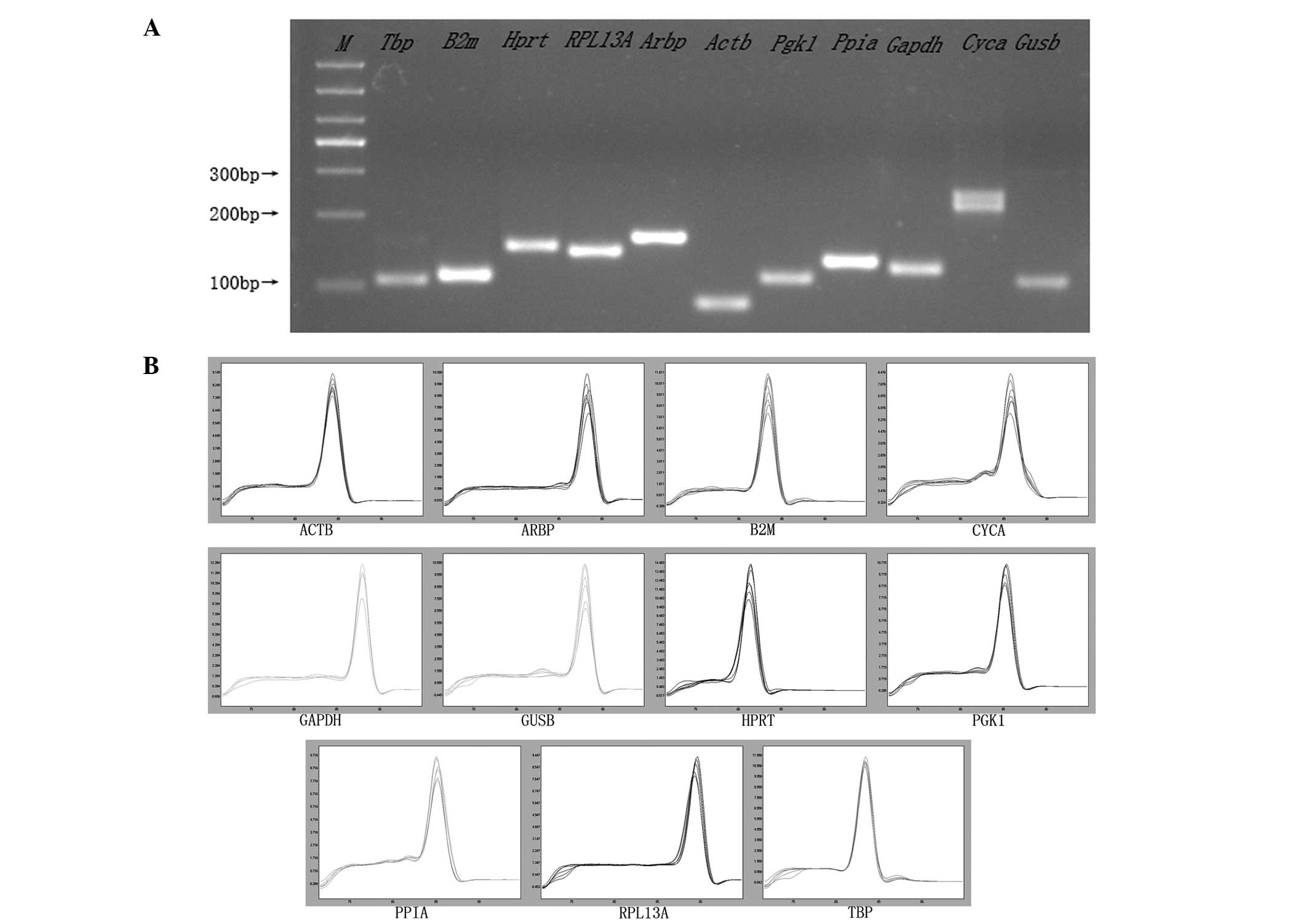
|
1
|
Bustin SA, Benes V, Garson JA, et al: The
MIQE guidelines: minimum information for publication of
quantitative real-time PCR experiments. Clin Chem. 55:611–622.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Huggett J, Dheda K, Bustin S and Zumla A:
Real-time RT-PCR normalisation; strategies and considerations.
Genes Immun. 6:279–284. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bustin SA and Nolan T: Pitfalls of
quantitative real-time reverse-transcription polymerase chain
reaction. J Biomol Tech. 15:155–166. 2004.PubMed/NCBI
|
|
4
|
Zhong Q, Zhang Q, Wang Z, et al:
Expression profiling and validation of potential reference genes
during Paralichthys olivaceus embryogenesis. Mar Biotechnol (NY).
10:310–318. 2008. View Article : Google Scholar
|
|
5
|
Bustin SA, Benes V, Nolan T and Pfaffl MW:
Quantitative real-time RT-PCR-a perspective. J Mol Endocrinol.
34:597–601. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dheda K, Huggett JF, Bustin SA, Johnson
MA, Rook G and Zumla A: Validation of housekeeping genes for
normalizing RNA expression in real-time PCR. Biotechniques.
37:112–114. 116118–119. 2004.PubMed/NCBI
|
|
7
|
Radonic A, Thulke S, Mackay IM, Landt O,
Siegert W and Nitsche A: Guideline to reference gene selection for
quantitative real-time PCR. Biochem Biophys Res Commun.
313:856–862. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Suzuki T, Kobayashi K and Sasaki O:
Real-time displacement measurement with a two-wavelength sinusoidal
phase-modulating laser diode interferometer. Appl Opt.
39:2646–2652. 2000. View Article : Google Scholar
|
|
9
|
Cordoba EM, Die JV, González-Verdejo CI,
Nadal S and Román B: Selection of reference genes in Hedysarum
coronarium under various stresses and stages of development. Anal
Biochem. 409:236–243. 2011. View Article : Google Scholar
|
|
10
|
Schmittgen TD and Zakrajsek BA: Effect of
experimental treatment on housekeeping gene expression: validation
by real-time, quantitative RT-PCR. J Biochem Biophys Methods.
46:69–81. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Santos AR and Duarte CB: Validation of
internal control genes for expression studies: effects of the
neurotrophin BDNF on hippocampal neurons. J Neurosci Res.
86:3684–3692. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sun C, Shao J, Su L, et al: Cholinergic
neuron-like cells derived from bone marrow stromal cells induced by
tricyclodecane-9-yl-xanthogenate promote functional recovery and
neural protection after spinal cord injury. Cell Transplant.
22:961–975. 2013. View Article : Google Scholar
|
|
13
|
Woodbury D, Schwarz EJ, Prockop DJ and
Black IB: Adult rat and human bone marrow stromal cells
differentiate into neurons. J Neurosci Res. 61:364–370. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sanchez-Ramos J, Song S, Cardozo-Pelaez F,
et al: Adult bone marrow stromal cells differentiate into neural
cells in vitro. Exp Neurol. 164:247–256. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xu C, Su G and Mu D: Retinal cells of rat
neonates inducing the bone marrow mesenchymal cells into RGC-like
cells. Chin Ophthal Res. 26:174–178. 2008.
|
|
16
|
Wang C, Su G, Qi S, et al: Study of GAP-43
expression on differentiation of bone mesenchymal stem cells to
neuron-like cells. Chin J Lab Diagn. 12:1206–1209. 2008.
|
|
17
|
Vandesompele J, De Preter K, Pattyn F, et
al: Accurate normalization of real-time quantitative RT-PCR data by
geometric averaging of multiple internal control genes. Genome
Biol. 3:RESEARCH00342002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Andersen CL, Jensen JL and Ørntoft TF:
Normalization of real-time quantitative reverse transcription-PCR
data: a model-based variance estimation approach to identify genes
suited for normalization, applied to bladder and colon cancer data
sets. Cancer Res. 64:5245–5250. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pfaffl MW, Tichopad A, Prgomet C and
Neuvians TP: Determination of stable housekeeping genes,
differentially regulated target genes and sample integrity:
BestKeeper-excel-based tool using pair-wise correlations.
Biotechnol Lett. 26:509–515. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Long X, Olszewski M, Huang W and Kletzel
M: Neural cell differentiation in vitro from adult human bone
marrow mesenchymal stem cells. Stem Cells Dev. 14:65–69. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Feng X, Xiong Y, Qian H, Lei M, Xu D and
Ren Z: Selection of reference genes for gene expression studies in
porcine skeletal muscle using SYBR green qPCR. J Biotechnol.
150:288–293. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Aramant RB, Seiler MJ and Ball SL:
Successful cotransplantation of intact sheets of fetal retina with
retinal pigment epithelium. Invest Ophthalmol Vis Sci.
40:1557–1564. 1999.PubMed/NCBI
|
|
23
|
Gouras P, Flood MT, Kjedbye H, Bilek MK
and Eggers H: Transplantation of cultured human retinal epithelium
to Bruch’s membrane of the owl monkey’s eye. Curr Eye Res.
4:253–265. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gimble JM, Zvonic S, Floyd ZE, Kassem M
and Nuttall ME: Playing with bone and fat. J Cell Biochem.
98:251–266. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dezawa M, Kanno H, Hoshino M, et al:
Specific induction of neuronal cells from bone marrow stromal cells
and application for autologous transplantation. J Clin Invest.
113:1701–1710. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gregory CA, Prockop DJ and Spees JL:
Non-hematopoietic bone marrow stem cells: molecular control of
expansion and differentiation. Exp Cell Res. 306:330–335. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gu S, Xing C, Han J, Tso MO and Hong J:
Differentiation of rabbit bone marrow mesenchymal stemcells into
corneal epithelial cells in vivo and ex vivo. Mol Vis. 15:99–107.
2009.
|
|
28
|
Liu C, Xin N, Zhai Y, et al: Reference
gene selection for quantitative real-time RT-PCR normalization in
the half-smooth tongue sole (cynoglossus semilaevis) at different
developmental stages, in various tissue types and on exposure to
chemicals. PLoS One. 9:e917152014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
McCurley AT and Callard GV:
Characterization of housekeeping genes in zebrafish: male-female
differences and effects of tissue type, developmental stage and
chemical treatment. BMC Mol Biol. 9:1022008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Farrokhi A, Eslaminejad MB, Nazarian H,
Moradmand A, Samadian A and Akhlaghi A: Appropriate reference gene
selection for real-time Pcr Data normalization during rat
mesenchymal stem cell differentiation. Cell Mol Biol
(Noisy-le-grand). 58(Suppl): OL1660–1670. 2012.
|
|
31
|
Swijsen A, Nelissen K, Janssen D, Rigo JM
and Hoogland G: Validation of reference genes for quantitative
real-time PCR studies in the dentate gyrus after experimental
febrile seizures. BMC Res Notes. 5:6852012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chang E, Shi S, Liu J, et al: Selection of
reference genes for quantitative gene expression studies in
Platycladus orientalis (Cupressaceae) using real-time PCR. PLoS
One. 7:e332782012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bonefeld BE, Elfving B and Wegener G:
Reference genes for normalization: a study of rat brain tissue.
Synapse. 62:302–309. 2008. View Article : Google Scholar : PubMed/NCBI
|