Introduction
Human cytomegalovirus (HCMV), a member of the herpes
virus family, is the leading viral cause of congenital
abnormalities and mental impairment in newborns, and contributes
significantly to morbidity and mortality rates in
immunocom-promised persons. HCMV can be transmitted via multiple
pathways, including saliva, sexual contact, placental transfer,
breast-feeding and blood transfusion (1). Over one-third of transplant
recipients are infected with HCMV, and the mortality rate of these
patients is as high as 85%, and 40% of patients with acquired
immunodeficiency syndrome, who have a secondary infection of HCMV
exhibit retinopathy, gastrointestinal damage and central nervous
system disorders (2,3). In addition, latent HCMV infection is
closely associated with cardiovascular disease, stroke, diabetes
and other chronic diseases (4,5).
However, the cellular and molecular mechanisms of the persistent
infection, latency and reactivation of HCMV remain to be fully
elucidated (2). Following the
widespread introduction of anti-HCMV drugs, including ganciclovir,
valganciclovir and foscarnet, there has been a significant
reduction in the number of HCMV infections, however, these drugs
have been demonstrated to cause potentially serious side effects,
exhibit low bioavailability and the inability to eliminate carrier
states, and issues with drug resistance, patient compliance and
cost, further limiting their effectiveness and wide application
(3). Therefore, the development of
safe and effective anti-HCMV drugs with novel mechanisms of action
is urgently required.
Curcumin is a traditional Chinese medicine, which is
extracted from the rhizome of the herb Curcuma longa
(6). Curcumin has a low toxicity
and has been widely used for the prevention of viral infection and
other diseases, including cancer (7–9). Our
previous study demonstrated that curcumin has significant anti-HCMV
activity and improves the survival rate of host cells by inhibiting
the HCMV DNA replication, apoptosis and inflammation induced by
HCMV infection (10). However, the
mechanism of anti-HCMV activity of curcumin remains to be fully
elucidated.
In the development of novel drugs, bioinformatics is
capable of identifying the key molecular targets in the
pathological process and confirming the association between the
structure and function of the drug. Bioinformatics has now become
an important biopharmaceutical tool, which reduces the requirement
for manpower and resources. The widespread application of
bioinformatics has significantly accelerated the understanding and
the development of novel drugs for several types of disease
(11). Our previous studies have
successfully applied bioinformatics to screen drugs for the
treatment of leukemia (12) and
aplastic anemia (13). In the
present study, the anti-HCMV mechanism of curcumin was determined
using bioinformatics, which was further confirmed by subsequent
experiments.
Materials and methods
Reagents and kits
Dulbecco's modified Eagle's medium (DMEM), fetal
bovine serum (FBS), SYBR Green, penicillin, streptomycin,
L-glutamine, trypsin, dimethyl sulfoxide (DMSO), Triton X-100,
phenylmethanesulfonyl fluoride (PMSF) and β-mercaptoethanol were
obtained from Sigma-Aldrich (St. Louis, MO, USA). Rabbit anti-heat
shock protein 90 (Hsp90) antibody was purchased from Cell Signaling
Technology (Boston, MA, USA). Goat anti-rabbit secondary antibody
was obtained from Invitrogen Life Technologies (Carlsbad, CA,
USA).
Cell culture and virus propagation
Human embryonic lung fibroblast (HELF) cells were
purchased from the National Platform of Experimental Cell Resources
for Sci-Tech (Beijing, China) and were cultured in DMEM,
supplemented with 10% FBS, 100 U/m penicillin, 100 µg/ml
streptomycin, and 2 mM L-glutamine at 37°C in a humidified
atmosphere of 95% air and 5% CO2. The HCMV laboratory
strain, AD169, was purchased from American Type Culture Collection
(Manassas, VA, USA) and prepared in the HELF cells maintained in
DMEM, containing 2% FBS (maintenance medium).
Antiviral compounds
Curcumin was obtained from the National Institute
for Food and Drug Control (Beijing, China) and stored as a 50 mM
stock solution in DMSO for in vitro use. The high, middle
and low doses of curcumin used in the present study were 2, 1 and
0.5 µM, respectively. Intravenous formulations of 0.2 mM
ganciclovir (GCV; Sigma-Aldrich) and 0.5 µM geldanamycin
(Sigma-Aldrich) in DMSO were used as references.
Drug genome similarity analysis of
transcription
The mechanism of action of curcumin was predicted
from pharmacogenomics databases, using bioinformatics to perform
genome-wide transcriptional analysis of similarity. Using curcumin
as a keyword, associated gene expression information was identified
in various gene expression databases, including Gene Expression
Omnious, BodyMap (Human and Mouse gene expression database),
ArrayExpress Database (Public repository for microarray-based gene
expression data), RNA Abundance Database, Stanford Microarray
Database, Human Gene Expression and ChipDB, a searchable database
of gene expression. These databases are open access database
resources and can be accessed online. According to the data in the
GenBank and internet platforms, the cloned drug genome sequences of
curcumin were transferred to the above online databases to
determine the expression information.
Molecular docking simulation
ChemBio Office (2010) was used to draw the molecular
structure of curcumin, and the resultant files were saved in the
standard delay format. MOE (2010) was applied to analyze the
three-dimensional minimum energy conformations. The docking
parameters in MOE were set as follows: The placement method, the
first scoring function rescoring 1 and the saved poses were
Triangle Matcher, London dG and 30, respectively. In addition, the
refinement, the second refinement scoring function rescoring 2 and
the saved poses were set to force field, none and 10, respectively
(7).
Western blotting
The HELF cells were seeded at a density of
1×106/ml in tissue culture flasks and cultured for 12 h.
The cells were subsequently infected with HCMV at 100 times the
median tissue culture infective dose/0.1 ml. Following incubation
at 37°C for 2 h under a humidified atmosphere of 95% air and 5%
CO2, the supernatant was removed and replaced with
maintenance medium, with or without curcumin. Following an
additional 48 h at 37°C, the cells were harvested and washed twice
with cold phosphate-buffered saline and lysed in cold
radioimmunoprecipitation buffer, containing 25 mM Tris-HCl (pH
7.6), 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 1 mM PMSF and
0.1% sodium dodecyl sulfate (SDS), on ice for 30 min, followed by
centrifugation at 9,600 × g at 4°C for 10 min. Protein
quantification was performed using a bicinchoninic acid protein
assay (Pierce, Rockford, IL, USA). Equal quantities of protein (50
µg) from the different groups were denatured in SDS sample
buffer and separated by 8% SDS-polyacrylamide gel electrophoresis.
The proteins were transferred onto polyvinylidene difluoride
membranes in buffer, containing 25 mM Tris, 192 mM glycine and 20%
methanol. The membranes were blocked with Tris-buffered saline,
containing TBS; 137 mM NaCl and 20 mM Tris-HCl (pH 7.5), containing
0.1% Tween 20 and 5% dried non-fat milk powder at room temperature
for 2 h. Following blocking, the blots were incubated with primary
antibodies against rabbit anti-human Hsp 90 (mAb; Cell Signaling
Technologies, Danvers, MA, USA; cat. no. 4877) and mouse anti-human
β-actin (mAb, ZSJB-Bio, Beijing, China; cat. no. 131023) at 1:1,000
in TBS containing 0.05% Tween-20 (TBST) at 4°C overnight. The
membranes were subsequently washed with TBST three times. The blots
were then incubated with Alexa Fluor®680 goat anti-mouse
IgG and goat anti-rabbit IgG at 1:10,000 (Invitrogen Life
Technologies, Grand Island, NY, USA) in TBST. After washing the
membranes with TBS, the signals were detected using Odyssey
infrared laser imaging system (LI-COR Biosciences, Inc,. Lincoln,
NE, USA), according to the manufacturer's instructions. For
densitometric analysis of western blotting images, density profiles
of the bands were measured using ImageJ software (National
Institute of Health, Bethesda, MA, USA).
Statistical analysis
The data were analyzed using Microsoft Office Excel
2003 (Microsoft, Redmond, WA, USA) and expressed as the mean ±
standard error of the mean. Group comparisons were evaluated using
one-way analysis of variance, and significant differences between
treatments were determined using the post-hoc test with
Tukey-Kramer's honestly significant difference simultaneous
pairwise mean comparison. P<0.05 was considered to indicate a
statistically significant difference.
Results
Curcumin genome similarity analysis of
transcription
To investigate the pharmacogenomics of curcumin, a
comparative analysis of 3,000 clinical drugs was performed, which
revealed that the gene expression profiles of curcumin had a
positive association with several drugs (Table I). Among these drugs, the
pharmacogenomics of curcumin were most similar to that of the
antiviral drug, geldanamycin. Geldanamycin exerts an antiviral
effect, predominantly by specifically binding to Hsp90 (14). This analysis suggested that
curcumin may inhibit HCMV by targeting Hsp90.
 | Table ICurcumin pharmacogenomics to analyze
the similarity of transcription. |
Table I
Curcumin pharmacogenomics to analyze
the similarity of transcription.
| Drug | Number of similar
gene expression profiles |
|---|
| Geldanamycin | 10 |
| Astemizole | 3 |
| Mefloquine | 3 |
| Thioridazine | 8 |
| Withaferin A | 2 |
| Vorinostat | 9 |
Molecular docking simulation of curcumin
with Hsp90
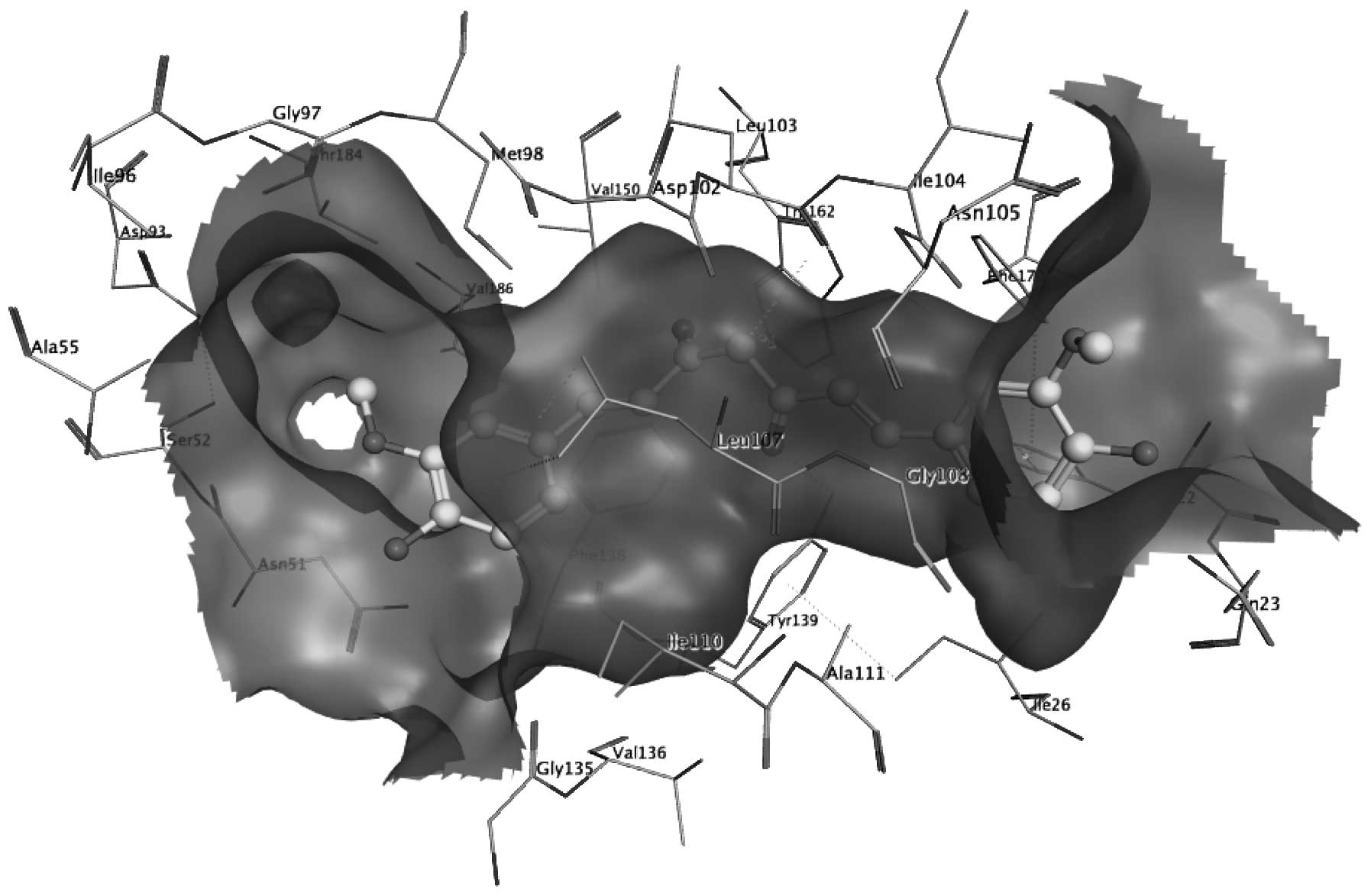
To confirm the above hypothesis, a molecular docking
simulation of curcumin with Hsp90 was performed, which revealed
that curcumin fit well into the binding pocket of Hsp90, among
which hydrogen bonds, hydrophobic interactions and conjugation
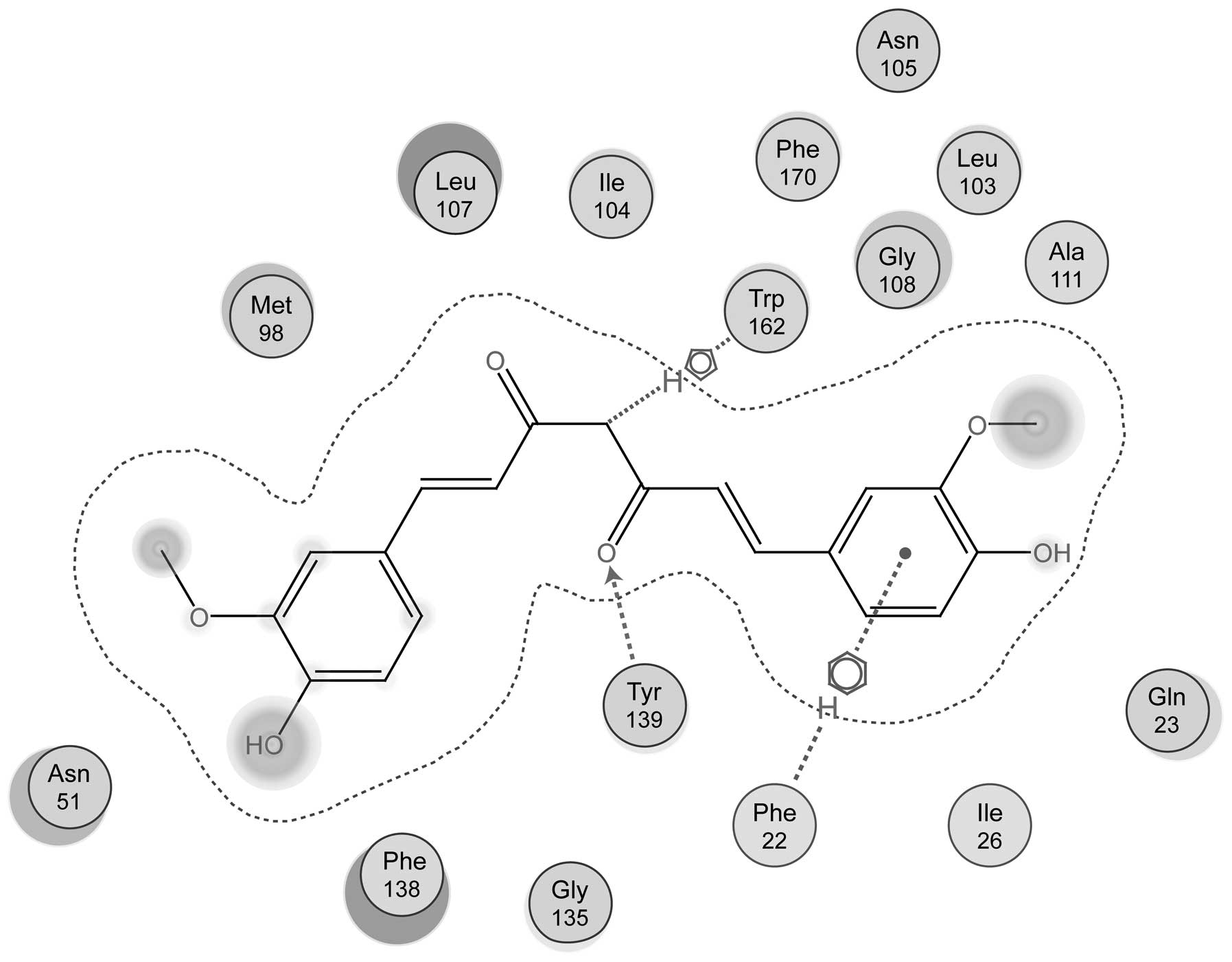
maintained the predominant driving force of adhesion (Fig. 1). As shown in Fig. 2, a carbonyl oxygen atom of curcumin
acted as a hydrogen bond acceptor to form a hydrogen bond with
Tyr139 of Hsp90. In addition, Trp162 and Phe22 of Hsp90 were
conjugated with curcumin (Fig. 2).
Furthermore, Ile26, Leu107, Ile104, Phe170, Leu103, Ala111 and
other hydrophobic amino acids of Hsp90 formed hydrophobic
interactions with curcumin (Fig.
2).
Curcumin reduces the protein expression
of Hsp90α
The present study investigated the effect of
curcumin on the protein expression of Hsp90α using western
blotting. In contrast to the mock-treated cells, infection with
HCMV in the curcumin-treated cells led to a significant increase in
the protein expression of Hsp90α (Fig.
3; P<0.05). As expected, treatment with geldanamycin, but
not GCV, markedly eliminated the induction of the protein
expression of Hsp90α by HCMV. Notably, curcumin also inhibited the
upregulated protein expression of Hsp90α by HCMV in a
dose-dependent manner (Fig.
3).
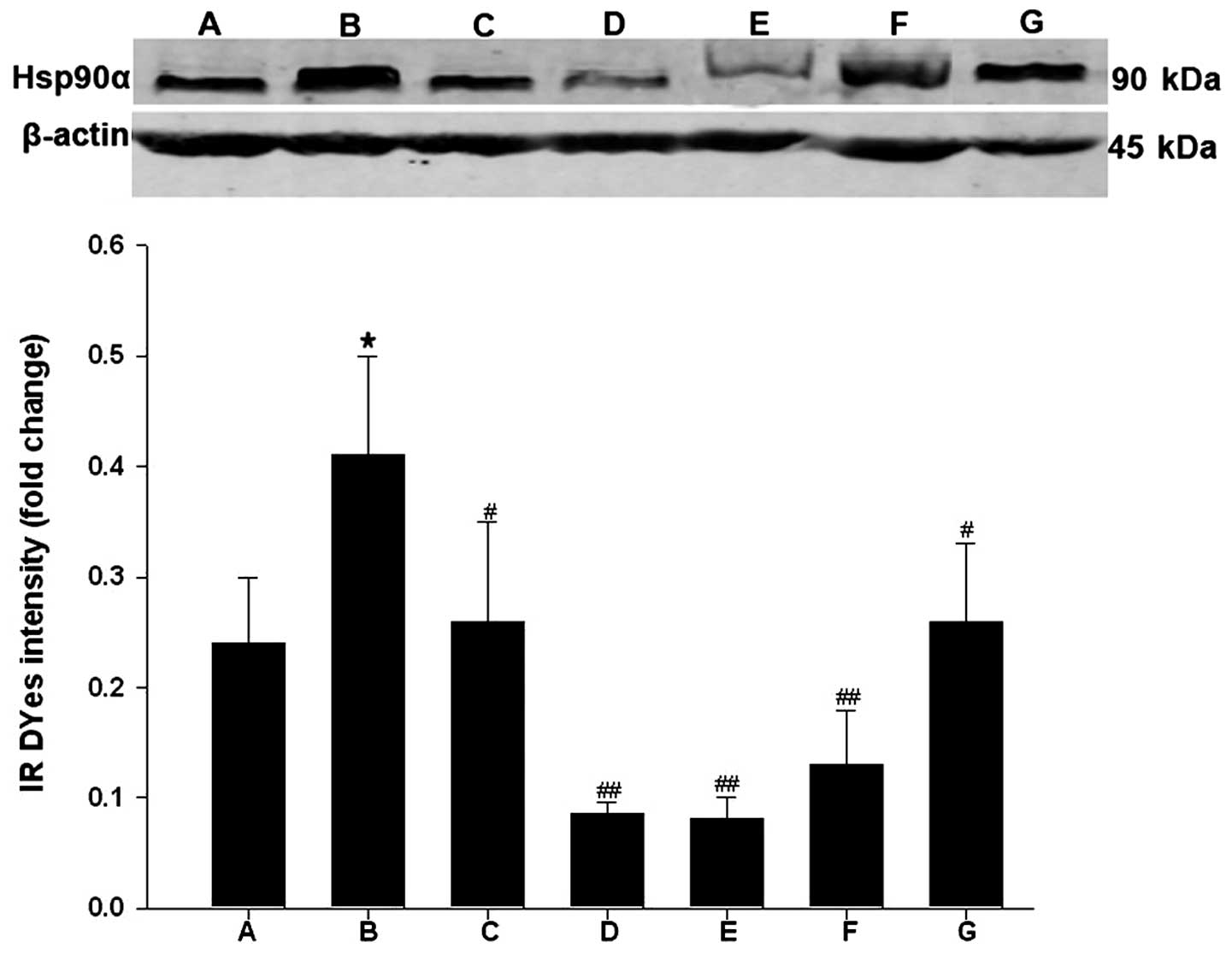 | Figure 3Curcumin decreases the protein
expression of Hsp90α. The HELF cells were treatment groups were as
follows: (A) HELF cells (control, mock treated); (B) HELF cells +
HCMV (model); (C) HELF cells + HCMV + GCV; (D) HELF cells + HCMV +
geldanamycin; (E) HELF cells + HCMV + curcumin (high dose, 0.8
µg/ml); (F) HELF cells + HCMV + curcumin (middle dose, 0.4
µg/ml); (G) HELF cells + HCMV + curcumin (low dose, 0.2
µg/ml). The protein expression of Hsp90α was determined
using western blotting, and relative protein expression levels were
normalized against β-actin and expressed as the fold change.
*P<0.05, compared with the control;
##P<0.01 and #P<0.05, compared with
HCMV infection. Data are expressed as the mean ± standard error of
the mean. HELF, Human embryonic lung fibroblast; HCMV, human
cytomegalovirus; Hsp90α, heat shock protein 90α. |
Discussion
In the present study, it was demonstrated that the
pharmacogenomics of curcumin were most similar to that of
geldanamycin, which is an inhibitor of Hsp90 and exerts a
wide-spectrum antiviral role (15). The in vitro experiments
revealed that curcumin significantly inhibited the protein
expression of Hsp90α. This data suggested that the inhibition of
Hsp90 may be one of the mechanisms by which curcumin inhibits HCMV
and improves the survival rate of the host cells.
Hsp90 is a highly conserved and essential molecular
chaperone at the center of a large protein-folding network
(16). It is one of the most
abundant chaperone proteins in eukaryotic and prokaryotic cells,
and is formed by Hsp90α and Hsp90β monomer polymerization in higher
eukaryotic organisms (17–19). Dysregulation of Hsp90α is closely
associated with the occurrence and development of different types
of cancer (17). Therefore,
targeting Hsp90 is a promising strategy for the development
anticancer drugs.
Hsp90 is a leading member of the heat shock protein
family and can form complexes with a variety of proteins. Hsp90 is
involved in improving cellular stress tolerance (18), regulating protein conformations
(19) and in client protein
maturation following transduction (20). Accumulating evidence has
demonstrated that Hsp90 and its client proteins cannot only alter
the micro-environment of a cell through involvement in cell
signaling transduction, cell cycle regulation and cell apoptosis
(21), but they also promote viral
infections of herpes simplex virus 1, Epstein-Barr virus or
intestinal virus by being involved in viral penetration (22), DNA polymerase positioning (14), expression of key proteins (23) and DNA injury repair (24). In agreement with these
observations, it was previously revealed that curcumin inhibits the
proliferation of HCMV and increases the survival rate of host cells
by inhibiting HCMV DNA replication, apoptosis and inflammation
(10). The present study used
molecular docking simulation analysis to demonstrate that curcumin
fit well into the binding pocket of Hsp90. Notably, curcumin
eliminated the induction of Hsp90α by HCMV infection in the HELF
cells, indicating that Hsp90 is involved in the process of viral
infection (25–27), and targeting Hsp90 offers a novel
promising strategy for the prevention and treatment of infection
with viruses, including HCMV.
In conclusion, the present study indicated that the
anti-HCMV activity of curcumin is possibly due to its inhibitory
effects on Hsp90α. These results suggested that Hsp90 may be a
novel target for the treatment of infectious diseases caused by
HCMV.
Acknowledgments
This study was supported by the Medjaden Academy
& Research Foundation for Young Scientists (grant. no.
MJR20140019) and the Beijing Natural Science Foundation (grant.
nos. 7151004 and 7153165). The authors would like to thank Medjaden
Bioscience Limited (Hong Kong, China) for assisting in the
preparation of this manuscript.
References
|
1
|
Weisblum Y, Panet A, Zakay-Rones Z,
Haimov-Kochman R, Goldman-Wohl D, Ariel I, Falk H, Natanson-Yaron
S, Goldberg MD, Gilad R, Lurain NS, Greenfield C, Yagel S and Wolf
DG: Modeling of human cytomegalovirus maternal-fetal transmission
in a novel decidual organ culture. J Virol. 85:13204–13213. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Reuter JD, Gomez DL, Wilson JH and Van Den
Pol AN: Systemic immune deficiency necessary for cytomegalovirus
invasion of the mature brain. J Virol. 78:1473–1487. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Imai Y, Shum C, Martin DF, Kuppermann BD,
Drew WL and Margolis TP: Emergence of drug-resistant
cytomegalovirus retinitis in the contralateral eyes of patients
with AIDS treated with ganciclovir. J Infect Dis. 189:611–615.
2004. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li S, Zhu J, Zhang W, Chen Y, Zhang K,
Popescu LM, Ma X, Lau WB, Rong R, Yu X, et al: Signature microRNA
expression profile of essential hypertension and its novel link to
human cytomegalovirus infection. Circulation. 124:175–184. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Huang ZR, Yu LP, Yang XC, Zhang F, Chen
YR, Feng F, Qian XS and Cai J: Human cytomegalovirus linked to
stroke in a Chinese population. CNS Neurosci Ther. 18:457–460.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shankar TN, Shantha NV, Ramesh HP, Murthy
IA and Murthy VS: Toxicity studies on turmeric (Curcuma longa):
Acute toxicity studies in rats, guineapigs & monkeys. Indian J
Exp Biol. 18:73–75. 1980.PubMed/NCBI
|
|
7
|
Vallee F, Carrez C, Pilorge F, Dupuy A,
Parent A, Bertin L, Thompson F, Ferrari P, Fassy F, Lamberton A,
Thomas A, et al: Tricyclic series of heat shock protein 90 (Hsp90)
inhibitors part I: Discovery of tricyclic imidazo [4,5-c] pyridines
as potent inhibitors of the Hsp90 molecular chaperone. J Med Chem.
54:7206–7219. 2011. View Article : Google Scholar
|
|
8
|
Stolina M, Sharma S, Lin Y, Dohadwala M,
Gardner B, Luo J, Zhu L, Kronenberg M, Miller PW, Portanova J, et
al: Specific inhibition of cyclooxygenase 2 restores antitumor
reactivity by altering the balance of IL-10 and IL-12 synthesis. J
Immunol. 164:361–370. 2000. View Article : Google Scholar
|
|
9
|
Pfaffl MW: A new mathematical model for
relative quantification in real-time RT-PCR. Nucleic Acids Res.
29:e452001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lv YL, Lan AJ, Lan YY, Li C, Lei N, Wu C
and Liu LH: Activity of curcumin against human cytomegalovirus in
vitro. Afr J Pharm Pharmacol. 6:30–35. 2012.
|
|
11
|
Austen M and Dohrmann C: Phenotype-first
screening for the identification of novel drug targets. Drug Discov
Today. 10:275–282. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lu XC, Chi XH, Yang B, Zhu HL, Liu LH,
Zhang F and Yan JW: Gene expression profile analysis of T
lymphocytes involved in pathogenesis of severe aplastic anemia by
using bioinformatics method as a novel way of drug screening.
Zhongguo Shi Yan Xue Ye Xue Za Zhi. 18:416–420. 2010.In Chinese.
PubMed/NCBI
|
|
13
|
Yang B, Lu XC, Liu LH, Zhu HL, Chi XH, Yao
SQ, Lou FD and Yu L: Experimental identification of drugs with
function of targeted up-regulating ID4 expression:
Bioinformatics-based prediction and preliminary validation.
Zhonghua Yi Xue Za Zhi. 89:1714–1716. 2009.In Chinese. PubMed/NCBI
|
|
14
|
Burch AD and Weller SK: Herpes simplex
virus type 1 DNA polymerase requires the mammalian chaperone hsp90
for proper localization to the nuleus. J Virol. 79:10740–10749.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Grenert JP, Sullivan WP, Fadden P,
Haystead TA, Clark J, Mimnaugh E, Krutzsch H, Ochel HJ, Schulte TW,
Sausville E, et al: The amino-terminal domain of heat shock protein
90 (hsp90) that binds geldanamycin is an ATP/ADP switch domain that
regulates hsp90 conformation. J Biol Chem. 272:23843–23850. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Geller R, Andino R and Frydman J: Hsp90
inhibitors exhibit resistance-free antiviral activity against
respiratory syncytial virus. PLoS One. 8:e567622013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cheng CF, Fan J, Zhao ZW, Woodley DT and
Li W: Secreted heat shock protein-90a: A more effective and safer
target and for anti-cancer drags. Curr Signal Tranduction Ther.
5:121–127. 2010. View Article : Google Scholar
|
|
18
|
Zhai QQ, Luo Y, Wang QD, Xia M, Xing GW,
Li YC, Jiang JH, Liu Z, Liu QY, Wang YF, et al: Determination of
SNX-2112, a selective Hsp90 inhibitor, in plasma samples by
hish-performance liquid chromatography and its application to
pharmacokineties in rats. J Pharm Biomed Anal. 53:1048–1052. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jiang S QL, Zhou F, Huang J, Guo Y and
Yang K: Molecular cloning and expression analysis of a heat shock
protein (Hsp90) gene from black tiger shrimp (Penaeus monodon). Mol
Biol Rep. 36:127–134. 2009. View Article : Google Scholar
|
|
20
|
Meng S SG and Li ZR: Recent progress in
the study of Hsp90 inhibitors. Chin J Antibiot. 36:241–248.
2011.
|
|
21
|
Wen KW and Damania B: Hsp90 and
Hsp40/Erdj3 are required for the expression and anti-apoptotic
function of KSHV K1. Oncogene. 29:3532–3544. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tsou YL, Lin YW, Chang HW, Lin HY, Shao
HY, Yu SL, Liu CC, Chitra E, Sia C and Chow YH: Heat shock protein
90: Role in enterovirus 71 entry and assembly and potential target
for therapy. PLoS One. 8:e771332013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Murata T, Iwata S, Siddiquey MN, Kanazawa
T, Goshima F, Kawashima D, Kimura H and Tsurumi T: Heat shock
protein 90 inhibitors repress latent membrane protein 1 (LMP1)
expression and proliferation of Epstein-Barr virus-positive natural
killer cell lymphoma. PLoS One. 8:e635662013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Luo Y, Lou S, Deng X, Liu Z, Li Y,
Kleiboeker S and Qiu J: Parvovirus B19 infection of human primary
erythroid progenitor cells triggers ATR-Chk1 signaling, which
promotes B19 virus replication. J Virol. 85:8046–8055. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Geller R, Vignuzzi M, Andino R and Frydman
J: Evolutionary constraints on chaperone-mediated folding provide
an antiviral approach refractory to development of drug resistance.
Genes Dev. 21:195–205. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Turnell AS and Grand RJ: DNA viruses and
the cellular DNA-damage response. J Gen Virol. 93:2076–2097. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xiao A, Wong J and Luo H: Viral
interaction with molecular chaperones: Role in regulating viral
infection. Arch Virol. 155:1021–1031. 2010. View Article : Google Scholar : PubMed/NCBI
|