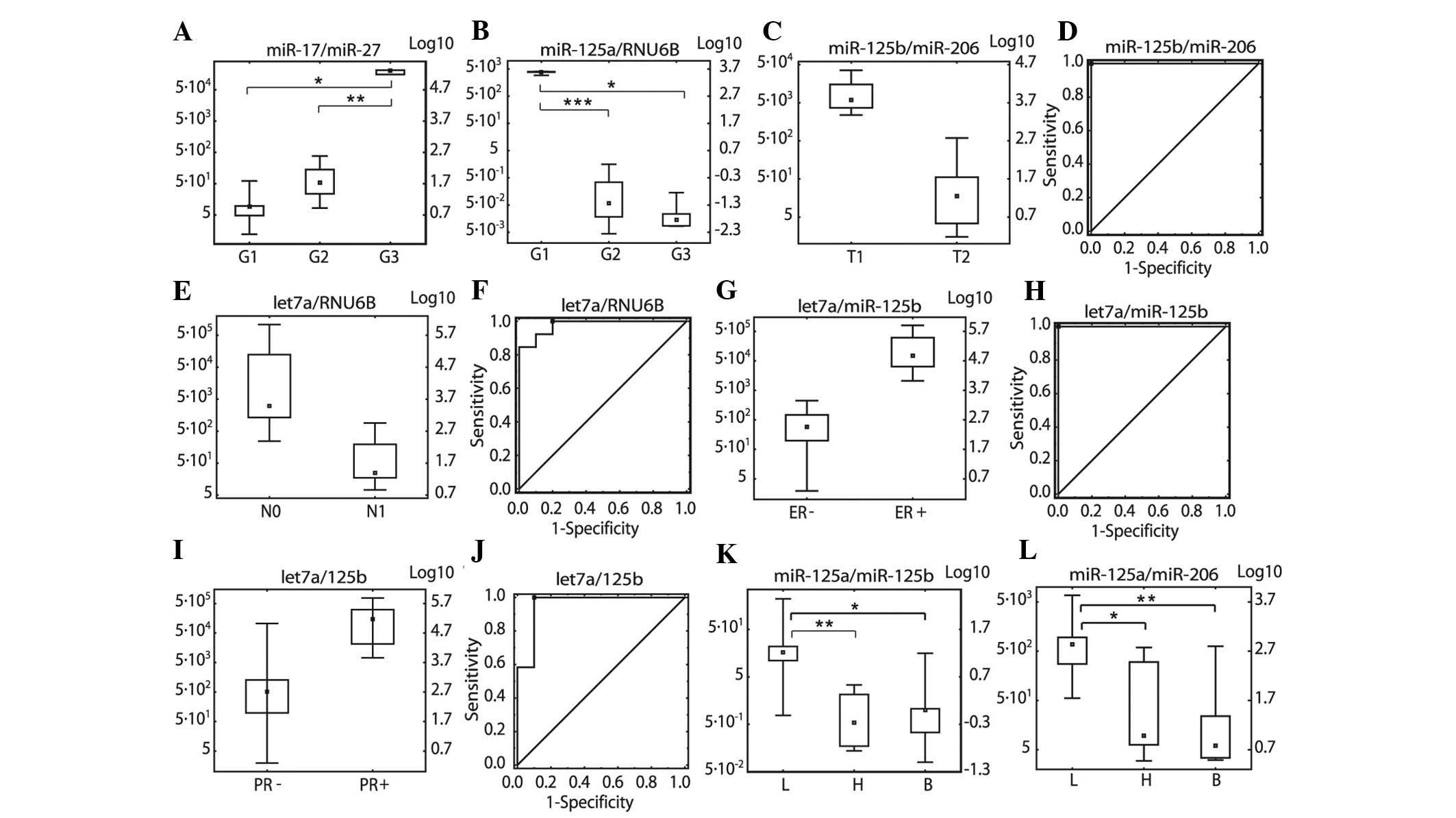
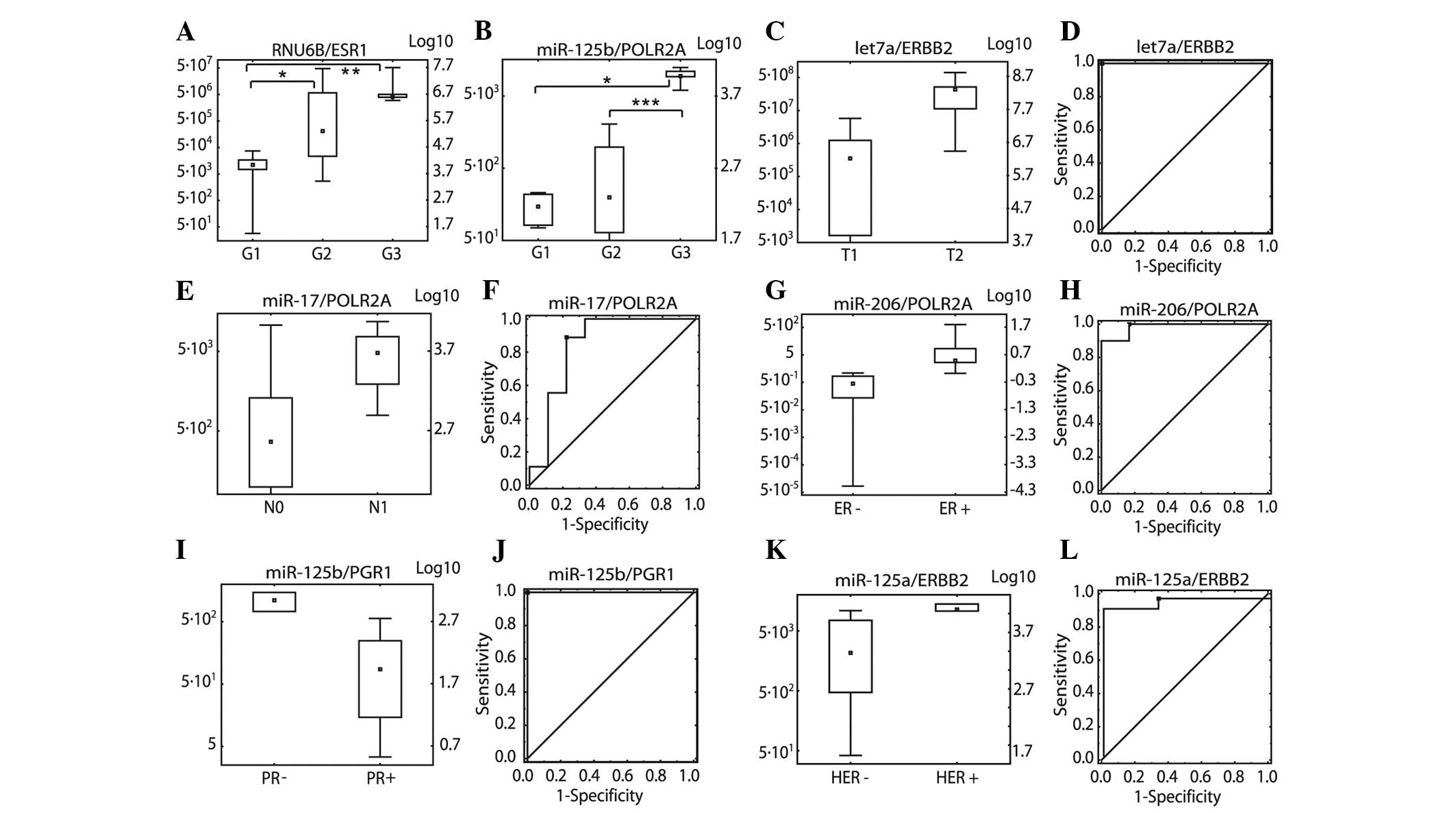
|
1
|
Ebert MS and Sharp PA: Roles for microRNAs
in conferring robustness to biological processes. Cell.
149:515–524. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dvinge H, Git A, Graf S, et al: The
shaping and functional consequences of the microRNA landscape in
breast cancer. Nature. 497:378–382. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mogilyansky E and Rigoutsos I: The
miR-17/92 cluster: a comprehensive update on its genomics,
genetics, functions and increasingly important and numerous roles
in health and disease. Cell Death Differ. 20:1603–1614. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Croft L, Szklarczyk D, Jensen LJ, et al:
Multiple independent analyses reveal only transcription factors as
an enriched functional class associated with microRNAs. BMC Syst
Biol. 6:902012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lehmann U: Aberrant DNA methylation of
microRNA genes in human breast cancer-a critical appraisal. Cell
Tissue Res. 356:657–664. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Castells A, Gusella JF, Ramesh V, et al: A
region of deletion on chromosome 22q13 is common to human breast
and colorectal cancers. Cancer Res. 60:2836–2839. 2000.PubMed/NCBI
|
|
7
|
Hu X, Guo J, Zheng L, et al: The
heterochronic microRNA let-7 inhibits cell motility by regulating
the genes in the actin cytoskeleton pathway in breast cancer. Mol
Cancer Res. 11:240–250. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim SJ, Shin JY, Lee KD, et al: MicroRNA
let-7a suppresses breast cancer cell migration and invasion through
downregulation of C-C chemokine receptor type 7. Breast Cancer Res.
14:R142012. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang X, Cao L, Wang Y, et al: Regulation
of let-7 and its target oncogenes (Review). Oncol Lett. 3:955–960.
2012.PubMed/NCBI
|
|
10
|
Volinia S, Calin GA, Liu CG, et al: A
microRNA expression signature of human solid tumors defines cancer
gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Santos-Jr GC, Goes AC, Vitto H, et al:
Genomic instability at the 13q31 locus and somatic mtDNA mutation
in the D-loop site correlate with tumor aggressiveness in sporadic
Brazilian breast cancer cases. Clinics (Sao Paulo). 67:1181–1190.
2012. View Article : Google Scholar
|
|
12
|
Olive V, Li Q and He L: mir-17-92: a
polycistronic oncomir with pleiotropic functions. Immunol Rev.
253:158–166. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hossain A, Kuo MT and Saunders GF:
Mir-17-5p regulates breast cancer cell proliferation by inhibiting
translation of AIB1 mRNA. Mol Cell Biol. 26:8191–8201. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yu Z, Wang C, Wang M, et al: A cyclin
D1/microRNA 17/20 regulatory feedback loop in control of breast
cancer cell proliferation. J Cell Biol. 182:509–517. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu Z, Willmarth NE, Zhou J, et al:
microRNA 17/20 inhibits cellular invasion and tumor metastasis in
breast cancer by heterotypic signaling. Proc Natl Acad Sci USA.
107:8231–8236. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sinha S, Singh RK, Alam N, et al:
Alterations in candidate genes PHF2, FANCC, PTCH1 and XPA at
chromosomal 9q22.3 region: pathological significance in early- and
late-onset breast carcinoma. Mol Cancer. 7:842008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tsuchiya Y, Nakajima M, Takagi S, et al:
MicroRNA regulates the expression of human cytochrome P450 1B1.
Cancer Res. 66:9090–9098. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jin L, Wessely O, Marcusson EG, et al:
Prooncogenic factors miR-23b and miR-27b are regulated by Her2/Neu,
EGF and TNF-alpha in breast cancer. Cancer Res. 73:2884–2896. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Iorio MV, Ferracin M, Liu CG, et al:
MicroRNA gene expression deregulation in human breast cancer.
Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gentile M, Olsen K, Dufmats M, et al:
Frequent allelic losses at 11q24.1-q25 in young women with breast
cancer: association with poor survival. Br J Cancer. 80:843–849.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Feliciano A, Castellvi J, Artero-Castro A,
et al: miR-125b acts as a tumor suppressor in breast tumorigenesis
via its novel direct targets ENPEP, CK2-alpha, CCNJ and MEGF9. PLoS
One. 8:e762472013. View Article : Google Scholar
|
|
22
|
Serpico D, Molino L and Di Cosimo S:
microRNAs in breast cancer development and treatment. Cancer Treat
Rev. 40:595–604. 2014. View Article : Google Scholar
|
|
23
|
Scott GK, Goga A, Bhaumik D, et al:
Coordinate suppression of ERBB2 and ERBB3 by enforced expression of
micro-RNA miR-125a or miR-125b. J Biol Chem. 282:1479–1486. 2007.
View Article : Google Scholar
|
|
24
|
Zhang Y, Yan LX, Wu QN, et al: miR-125b is
methylated and functions as a tumor suppressor by regulating the
ETS1 proto-oncogene in human invasive breast cancer. Cancer Res.
71:3552–3562. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nexo BA, Vogel U, Olsen A, et al: Linkage
disequilibrium mapping of a breast cancer susceptibility locus near
RAI/PPP1R13 L/iASPP. BMC Med Genet. 9:562008. View Article : Google Scholar
|
|
26
|
Nohata N, Hanazawa T, Enokida H, et al:
microRNA-1/133a and microRNA-206/133b clusters: dysregulation and
functional roles in human cancers. Oncotarget. 3:9–21.
2012.PubMed/NCBI
|
|
27
|
Adams BD, Furneaux H and White BA: The
micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen
receptor-alpha (ERalpha) and represses ERalpha messenger RNA and
protein expression in breast cancer cell lines. Mol Endocrinol.
21:1132–1147. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Song G, Zhang Y and Wang L: MicroRNA-206
targets notch3, activates apoptosis and inhibits tumor cell
migration and focus formation. J Biol Chem. 284:31921–31927. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yan D, Dong Xda E, Chen X, et al:
MicroRNA-1/206 targets c-Met and inhibits rhabdomyosarcoma
development. J Biol Chem. 284:29596–29604. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhou J, Tian Y, Li J, et al: miR-206 is
down-regulated in breast cancer and inhibits cell proliferation
through the up-regulation of cyclinD2. Biochem Biophys Res Commun.
433:207–212. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Applied Biosystems: Endogenous Controls
for Real-Time Quantitation of miRNA Using TaqMan MicroRNA Assays.
2010.
|
|
32
|
Matthaei H, Wylie D, Lloyd MB, et al:
miRNA biomarkers in cyst fluid augment the diagnosis and management
of pancreatic cysts. Clin Cancer Res. 18:4713–4724. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pfaffl MW, Tichopad A, Prgomet C, et al:
Determination of stable housekeeping genes, differentially
regulated target genes and sample integrity: BestKeeper-Excel-based
tool using pair-wise correlations. Biotechnol Lett. 26:509–515.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Escobar PF, Patrick RJ, Rybicki LA, et al:
The 2003 revised TNM staging system for breast cancer: Results of
stage re-classification on survival and future comparisons among
stage groups. Ann Surg Oncol. 14:143–147. 2007. View Article : Google Scholar
|
|
35
|
Elston CW and Ellis IO: Pathological
prognostic factors in breast cancer. I. The value of histological
grade in breast cancer: Experience from a large study with
long-term follow-up. Histopathology. 19:403–410. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Corcoran C, Friel AM, Duffy MJ, et al:
Intracellular and extracellular microRNAs in breast cancer. Clin
Chem. 57:18–32. 2011. View Article : Google Scholar
|
|
37
|
Calvano Filho CM, Calvano-Mendes DC,
Carvalho KC, et al: Triple-negative and luminal A breast tumors:
differential expression of miR-18a-5p, miR-17-5p and miR-20a-5p.
Tumour Biol. 35:7733–7741. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hannafon BN, Sebastiani P, de las Morenas
A, et al: Expression of microRNA and their gene targets are
dysregulated in preinvasive breast cancer. Breast Cancer Res.
13:R242011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang Y, Rathinam R, Walch A, et al: ST14
(suppression of tumorigenicity 14) gene is a target for miR-27b and
the inhibitory effect of ST14 on cell growth is independent of
miR-27b regulation. J Biol Chem. 284:23094–23106. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Baffa R, Fassan M, Volinia S, et al:
MicroRNA expression profiling of human metastatic cancers
identifies cancer gene targets. J Pathol. 219:214–221. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bockmeyer CL, Christgen M, Muller M, et
al: MicroRNA profiles of healthy basal and luminal mammary
epithelial cells are distinct and reflected in different breast
cancer subtypes. Breast Cancer Res Treat. 130:735–745. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li Y, Hong F and Yu Z: Decreased
expression of microRNA-206 in breast cancer and its association
with disease characteristics and patient survival. J Int Med Res.
41:596–602. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Falkenberg N, Anastasov N, Rappl K, et al:
MiR-221/-222 differentiate prognostic groups in advanced breast
cancers and influence cell invasion. Br J Cancer. 109:2714–2723.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lerebours F, Cizeron-Clairac G, Susini A,
et al: miRNA expression profiling of inflammatory breast cancer
identifies a 5-miRNA signature predictive of breast tumor
aggressiveness. Int J Cancer. 133:1614–1623. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Davoren PA, McNeill RE, Lowery AJ, et al:
Identification of suitable endogenous control genes for microRNA
gene expression analysis in human breast cancer. BMC Mol Biol.
9:762008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Meyer SU, Pfaffl MW and Ulbrich SE:
Normalization strategies for microRNA profiling experiments: a
'normal' way to a hidden layer of complexity? Biotechnol Lett.
32:1777–1788. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Qureshi R and Sacan A: A novel method for
the normalization of microRNA RT-PCR data. BMC Med Genomics.
6(Suppl 1): 142013. View Article : Google Scholar
|
|
48
|
Sheinerman KS, Tsivinsky VG, Crawford F,
et al: Plasma microRNA biomarkers for detection of mild cognitive
impairment. Aging (Albany NY). 4:590–605. 2012.
|
|
49
|
Hennessey PT, Sanford T, Choudhary A, et
al: Serum microRNA biomarkers for detection of non-small cell lung
cancer. PLoS One. 7:e323072012. View Article : Google Scholar : PubMed/NCBI
|