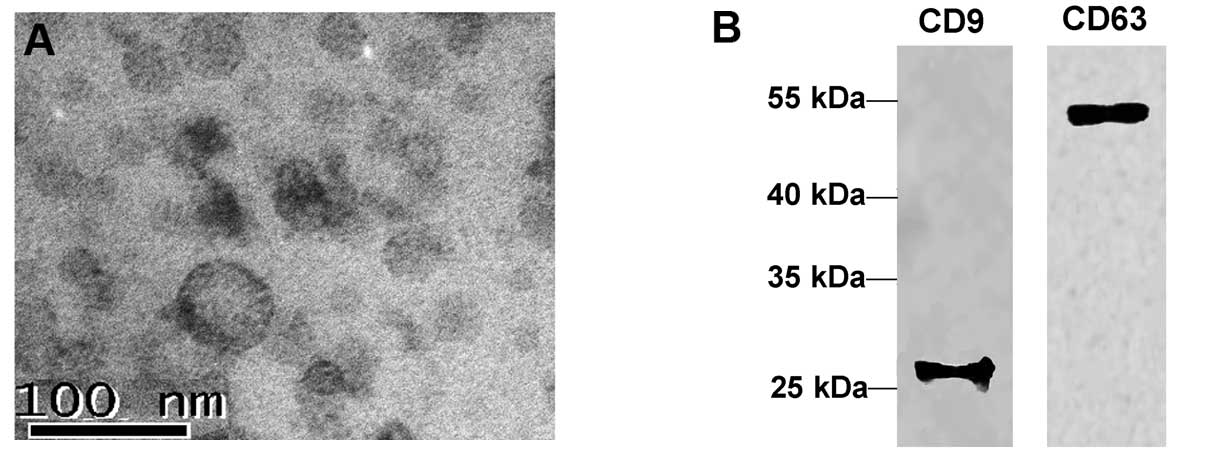
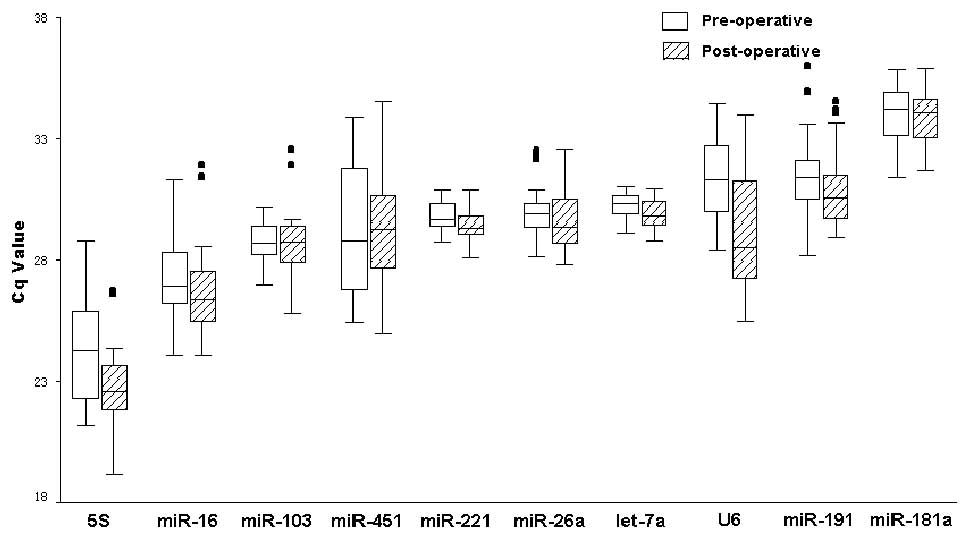
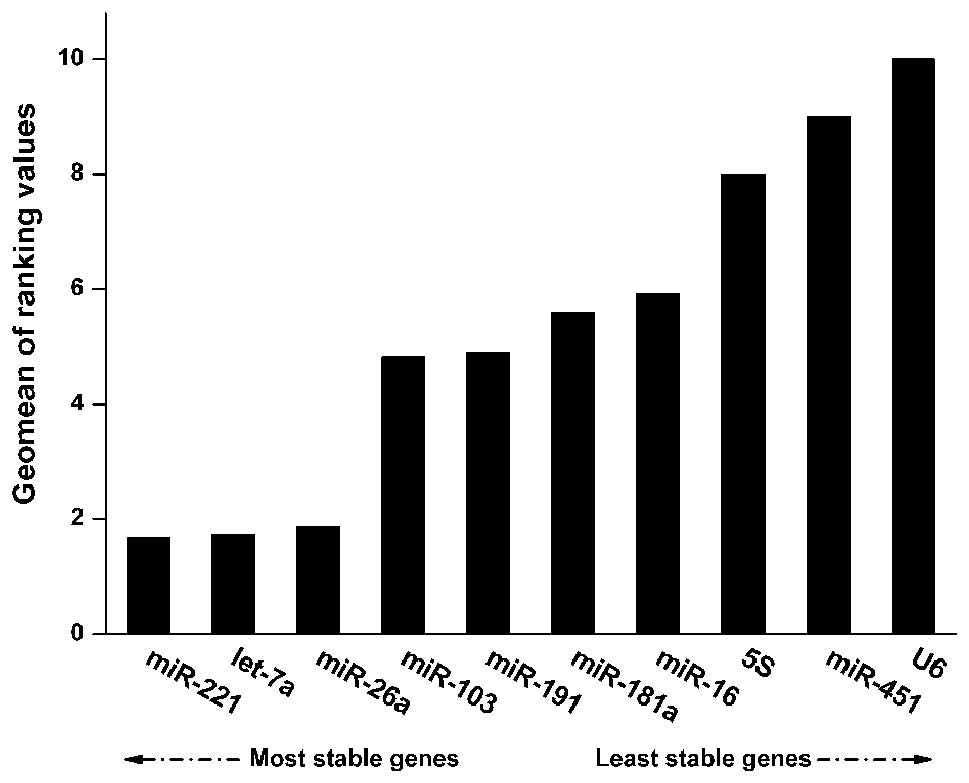
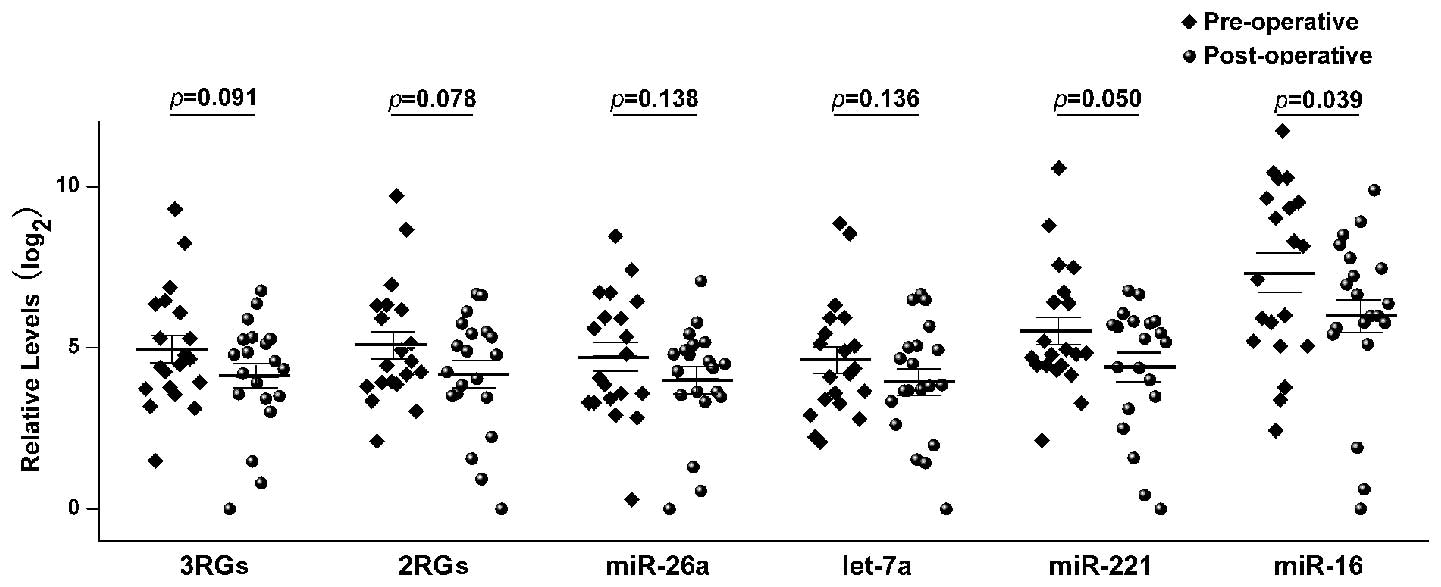
|
1
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li W, Chen YQ, Shen YB, et al: HIF-1α
knockdown by miRNA decreases survivin expression and inhibits A549
cell growth in vitro and in vivo. Int J Mol Med. 32:271–280.
2013.PubMed/NCBI
|
|
3
|
Vlassov AV, Magdaleno S, Setterquist R and
Conrad R: Exosomes: Current knowledge of their composition,
biological functions, and diagnostic and therapeutic potentials.
Biochim Biophys Acta. 1820:940–948. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Giusti I, D'Ascenzo S and Dolo V:
Microvesicles as potential ovarian cancer biomarkers. BioMed Res
Int. 2013:7030482013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gallo A, Tandon M, Alevizos I and Illei
GG: The majority of microRNAs detectable in serum and saliva is
concentrated in exosomes. PLoS One. 7:e306792012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Michael A, Bajracharya SD, Yuen PS, et al:
Exosomes from human saliva as a source of microRNA biomarkers. Oral
Dis. 16:34–38. 2010. View Article : Google Scholar :
|
|
7
|
Keller S, Ridinger J, Rupp AK, Janssen JW
and Altevogt P: Body fluid derived exosomes as a novel template for
clinical diagnostics. J Transl Med. 9:862011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wahlgren J, De L, Karlson T, Brisslert M,
et al: Plasma exosomes can deliver exogenous short interfering RNA
to monocytes and lymphocytes. Nucleic Acids Res. 40:e1302012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Taylor DD and Gercel-Taylor C: MicroRNA
signatures of tumor-derived exosomes as diagnostic biomarkers of
ovarian cancer. Gynecol Oncol. 110:13–21. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rabinowits G, Gerçel-Taylor C, Day JM,
Taylor DD and Kloecker GH: Exosomal microRNA: A diagnostic marker
for lung cancer. Clin Lung Cancer. 10:42–46. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bala S, Petrasek J, Mundkur S, et al:
Circulating microRNAs in exosomes indicate hepatocyte injury and
inflammation in alcoholic, drug-induced, and inflammatory liver
diseases. Hepatology. 56:1946–1957. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Murakami Y, Toyoda H, Tanahashi T, et al:
Comprehensive miRNA expression analysis in peripheral blood can
diagnose liver disease. PLoS One. 7:e483662012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tang ZY, Ye SL, Liu YK, et al: A decade's
studies on metastasis of hepatocellular carcinoma. J Cancer Res
Clin Oncol. 130:187–196. 2004. View Article : Google Scholar
|
|
15
|
Lin Q, Mao W, Shu Y, et al: A cluster of
specified microRNAs in peripheral blood as biomarkers for
metastatic non-small-cell lung cancer by stem-loop RT-PCR. J Cancer
Res Clin Oncol. 138:85–93. 2012. View Article : Google Scholar :
|
|
16
|
Peltier HJ and Latham GJ: Normalization of
microRNA expression levels in quantitative RT-PCR assays:
Identification of suitable reference RNA targets in normal and
cancerous human solid tissues. RNA. 14:844–852. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhu HT, Dong QZ, Wang G, et al:
Identification of suitable reference genes for qRT-PCR analysis of
circulating microRNAs in hepatitis B virus-infected patients. Mol
Biotechnol. 50:49–56. 2012. View Article : Google Scholar
|
|
18
|
Lardizábal MN, Nocito AL, Daniele SM,
Ornella LA, Palatnik JF and Veggi LM: Reference genes for real-time
PCR quantification of microRNAs and messenger RNAs in rat models of
hepato-toxicity. PLoS One. 7:e363232012. View Article : Google Scholar
|
|
19
|
Xu J, Wu C, Che X, et al: Circulating
microRNAs, miR-21, miR-122, and miR-223, in patients with
hepatocellular carcinoma or chronic hepatitis. Mol Carcinog.
50:136–142. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Blondal T, Jensby Nielsen S, Baker A, et
al: Assessing sample and miRNA profile quality in serum and plasma
or other biofluids. Methods. 59:S1–S6. 2013. View Article : Google Scholar
|
|
21
|
Qi P, Cheng SQ, Wang H, Li N, Chen YF and
Gao CF: Serum microRNAs as biomarkers for hepatocellular carcinoma
in Chinese patients with chronic hepatitis B virus infection. PLoS
One. 6:e284862011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Théry C, Amigorena S, Raposo G and Clayton
A: Isolation and characterization of exosomes from cell culture
super-natants and biological fluids. Curr Protoc Cell Biol Chapter
3. Unit 3.22. 2006. View Article : Google Scholar
|
|
23
|
Pfaffl MW: A new mathematical model for
relative quantification in real-time RT-PCR. Nucleic Acids Res.
29:e452001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bustin SA, Benes V, Garson JA, et al: The
MIQE guidelines: Minimum information for publication of
quantitative real-time PCR experiments. Clin Chem. 55:611–622.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Vandesompele J, De Preter K, Pattyn F, et
al: Accurate normalization of real-time quantitative RT-PCR data by
geometric averaging of multiple internal control genes. Genome
Biol. 3:RESEARCH0034. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Andersen CL, Jensen JL and Ørntoft TF:
Normalization of real-time quantitative reverse transcription-PCR
data: A model-based variance estimation approach to identify genes
suited for normalization, applied to bladder and colon cancer data
sets. Cancer Res. 64:5245–5250. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pfaffl MW, Tichopad A, Prgomet C and
Neuvians TP: Determination of stable housekeeping genes,
differentially regulated target genes and sample integrity:
BestKeeper - Excel-based tool using pair-wise correlations.
Biotechnol Lett. 26:509–515. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Silver N, Best S, Jiang J and Thein SL:
Selection of housekeeping genes for gene expression studies in
human reticulocytes using real-time PCR. BMC Mol Biol. 7:332006.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xie F, Xiao P, Chen D, Xu L and Zhang B:
miRDeepFinder: A miRNA analysis tool for deep sequencing of plant
small RNAs. Plant Mol Biol. 80:75–84. 2012. View Article : Google Scholar
|
|
30
|
Silverman JM and Reiner NE: Exosomes and
other microvesicles in infection biology: Organelles with
unanticipated phenotypes. Cell Microbiol. 13:1–9. 2011. View Article : Google Scholar
|
|
31
|
Pols MS and Klumperman J: Trafficking and
function of the tetraspanin CD63. Exp Cell Res. 315:1584–1592.
2009. View Article : Google Scholar
|
|
32
|
Mathivanan S and Simpson RJ: ExoCarta: A
compendium of exosomal proteins and RNA. Proteomics. 9:4997–5000.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mathivanan S, Ji H and Simpson RJ:
Exosomes: Extracellular organelles important in intercellular
communication. J Proteomics. 73:1907–1920. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Song J, Bai Z, Han W, et al:
Identification of suitable reference genes for qPCR analysis of
serum microRNA in gastric cancer patients. Dig Dis Sci. 57:897–904.
2012. View Article : Google Scholar
|
|
35
|
Li J, Wei H, Li Y, Li Q and Li N:
Identification of a suitable endogenous control gene in porcine
blastocysts for use in quantitative PCR analysis of microRNAs. Sci
China Life Sci. 55:126–131. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bustin SA and Nolan T: Pitfalls of
quantitative real-time reverse-transcription polymerase chain
reaction. J Biomol Tech. 15:155–166. 2004.PubMed/NCBI
|
|
37
|
Dheda K, Huggett JF, Chang JS, et al: The
implications of using an inappropriate reference gene for real-time
reverse transcription PCR data normalization. Anal Biochem.
344:141–143. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Qu KZ, Zhang K, Li HR, Afdhal NH and
Albitar M: Circulating microRNAs as biomarkers for hepatocellular
carcinoma. J Clin Gastroenterol. 45:355–360. 2011. View Article : Google Scholar : PubMed/NCBI
|