Introduction
Osteosarcoma (OS) is the most common type of primary
malignant tumor of the bone in young adolescents and infants
(1). Pulmonary metastasis is the
predominant cause of mortality in patients with OS. Studies have
demonstrated that the five-year survival rate of patients with
metastatic diseases was <20% (1-3).
Clearly, a comprehensive understanding of the biological mechanisms
of this malignancy is required for the management of OS.
Accumulating studies have demonstrated that miRNAs
are critical in cell proliferation, apoptosis and metastasis
(4-6). Let-7 is one of the most extensively
investigated miRNAs in the Caenorhabditis elegans genome. It
regulated seam cell terminal differentiation, possibly by acting as
a regulator of multiple genes required for cell cycle and
proliferation. Eleven members of the let-7 cluster have been
identified in the human genome. Notably, the let-7 family is one of
the first reported tumor suppressor miRNAs in cancer, which
negatively regulates RAS and is expressed at lower levels in lung
tumors than in normal lung tissue (7). The let-7 cluster has been shown to be
significantly correlated with the occurrence and development of
cancer, suggesting that it is involved in the regulation of
oncogenic pathways in numerous types of tumors (8,9).
Reduced expression of let-7 has also been associated with shortened
postoperative survival in patients with lung cancer (8). In addition, enhanced expression of
let-7 family members is able to inhibit malignant tumor cell growth
(9,10). Let-7i is one member of the let-7
family. Recently, studies have indicated that let-7i is involved in
cancer metastasis (11). However,
it is unknown whether let-7i is essential in OS development and
metastasis.
In the present study, the association between let-7i
and Aurora-B in OS tissue and cell lines were investigated, in
order to elucidate the possible molecular mechanisms of metastasis
of OS and improve the therapeutic strategies for the management of
OS.
Materials and methods
Patients and clinical samples
Twenty-one OS specimens were collected prior to
neoadjuvant chemotherapy in the Department of Orthopedics, The
First Affiliated Hospital of Nanchang University and The Cancer
Hospital of Jiangxi Province (Nanchang, China) between 2009 and
2012. The matched normal tissues obtained from an area 5 cm from
the tumor margin, were used as negative controls. The diagnosis was
confirmed by two pathologists. The study protocol and operational
procedures were approved by the Human Ethics Committee of Nanchang
University, and a signed informed consent form was obtained from
all patients or patients' family members.
Cell lines and cell culture
The U2-OS and HOS human osteosarcoma cell lines and
the human osteoblast cell line HOB was obtained from American Type
Culture Collection (Manassas, VA, USA), and routinely cultured in
Dulbecco's modified Eagle's medium (Hyclone, Logan, UT, USA)
supplemented with 10% fetal bovine serum (FBS; Sigma Aldrich, St.
Louis, MO, USA) in a humidified 37°C incubator containing 5%
CO2.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
The formalin-fixed paraffin-embedded (FFPE) OS
tissues and adjacent tumorous tissues were obtained for microRNA
isolation using the Qiagen RN easy FFPE protocol (Qiagen, Valencia,
CA, USA) according to Kelly et al (12). The let-7i expression levels were
evaluated by RT-qPCR (StepOne™; Bio-Rad Laboratories, Inc.,
Hercules, CA, USA), using U6 snRNA as the endogenous reference
gene. Total RNA from OS cells was extracted using 72 h culture with
TRIzol (Invitrogen Life Technologies, Carlsbad, CA, USA). Reverse
transcription was performed with 2 mg total RNA using PrimeScript
RT Reagent Kit (Takara Bio, Inc., Otsu, Japan). Then each sample
was analyzed by qPCR under the conditions described in the
manufacturer's instructions for SYBR Premix Ex Tap II (Invitrogen
Life Technologies): 50°C for 2 min, 95°C for 2 min, followed by 40
cycles of 95°C for 15 sec and 60°C for 30 sec. Relative expression
was calculated using the 2-ΔΔCt method. All procedures were
conducted according to the manufacturer's instructions. The primer
sequences can be found in Table
I.
 | Table IPrimer sequences. |
Table I
Primer sequences.
| Primer | Sequences (5′ to
3′) |
|---|
| let-7i-RT |
GTCGTATCCAGTGCAGGGTCCG |
|
AGGTATTCGCACTGGAACAGCA |
| Let-7i-Q-F |
GCGTGAGGTAGTAGTTTG |
| U6-RT |
GTCGTATCCAGTGCAGGGTCCGAGG |
|
TATTCGCACTGGATACGACAAAAAT |
| U6-Q-F |
GCACTGGACTTGGAGTCA |
| Q-miR-5p-R |
CAGTGCAGGGTCCGAGGT |
| Aurora-B-196
bp-F |
AAGGAGAACTCCTACCCCTGG |
| Aurora-B-196
bp-R |
TTAAGATGTCGGGTGTCCCAC |
| β-actin-295 bp-F |
TCACCCACACTGTGCCATCATCGA |
| β-actin-295 bp-R |
CAGCGGAACCGCTCATTGCCAATGG |
Cell growth assay
OS cells were cultured into five 96-well tissue
culture plates at a cell density of 5,000 cells/ml in a volume of
200 µl culture medium. A total of 20 µl
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT;
0.5 mg/ml; Beyotime Institute of Biotechnology, Beijing, China) was
added to each well, and the wells were incubated at 37°C for 4 h.
Subsequently, the supernatant was removed, and 20 µl dimethyl
sulfoxide was added to each well to dissolve the formazan crystals
at 37°C for 20 min. The optical density values were measured at 490
nm wave length in triplicate using Synergy™ HT (BioTek Instruments,
Inc., Winooski, VT, USA).
Western blot analysis
Total protein from cells was extracted using
radioimmunoprecipitation lysis buffer (Beyotime Institute of
Biotechnology) containing 60 µg/ml PMSF (Solarbio, Beijing, China).
Protein concentration was determined by a Bradford assay (Bio-Rad
Laboratories, Inc.). Western blot analysis was conducted using
antibodies against Aurora-B (EP1009Y; Abcam, Cambridge, UK) and
β-actin (sc-130657; Santa Cruz Biotechnology Inc., Santa Cruz, CA,
USA). The immune complexes were detected with a pro-light
horseradish peroxidase kit (Life Technologies, Carlsbad, CA, USA).
Six independent experiments were performed over multiple days.
Migration assay
Cell migration was assessed by a wound healing
assay, which determined the ability of the cells to move into a
cellular space in two-dimension in vitro. In brief, cells
were grown to 100% confluence in 6-well tissue culture plastic
dishes to a density of ~5×106 cells/well. The cells were
denuded by dragging a rubber policeman (Fisher Scientific, Hampton,
NH, USA) through the center of the plate. Cultures were rinsed with
phosphate-buffered saline and replaced with fresh quiescent medium
alone or containing 10% FBS, following which the cells were
incubated at 37°C for 24 h. Images were captured at 0 and 24 h
using an ECLIPSE-TS-100 microscope (magnification, x200; Nikon,
Tokyo, Japan), and the migrated distance was measured by Image J,
version 1.48 (National Center for Biotechnology Information,
Bethesda, MD, USA). The cell migration rate was obtained by
counting three fields per area and are represented as the average
of six independent experiments conducted over multiple days.
Transwell invasion assays
Invasion of OS cells was measured using the BD
BioCoatTM BD Matrigel™ Invasion Chamber (BD Biosciences, Franklin
Lakes, NJ, USA) according to the manufacturer's instructions. The
medium in the lower chamber contained 5% fetal calf serum as a
source of chemoattractants. Cells were suspended in serum-free
medium and added to the upper chambers at the same time. Cells that
passed through the Matrigel-coated membrane were stained with
Diff-Quik (Sysmex, Kobe, Japan) and photographed (magnification,
x400). Images were captured at 24 h, and cell counting was measured
by Image J software. The values for invasion were obtained by
counting three fields per membrane and represented as the average
of six independent experiments conducted over multiple days.
Lentivirus-vector construction and cell
transfection
To construct vectors for downregulating Aurora-B,
the sequences of interfering microRNA targeting Aurora-B (5′-CCG
GCTCCAAACTGCTCAGGCATAACTCGAGTTATGCCTGA GCAGTTTGGAGTTTTTG-3′) were
inserted into lentivirus vector GV115 (GeneChem Co., Ltd.,
Shanghai, China). U2-OS and HOS cells were transfected with
lentivirus vectors (GeneChem Co., Ltd.) of downregulating Aurora-B
(LV-shAurora-B) and negative lentivirus vectors (LV-Neg),
respectively (MOI=20). The let-7 mimics and negative mimics
(mimics-Neg) were transfected with Lipofectamine 2000 (Life
Technologies). The transfection efficiency was evaluated under the
fluorescence microscope (BX61; Olympus, Tokyo, Japan).
Target prediction
Prediction of the Aurora-B 3′-UTR as a miRNA binding
target was determined using TargetScan (http://www.targetscan.org), microRNA (http://www.microrna.org), and PicTar
(pictar.mdc-berlin.de). MiRNAs that were simultaneously predicted
by all 3 programs were selected for the present study.
Vector construction and luciferase
reporter assay
To generate a luciferase reporter construct, 3′UTR
and mutant 3′UTR of Aurora-B were inserted downstream of firefly
luciferase in pGL3 (Promega Corporation, Madison, WI, USA). Cells
were cotransfected with let-7i and 3′UTR or mutant 3′UTR luciferase
reporters, using pRL-TK (Promega Corporation) as the control
vector. Luciferase activity was measured using the Dual-Luciferase
Assay kit (Promega Corporation) with a beta-counter luminometer
(Promega Corporation). Relative luciferase activity was calculated
as ratio of the raw firefly luciferase activity to the renilla
luciferase activity.
Statistical analysis
All data are presented as the mean ± standard
deviation, an independent-samples t-test was performed for
statistical analysis, and P<0.05 was considered to indicate a
statistically significant difference. All analyses were performed
using SPSS Version 13.0 (SPSS Inc., Chicago, IL, USA).
Results
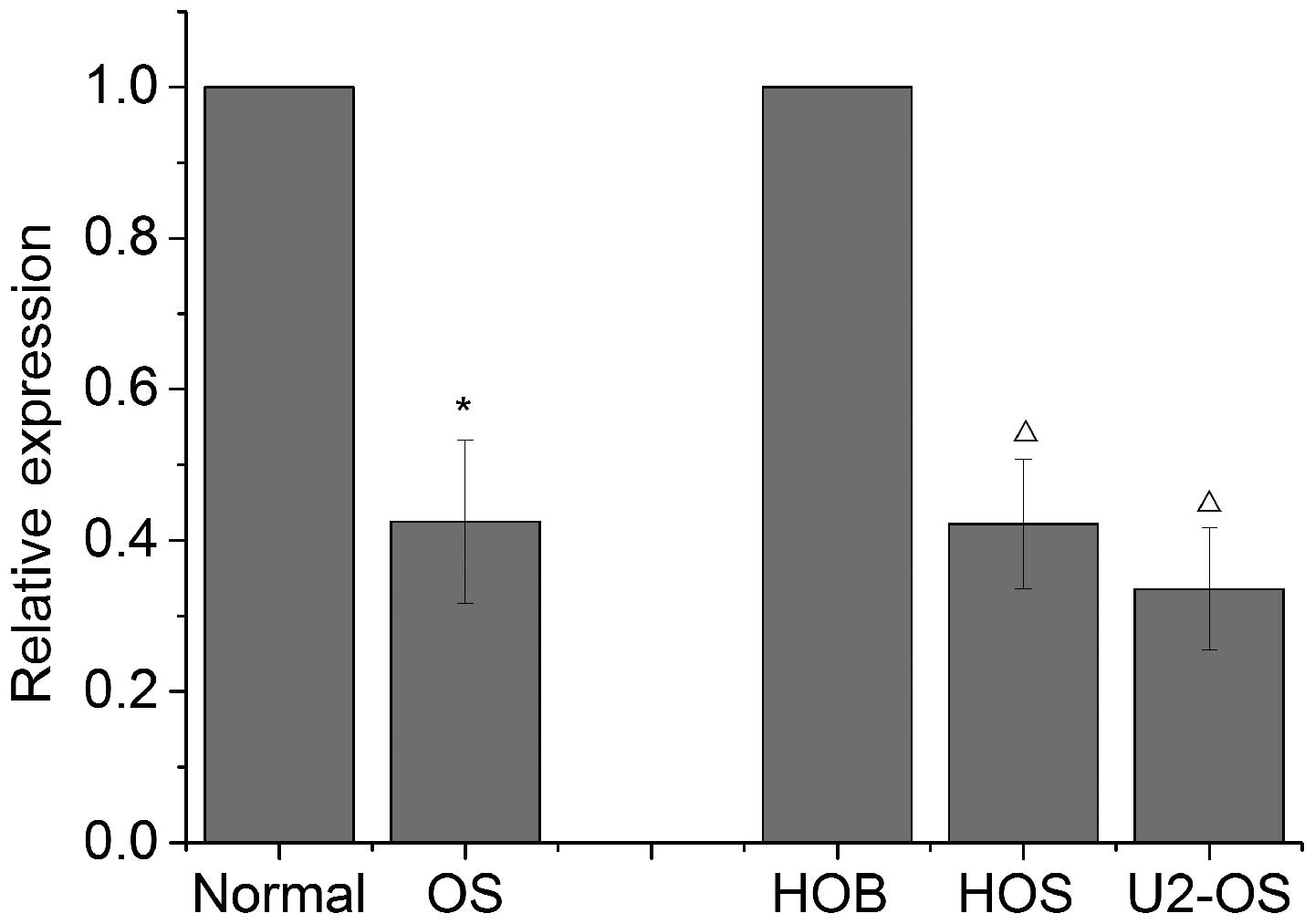
Let-7i is downregulated in OS tissues and
cell lines
In order to investigate the correlation between
let-7i dysregulation and OS, RT-qPCR was conducted to measure
let-7i expression in 21 human OS and normal tissues (adjacent
tumorous tissues), U2-OS and HOS cell lines and osteoblast cell
lines. The results revealed that expression of let-7i was
significantly downregulated in OS tissues compared with that in
normal tissues (Fig. 1).
Furthermore, a decreased expression of let-7i was also observed in
OS cells when compared with that in osteoblast cells (Fig. 1). These results suggest that the
let-7i may be essential in OS.
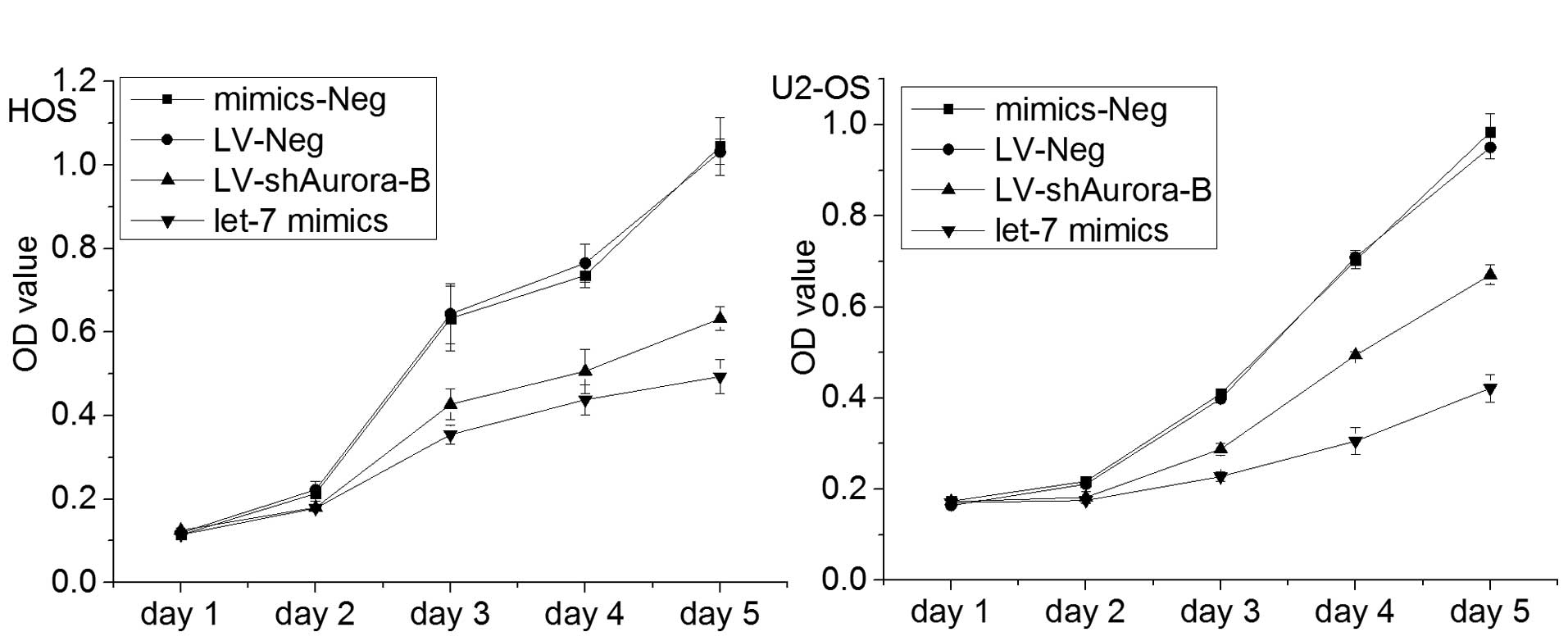
Enhanced let-7i expression suppresses
cell viability of OS cells in vitro
In order to investigate the involvement of let-7i in
the OS malignant phenotype, the expression of let-7i in U2-OS and
HOS cells was restored by infection with let-7i mimic. Cell
viability was investigated through evaluation of proliferation by
MTT assays. The results revealed that the viability of OS cells was
inhibited by restoration of expression of let-7i in OS cells
(Fig. 2). These data indicated
that let-7i decreased OS cell viability in vitro.
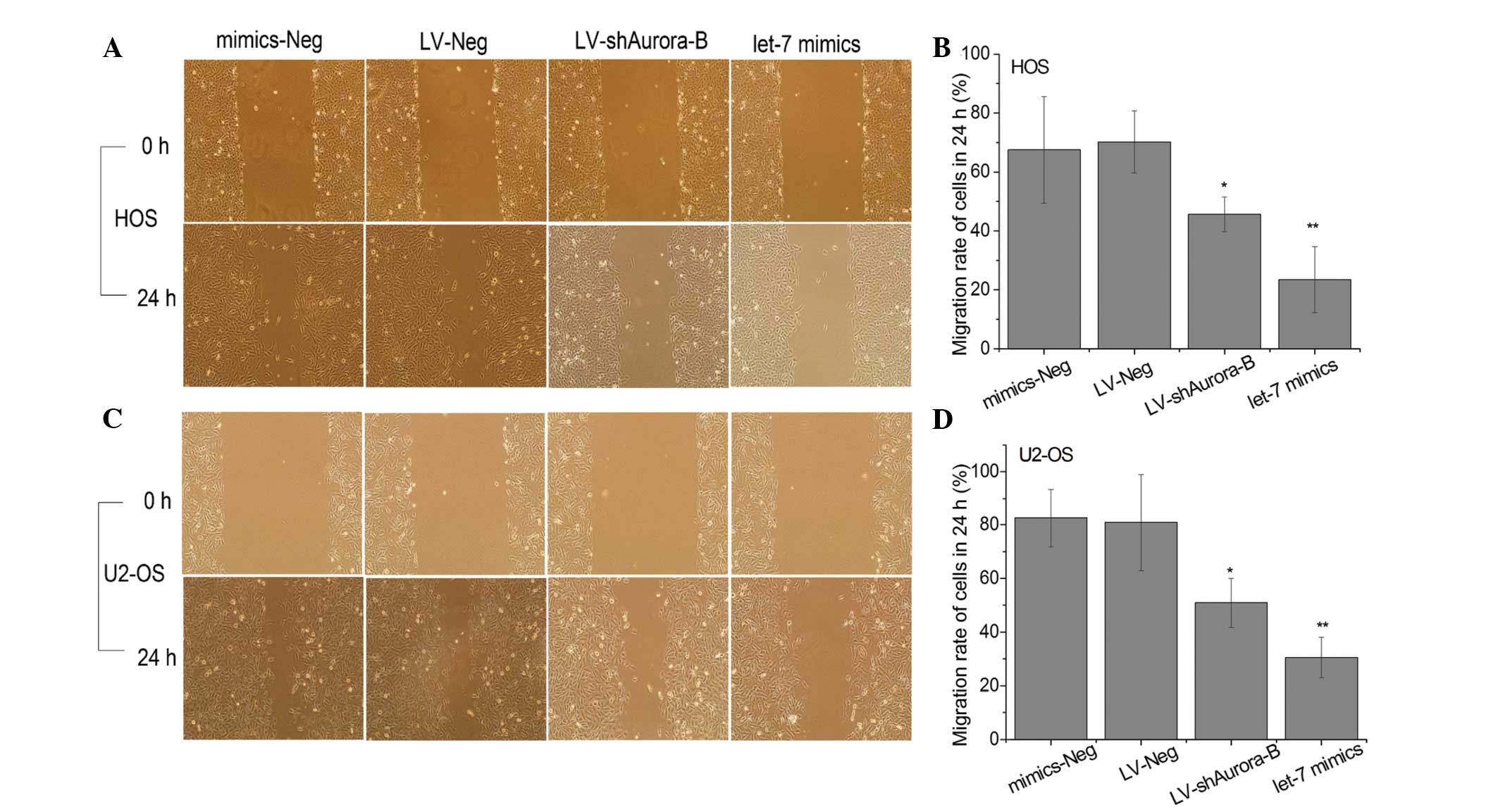
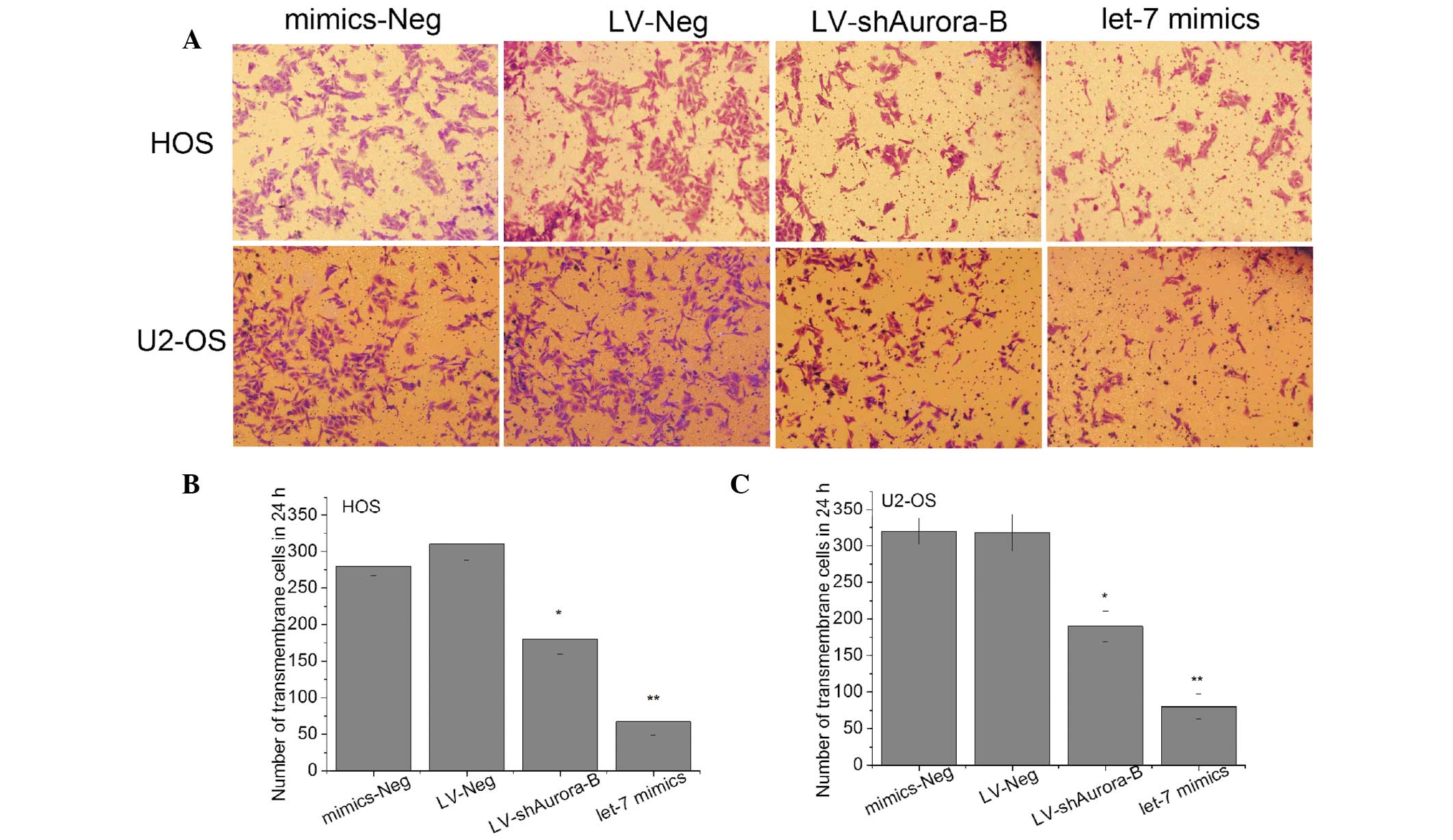
Enhanced let-7i expression suppresses
cell migration and invasion of OS cells in vitro
For investigating the effect of let-7i on migration
and invasion of OS cells, the let-7i mimic was used to restore
let-7i expression in U2-OS and HOS cells, and the migratory and
invasive ability of cells were measured by wound healing and
Transwell assays. The migratory rate and number of invasive cells
were significantly lower in cells infected with let-7i mimics than
that in cells infected with negative mimics (Figs. 3 and 4). These results suggest that enhanced
expression of let-7i inhibited OS cell migration and invasion in
vitro.
Let-7i negatively regulates Aurora-B
expression in OS cells
Our previous study indicated that Aurora-B is
involved in OS cell invasion and metastasis (13), and recent studies have demonstrated
that Aurora-B is a target of let-7a (14). Therefore, in order to explore
whether Aurora-B is a target of let-7i, the prediction was
performed by two target prediction websites [Pictar and Targetscan
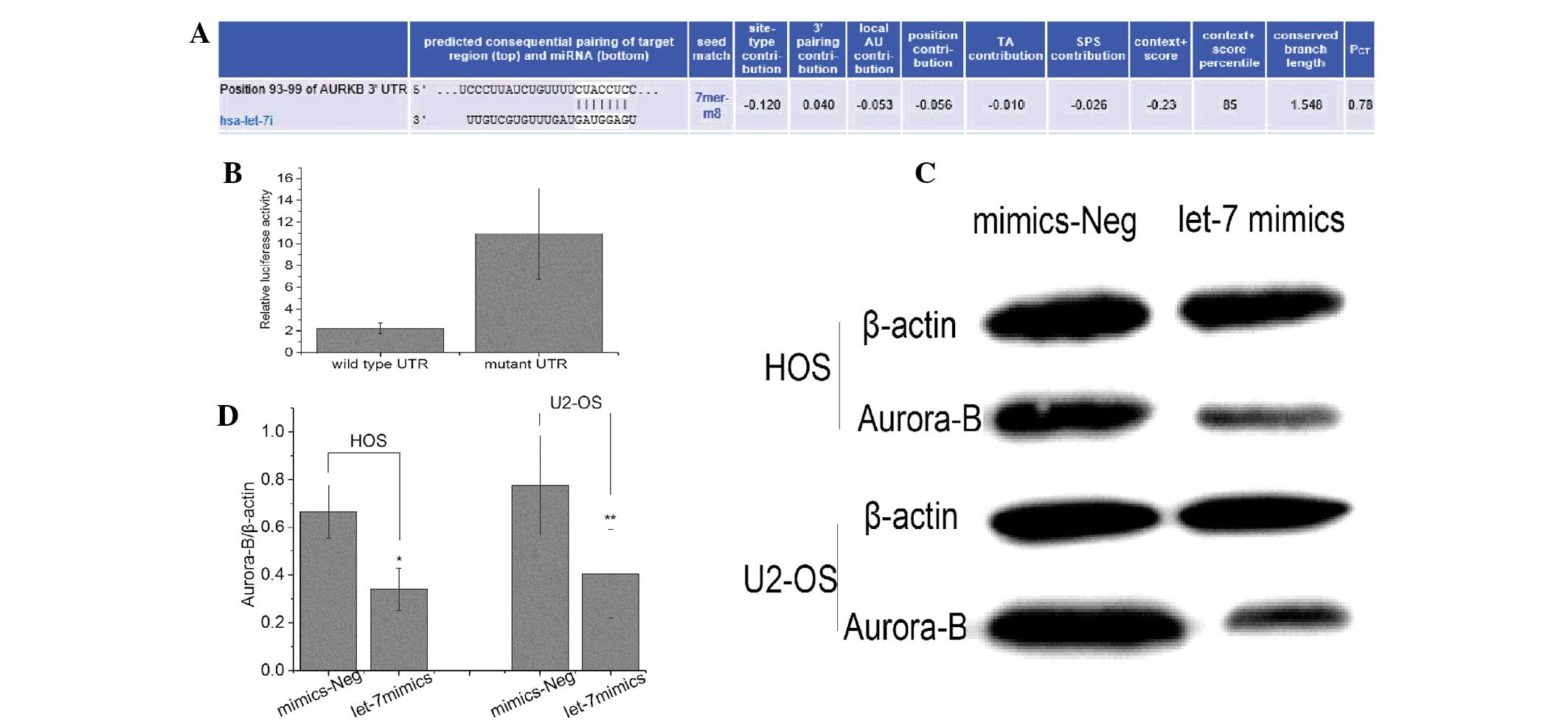
(15)]. It was found that Aurora-B
may be the target gene of let-7i (Fig.
5A). In addition, to investigate whether Aurora-B was regulated
by let-7i through direct binding to its 3′ untranslated region
(UTR), the full-length wild-type and mutant fragments of Aurora-B
mRNA 3′-UTR were constructed, and inserted into the region
immediately downstream of a luciferase reporter gene. Subsequently,
let-7i mimic oligos were co-transfected with different luciferase
3′-UTR constructs into U2-OS cells. Results revealed that let-7i
decreased the relative luciferase activity in the wild-type 3′-UTR
of Aurora-B. Furthermore, luciferase activity was not significantly
decreased in the UTRs with mutant binding sites compared with the
mut-type counterparts (Fig. 5B).
The results suggested that Aurora-B may be targeted by let-7i in OS
cell.
To further determine let-7i negative regulation of
Aurora-B expression in OS cells, the let-7i expression in U2-OS and
HOS cells was restored by infection with let-7i mimic, and Aurora-B
mRNA and protein expression was analyzed by RT-qPCR and western
blot analysis. The results show that expression levels of Aurora-B
protein and mRNA were significantly lower in cells infected with
let-7i mimic than that in cells infected with negative mimic,
suggesting that let-7i can negatively regulate Aurora-B expression
in OS cells (Fig. 5C and D).
Let-7i inhibits OS cell malignant
phenotype partly by targeting Aurora-B
To explore the functional correlation between let-7i
and Aurora-B in OS. The U2-OS and HOS cells were infected with
let-7i mimic and LV-shAurora-B, respectively. It was found that the
level of Aurora-B protein expression significantly decreased in
cells infected with LV-shAurora-B or let-7i mimic. Furthermore, the
effect of enhancing let-7i and silencing Aurora-B on cell
proliferation, migration and invasion was investigated. Results
revealed that the inhibitory effect of silencing Aurora-B by
LV-shAurora-B on cell proliferation, migratory and invasive ability
was significantly lower than that by let-7i mimic (Figs. 2Figure 3–4). These results indicated that let-7i
inhibits the malignant phenotype partially by targeting Aurora-B in
OS cells.
Discussion
In the present study, it was demonstrated that
let-7i expression in OS tissues and cell lines was downregulated
compared with that in normal tissues and osteoblast cell lines.
Restored expression of let-7i inhibited the malignant phenotype of
OS cells in vitro. Furthermore, it was shown that Aurora-B
is a direct target of let-7i and it mediated the suppression of
Aurora-B by binding to its 3′-UTR. Therefore, the present findings
highlight the significance of let-7i as a tumor suppressor in cell
malignant phenotype by targeting Aurora-B in OS.
The hsa-let-7i gene is a novel member of the let-7
miRNA family, and is located at 12q14.1. Although little is known
regarding its function, recent studies have indicated that let-7i
is a novel biomarker and therapeutic target in human epithelial
ovarian cancer (16).
Balakathiresan et al (17)
revealed that its expression is elevated in the serum and
cerebrospinal fluid of individuals who exposure to blast wave,
suggesting it involve in blast-induced traumatic brain injury.
However, let-7i expression is decreased in several types of
malignancies (8,11). Previous studies have shown that
let-7i is downregulated in ovarian cancer and may be used as a
therapeutic target to modulate platinum-based chemotherapy and as a
biomarker to predict chemotherapy response and survival (16,18).
Lai et al (19) showed that
aberrant expression of let-7i in T cells contributes to
immunopathogenesis. Recently, a study showed that repression of
bone morphogenetic protein 4 by let-7i attenuates mesenchymal
migration of head and neck cancer cells (11). Notably, Zhang et al
(20) showed that mature
hsa-let-7i expression was elevated and correlated with colorectal
cancer metastasis. In the present study, it was demonstrated that
let-7i was aberrantly expressed in OS tissues with pulmonary
metastatic disease and cell lines, and restoration of let-7i
inhibits the OS cell malignant phenotype in vitro.
Aurora-B is located on chromosome 17p13.1, a region
that is not typically amplified in human malignancies. Increasing
evidence shows that Aurora B is hypothesized to be an important
antitumor target (21). Recently,
studies have revealed that nuclear Aurora-B expression is strongly
associated with tumor metastasis (22-25).
Our previous study demonstrated that inhibition of Aurora-B
suppress cell migration and invasion in OS cells (13). A recent study implicated Aurora-B
as a target of let-7a, which contributes to the growth of
endometrial carcinoma cells (14).
Our results indicated that Aurora-B is the direct target of let-7i,
which inhibited invasion and metastasis by binding the 3′-UTR of
Aurora-B.
In conclusion, the present study demonstrated that
let-7i can inhibit the malignant phenotype of OS cells by directly
binding to the 3′-UTR of Aurora-B in vitro, suggesting that
let-7i may be a novel potential target for OS treatment. However,
tumor microenvironment may also be important in tumor development
and metastasis. Thus, further experiments in vivo are
required to be performed to investigate whether let-7i could act as
a tumor inhibitor in OS.
Acknowledgments
The present study was supported by grants from the
National Natural Science Foundation of China (grant no. 81360399),
the Natural Science Foundation of Jiangxi Province (grant no.
2012ZBAB205016) and Jiangxi Province Education Department of
Science and Technology (grant no. GJJ12097).
References
|
1
|
Hegyi M, Semsei AF, Jakab Z, et al: Good
prognosis of localized osteosarcoma in young patients treated with
limb-salvage surgery and chemotherapy. Pediatr Blood Cancer.
57:415–422. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mialou V, Philip T, Kalifa C, et al:
Metastatic osteosarcoma at diagnosis: prognostic factors and
long-term outcome-the French pediatric experience. Cancer.
104:1100–1109. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stokkel MP, Linthorst MF, Borm JJ,
Taminiau AH and Pauwels EK: A reassessment of bone scintigraphy and
commonly tested pretreatment biochemical parameters in newly
diagnosed osteosarcoma. J Cancer Res Clin Oncol. 128:393–399. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhao H, Guo M, Zhao G, Ma Q, Ma B, Qiu X,
et al: miR-183 inhibits the metastasis of osteosarcoma via
downregulation of the expression of Ezrin in F5M2 cells. Int J Mol
Med. 30:1013–1020. 2012.PubMed/NCBI
|
|
5
|
Wu X, Zhong D, Gao Q, Zhai W, Ding Z and
Wu J: MicroRNA-34a inhibits human osteosarcoma proliferation by
downregulating ether a go-go 1 expression. Int J Med Sci.
10:676–682. 2013. View Article : Google Scholar
|
|
6
|
Dong Q, Meng P, Wang T, Qin W, Wang F,
Yuan J, et al: MicroRNA let-7a inhibits proliferation of human
prostate cancer cells in vitro and in vivo by targeting E2F2 and
CCND2. PLoS One. 5:e101472010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Johnson SM, Grosshans H, Shingara J, Byrom
M, Jarvis R, Cheng A, et al: RAS is regulated by the let-7 microRNA
family. Cell. 120:635–647. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Takamizawa J, Konishi H, Yanagisawa K,
Tomida S, Osada H, Endoh H, et al: Reduced expression of the let-7
microRNAs in human lung cancers in association with shortened
postoperative survival. Cancer Res. 64:3753–3756. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nadiminty N, Tummala R, Lou W, Zhu Y, Shi
XB, Zou JX, et al: MicroRNA let-7c is downregulated in prostate
cancer and suppresses prostate cancer growth. PLoS One.
7:e328322012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
De Vito C, Riggi N, Suva ML, Janiszewska
M, Horlbeck J, Baumer K, et al: Let-7a is a direct EWS-FLI-1 target
implicated in Ewing's sarcoma development. PLoS One. 6:e235922011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang WH, Lan HY, Tai SK and Yang MH:
Repression of bone morphogenetic protein 4 by let-7i attenuates
mesenchymal migration of head and neck cancer cells. Biochem
Biophys Res Commun. 433:24–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kelly AD, Haibe-Kains B, Janeway KA, Hill
KE, Howe E, Goldsmith J, et al: MicroRNA paraffin-based studies in
osteosarcoma reveal reproducible independent prognostic profiles at
14q32. Genome Med. 5:22013. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhu XP, Liu ZL, Peng AF, Zhou YF, Long XH,
Luo QF, et al: Inhibition of Aurora-B suppresses osteosarcoma cell
migration and invasion. Exp Ther Med. 7:560–564. 2014.PubMed/NCBI
|
|
14
|
Liu P, Qi M, Ma C, Lao G and Liu Y: Let7a
inhibits the growth of endometrial carcinoma cells by targeting
Aurora-B. FEBS Lett. 587:2523–2529. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xie D, Shang C, Zhang H, Guo Y and Tong X:
Up-regulation of miR-9 target CBX7 to regulate invasion ability of
bladder transitional cell carcinoma. Med Sci Monit. 21:225–230.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang N, Kaur S, Volinia S, Greshock J,
Lassus H, Hasegawa K, et al: MicroRNA microarray identifies Let-7i
as a novel biomarker and therapeutic target in human epithelial
ovarian cancer. Cancer Res. 68:10307–10314. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Balakathiresan N, Bhomia M, Chandran R,
Chavko M, McCarron RM and Maheshwari RK: MicroRNA let-7i is a
promising serum biomarker for blast-induced traumatic brain injury.
J Neurotrauma. 29:1379–1387. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu K, Qian T, Tang L, Wang J, Yang H and
Ren J: Decreased expression of microRNA let-7i and its association
with chemo-therapeutic response in human gastric cancer. World J
Surg Oncol. 10:2252012. View Article : Google Scholar
|
|
19
|
Lai NS, Yu HC, Chen HC, Yu CL, Huang HB
and Lu MC: Aberrant expression of microRNAs in T cells from
patients with ankylosing spondylitis contributes to the
immunopathogenesis. Clin Exp Immunol. 173:47–57. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang P, Ma Y, Wang F, Yang J, Liu Z, Peng
J, et al: Comprehensive gene and microRNA expression profiling
reveals the crucial role of hsa-let-7i and its target genes in
colorectal cancer metastasis. Mol Biol Rep. 39:1471–1478. 2012.
View Article : Google Scholar
|
|
21
|
Zhang L and Zhang S: ZM447439, the Aurora
kinase B inhibitor, suppresses the growth of cervical cancer SiHa
cells and enhances the chemosensitivity to cisplatin. J Obstet
Gynaecol Res. 37:591–600. 2011. View Article : Google Scholar
|
|
22
|
Takeshita M, Koga T, Takayama K, Ijichi K,
Yano T, Maehara Y, et al: Aurora-B overexpression is correlated
with aneuploidy and poor prognosis in non-small cell lung cancer.
Lung Cancer. 80:85–90. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tuncel H, Shimamoto F, Kaneko Guangying Qi
H, Aoki E, Jikihara H, Nakai S, et al: Nuclear Aurora B and
cytoplasmic Survivin expression is involved in lymph node
metastasis of colorectal cancer. Oncol Lett. 3:1109–1114.
2012.PubMed/NCBI
|
|
24
|
Pohl A, Azuma M, Zhang W, Yang D, Ning Y,
Winder T, et al: Pharmacogenetic profiling of Aurora kinase B is
associated with overall survival in metastatic colorectal cancer.
Pharmacogenomics J. 11:93–99. 2011. View Article : Google Scholar
|
|
25
|
Qi G, Ogawa I, Kudo Y, Miyauchi M,
Siriwardena BS, Shimamoto F, et al: Aurora-B expression and its
correlation with cell proliferation and metastasis in oral cancer.
Virchows Arch. 450:297–302. 2007. View Article : Google Scholar : PubMed/NCBI
|