Introduction
Fabry disease (OMIM 301500) is an X-linked lysosomal
storage disorder, which is caused by mutations in the gene encoding
the lysosomal enzyme α-galactosidase A (α-Gal A; EC 3.2.1.22)
(1,2). In Fabry disease,
globotriaosylceramide (Gb3) accumulation occurs in lysosomes and
other cellular compartments, and results in disease manifestation
and organ failure. Among the types of cell in the kidney in which
Gb3 accumulates and Gb3 inclusions are found, podocytes are most
prominently involved, followed by distal tubular cells and parietal
epithelial cells. Prominent Gb3 accumulation occurs in hemizygotes
at or even prior to birth, prior to the development of any clinical
symptoms (3). The initial symptoms
of Fabry disease, including angiokeratoma, neuropathic pain,
hypohidrosis or hyperhidrosis, and digestive tract symptoms,
usually appear in childhood in heterozygotes. During the second
decade of life, potentially life-threatening involvement may
develop, including stroke, chronic kidney disease associated with
fibrosis, proteinuria and progressive decrease in glomerular
filtration rate, left ventricular hypertrophy and heart failure
(3). Organ fibrosis is a key
feature of Fabry disease (4).
Progressive podocyte injury can lead to podocyte loss and segmental
glomerulosclerosis, the latter of which is a common late finding in
Fabry nephropathy.
The presence of deacylated Gb3,
globotriaosylsphingosine (lyso-Gb3), in the plasma of disease
patients with Fabry has been noted as a hallmark of Fabry disease
(5). Aerts et al (5) reported that the plasma of Fabry
patients contains markedly increased concentrations of lyso-Gb3,
and the increase in the plasma concentration of this soluble
glycolipid exceeds that of Gb3. Lyso-Gb3 promotes Gb3 storage and
induces proliferation of smooth muscle cells in vitro, which
is suggestive of a causative role of lyso-Gb3 in the pathogenesis
of Fabry disease (5). In cultured
human podocytes, lyso-Gb3 recruits secondary mediators of
inflammation and fibrosis. Even in normal human podocytes, lyso-Gb3
increases the expression of the fibrogenic cytokine transforming
growth factor β1 (TGFβ1) in a dose- and time-dependent manner, and
increases the synthesis of extracellular matrix (ECM) proteins,
including fibronectin and type IV collagen, in a TGFβ1-dependent
manner (6). Fibrosis is
characterized by an increased accumulation of the ECM (7–9). In
addition, through potential direct effects of glycolipids on
tubular cells, proteinuria itself may lead to tubular cell
activation, inflammatory responses and interstitial fibrosis
(4). Activation of interstitial
fibroblasts gives rise to collagen-secreting myofibroblasts.
However, different studies have demonstrated that myofibroblasts
can also originate from renal tubular epithelial and endothelial
cells, which undergo the epithelial-mesenchymal transition (EMT),
in mouse models of renal fibrosis (10,11).
The present study aimed to identify changes in the
gene expression profiles of epithelial and mesangial cells grown in
culture and exposed to Gb3 or lyso-Gb3, as a model of Fabry
disease. Microarrays were used to identify the gene expression
profiles, which contribute to the EMT process and leads to kidney
fibrosis. In addition, the expression profiles in different tissues
and cells were compared to discern the cell-specific activation
patterns in the epithelial and mesangial cells.
Materials and methods
Animals
Fabry mice were bred to acquire a sufficient number
of mice (12). Mice were
maintained on a diurnal light cycle of 12 h in a temperature
(21±2°C) and humidity (55±5%) controlled room. Mice were fed an
autoclaved diet and water ad libitum. The present study was
approved by the Institutional Animal Care and Use Committee of the
Ewha Womans University School of Medicine (Seoul, Korea). All mice
were treated in accordance with the Animal Care Guidelines of the
Ewha Womans University School of Medicine, and the National
Research Council (US) Guide for the Care and Use of Laboratory
Animals (13). All mice were
genotyped using polymerase chain reaction (PCR; Bio-Rad
Laboratories, Inc., Johannesburg, South Africa). The mice were all
16-week-old males and were grouped into wild-type and hemizygous
Fabry mice. A minimum of 3 age-matched mice were used for each
group. Animals were sacrificed by CO2 inhalation
euthanasia.
Cell culture
For the in vitro investigation, the HK-2
Homo sapiens kidney tubular epithelial cell line and SV40
MES 13 mouse kidney glomerular mesangial cell line were purchased
from American Type Culture Collection (Manassas, VA, USA). The HK-2
cells were cultured in complete growth media; keratinocyte
serum-free medium supplemented with 0.05 mg/ml bovine pituitary
extract, 5 ng/ml human recombinant epidermal growth factor (EGF)
and 5 µg/ml gentamicin (Invitrogen Life Technologies,
Carlsbad, CA, USA). The SV40 MES 13 cells were maintained in a 3:1
mixture of Dulbecco's modified Eagle's medium and Ham's F12 medium
(Welgene, Daegu, Korea) with 14 mM HEPES (Welgene) supplemented
with 5% fetal bovine serum (Invitrogen Life Technologies) and
penicillin/streptomycin (100 U/ml; Invitrogen Life Technologies) at
37°C in 5% CO2 in a humidified incubator.
Gb3 and lyso-Gb3 treatment
The HK-2 cells (1×105) and SV40 MES 13
(4×104) cells were seeded into 60 mm tissue culture
plates (SPL Life Sciences, Pocheon-si, Korea), and proportion of
which were treated with Gb3 (Matreya, Pleasant Gap, PA, USA) or
lyso-Gb3 (Sigma-Aldrich, St. Louis, MO, USA) in serum-free culture
medium. The final concentration of treatment was 30 µM for
Gb3 and 400 nM for lyso-Gb3, which were dissolved in 100% dimethyl
sulfoxide (Sigma-Aldrich). The cells were treated with Gb3 or
lyso-Gb3 for 72 h.
Microarray analysis
The HK-2 cells and SV40 MES 13 cells were assigned
to a control and a Gb3- or lyso-Gb3-treated group. The cells from
each group were pooled, and the total RNA was extracted using an
RNeasy Plus Mini Kit (Qiagen, Valencia, CA, USA), according to the
manufacturer's instructions. The quality of RNA was analyzed using
a 2100 Bioanalyzer (Agilent Technologies, Inc., Palo Alto, CA,
USA). The total RNA from each group was amplified separately using
a Low RNA Input Linear Amplification kit PLUS (Agilent
Technologies). The Cy3 (control)- or Cy5 (Gb3 or lyso-Gb3
treated)-labeled targets were hybridized to an Agilent 44K human or
mouse oligo microarray at 60°C for 17 h. Following hybridization,
the arrays were washed in three consecutive steps using an Agilent
Gene Expression Wash Buffer kit (Agilent Technologies, Inc.). The
microarrays were scanned using a DNA Microarray scanner using
SureScan High-Resolution Technology (G2565CA; Agilent Technologies,
Inc.) and were analyzed using GeneSpring GX software (Agilent
Technologies, Inc.). For normalization and hierarchical clustering
analysis, the data obtained from the Gb3- and lyso-Gb3-treated
cells were corrected using the values for the appropriate control
and then imported into the GeneSpring GX software. Significant
genes were selected for probes that exhibited a difference of
>2-fold in the normalization ratio. In the functional
investigation, analysis of gene ontology was used to predict the
function of each gene, classified by the biological process. Genes
specifically expressed in each cluster of Gb3 or lyso-Gb3 treated
cells were identified by comparing with non-treated control cells.
Differentially expressed genes were categorized in Gene Ontology
groups using the DAVID tool (http://david.abcc.ncifcrf.gov/).
Reverse transcription-quantitative
(RT-qPCR)
Using the results from the microarray analysis, the
present study identified genes, which were either upregulated or
downregulated by >2-fold as significant genes. RT-qPCR was
performed using SYBR® Premix Ex Taq™ (Takara,
Bio, Inc., Shiga, Japan) on an ABI 7500 Fast Real-Time PCR system
(PE Applied Biosystems, Foster City, CA, USA) to confirm the
relative expression levels of the genes in the control, Gb3 and
lyso-Gb3 groups. RNA was reverse transcribed using the High
Capacity cDNA Archive kit (Applied Biosystems). The total volume of
the PCR reaction was 20 µl, containing 0.8 µl of each
primer (5 µM), 1 µl cDNA, 10 µl 2X
SYBR® Premix Ex Taq™ II, 0.4 µl Rox dye
and 7.8 µl sterile ddH2O. PCR cycling conditions
were as follows: Initial 30 sec denaturation at 95°C, followed by
40 cycles of amplification at 95°C for 3 sec and 60°C for 30 sec,
and a subsequent melting curve analysis, where the temperature was
increased from 60 to 95°C. In order to quantify each candidate gene
expression, the mRNA expression levels were normalized to the
GAPDH mRNA level. Relative gene expression was analyzed
using the comparative cycle threshold (Ct) method
(2−ΔΔCt) (14). RT-qPCR
was performed in triplicate for each sample and was repeated three
times for each assay.
Western blot analysis
The Gb3- or lyso-Gb3-treated cells were washed with
ice-cold phosphate-buffered saline and resuspended in PRO-PREP
buffer (iNtRON Biotechnology, Inc.) supplemented with phosphatase
inhibitor cocktail solution for 30 min on ice. Insoluble material
was removed by centrifugation at 13,000 x g for 20 min at 4°C.
Total protein concentration was determined using bicinchoninic acid
assay (Pierce Biotechnology, Inc., Rockford, IL, USA). The protein
was electrophoresed on a 10% sodium dodecyl sulfate-polyacrylamide
gel, and transferred onto a polyvinylidene membrane (EMD Millipore,
Billerica, MA, USA). The membrane was blocked with 5% nonfat dry
milk in Tris-buffered saline (Elpis Biotech, Inc., Taejeon, Korea)
containing 0.1% Tween-20 (TBST; Sigma-Aldrich) for 1 h and then
incubated overnight at 4°C with anti-WT1 polyclonal antibody (cat.
no. sc-192; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA).
The primary antibody was removed, and the blots were washed three
times with TBST and then incubated for 1 h at room temperature with
goat anti-rabbit IgG (cat. no. sc-2054; Santa Cruz Biotechnology,
Inc.) as the secondary antibody. Following removal of the secondary
antibody, the blots were washed and the specific bands were
detected using enhanced chemiluminescence with a WestSave GOLD™
Western Blot Detection kit (AbFronteir, Seoul, Korea), according to
the manufacturer's instructions. All signals were analyzed using
densitometric scanning (LAS-3000; Fujifilm, Tokyo, Japan).
Statistical analysis
The values are presented as either the mean ±
standard deviation or ± standard error of the mean. Data were
analyzed using Prism 4.0 software (GraphPad Software. Inc., La
Jolla, CA, USA). The significance of differences between groups was
determined using Student's t-test. P<0.05 was considered to
indicate a statistically significant difference.
Results
Gene expression profile from microarrays
of SV40 MES 13 mesangial cells and HK-2 tubular epithelial
cells
The present study examined the gene expression
profiles in the kidney cells following treatment with Gb3 or
lyso-Gb3 (Table. I and II). If the gene expression value was
upregulated or downregulated >2-fold, compared with the control
group, the change was considered to be significant.
 | Table IGene expression patterns in SV40 MES
13 cells treated with Gb3 or lyso-Gb3. |
Table I
Gene expression patterns in SV40 MES
13 cells treated with Gb3 or lyso-Gb3.
| Gene symbol | GenBank ID | Gene name | Fold change
|
|---|
| Gb3 | Lyso-Gb3 |
|---|
| Cytoskeleton | | | | |
| FSCN2 | NM_172802 | Fascin homolog 2,
actin-bundling protein, retinal | 15.97 | −1.74 |
|
NCKAP1L | NM_153505 | Hematopoietic
protein 1 | 5.91 | 6.72 |
| VIL1 | NM_009509 | Villin 1 | 3.57 | 1.51 |
| EGF | NM_010113 | Epidermal growth
factor | 2.10 | −1.35 |
| SCIN | NM_009132 | Scinderin | 2.10 | −1.50 |
| Extracellular
matrix | | | | |
| MMP9 | NM_013599 | Matrix
metalloproteinase 9 | 5.59 | −1.13 |
| COL4A3 | NM_007734 | Collagen, type IV,
alpha 3 (Goodpasture antigen) | 2.77 | −1.33 |
|
COL15A1 | NM_009928 | Collagen, type XV,
alpha 1 | 2.29 | 2.61 |
| NTN4 | NM_021320 | Netrin 4 | 2.44 | 1.84 |
| RELN | NM_011261 | Reelin
precursor | −1.08 | 27.84 |
| IMPG2 | NM_174876 | Interphotoreceptor
matrix proteoglycan 2 | −2.34 | −4.13 |
| KERA | NM_008438 | Keratocan | −3.39 | −11.52 |
| Cell
proliferation/differentiation | | | | |
| PRM2 | NM_008933 | Protamine 2 | 22.61 | 1.49 |
| DNER | NM_152915 | Delta/notch-like
EGF-related receptor | 9.12 | 7.55 |
| WT1 | NM_144783 | Wilms' tumor 1 | 6.36 | 2.60 |
| FOXP2 | NM_053242 | Forkhead box
P2 | 2.89 | 45.68 |
| HOXA11 | NM_010450 | Homeobox A11 | −2.51 | −2.19 |
| DLL1 | NM_007865 | Delta-like 1 | −5.14 | −6.89 |
| DMRT2 | NM_145831 | Doublesex and mab-3
related 1 transcription factor 2 isoform | −5.79 | 3.05 |
| Cell migration | | | | |
| CYBB | NM_007807 | Cytochrome b-245,
beta polypeptide | 8.20 | 2.36 |
| ISL1 | NM_021459 | ISL1 transcription
factor, LIM/homeodomain (islet 1) | 6.27 | 1.08 |
| TNF | NM_013693 | Tumor necrosis
factor alpha | 4.72 | 1.57 |
| NM_009382 | Thymus cell antigen
1, theta | 1.32 | 16.51 |
| PRRXL1 | NM_001001796 | Paired related
homeobox protein-like 1 | −1.76 | −5.97 |
| F8 | NM_007977 | Coagulation factor
VIII | −2.90 | −3.33 |
 | Table IIGene expression patterns in HK-2
cells treated with Gb3 or lyso-Gb3. |
Table II
Gene expression patterns in HK-2
cells treated with Gb3 or lyso-Gb3.
| Gene symbol | GenBank ID | Gene name | Fold change
|
|---|
| Gb3 | Lyso-Gb3 |
|---|
| Cytoskeleton |
| AKAP4 | NM_003886 | A kinase (PRKA)
anchor protein 4 | 3.84 | −4.89 |
| EML1 | NM_001008707 | Echinoderm
microtubule associated protein like 1 | 2.62 | −1.17 |
| FGF23 | NM_020638 | Fibroblast growth
factor 23 | 1.21 | −13.61 |
| ERMN | NM_020711 | Ermin, ERM-like
protein | −13.79 | −15.86 |
| SGCD | NM_000337 | Sarcoglycan, delta
(35 kDa dystrophin-associated glycoprotein) | −17.57 | 1.19 |
| Extracellular
matrix |
| IL19 | NM_153758 | Interleukin 19 | 24.90 | 1.20 |
|
SERPING1 | NM_000062 | Serpin peptidase
inhibitor, clade G (C1 inhibitor), member 1 | 10.25 | 1.09 |
| MYH11 | NM_001040113 | Smooth muscle
myosin heavy chain 11 isoform SM2A | 3.59 | 5.75 |
|
ADAMTS6 | NM_197941 | ADAM
metallopeptidase with thrombospondin type 1 motif, 6
preproprotein | 2.76 | 9.00 |
|
COL15A1 | NM_001855 | Collagen, type XV,
alpha 1 | 3.16 | −1.06 |
| CRIM1 | NM_016441 | Cysteine rich
transmembrane BMP regulator 1 (chordin-like) | −9.69 | −13.96 |
| SEPP1 | NM_005410 | Selenoprotein P,
plasma, 1 | −13.32 | −15.61 |
| Cell
proliferation/differentiation |
| RARB | NM_000965 | Retinoic acid
receptor, beta | 23.54 | −1.06 |
| IL4 | NM_000589 | Interleukin 4 | 22.44 | 3.47 |
| GKN1 | NM_019617 | Gastrokine 1 | 11.04 | −1.75 |
| INS | NM_000207 | Insulin | −2.33 | −3.06 |
| TFAP2A | M61156 | Transcription
factor AP-2 alpha (activating enhancer binding protein 2
alpha) | −2.47 | −2.96 |
| IL2 | NM_000586 | Interleukin 2 | −16.42 | −18.89 |
| Cell migration |
| CSF1 | NM_172210 | Colony stimulating
factor 1 (macrophage) | 3.69 | 1.17 |
| HES1 | NM_005524 | Hairy and enhancer
of split 1, (Drosophila) | 2.24 | 1.42 |
| EPHB3 | NM_004443 | EPH receptor
B3 | 1.36 | 2.31 |
| PROP1 | NM_006261 | PROP paired-like
homeobox 1 | 1.24 | 2.11 |
SV40 MES 13 cells
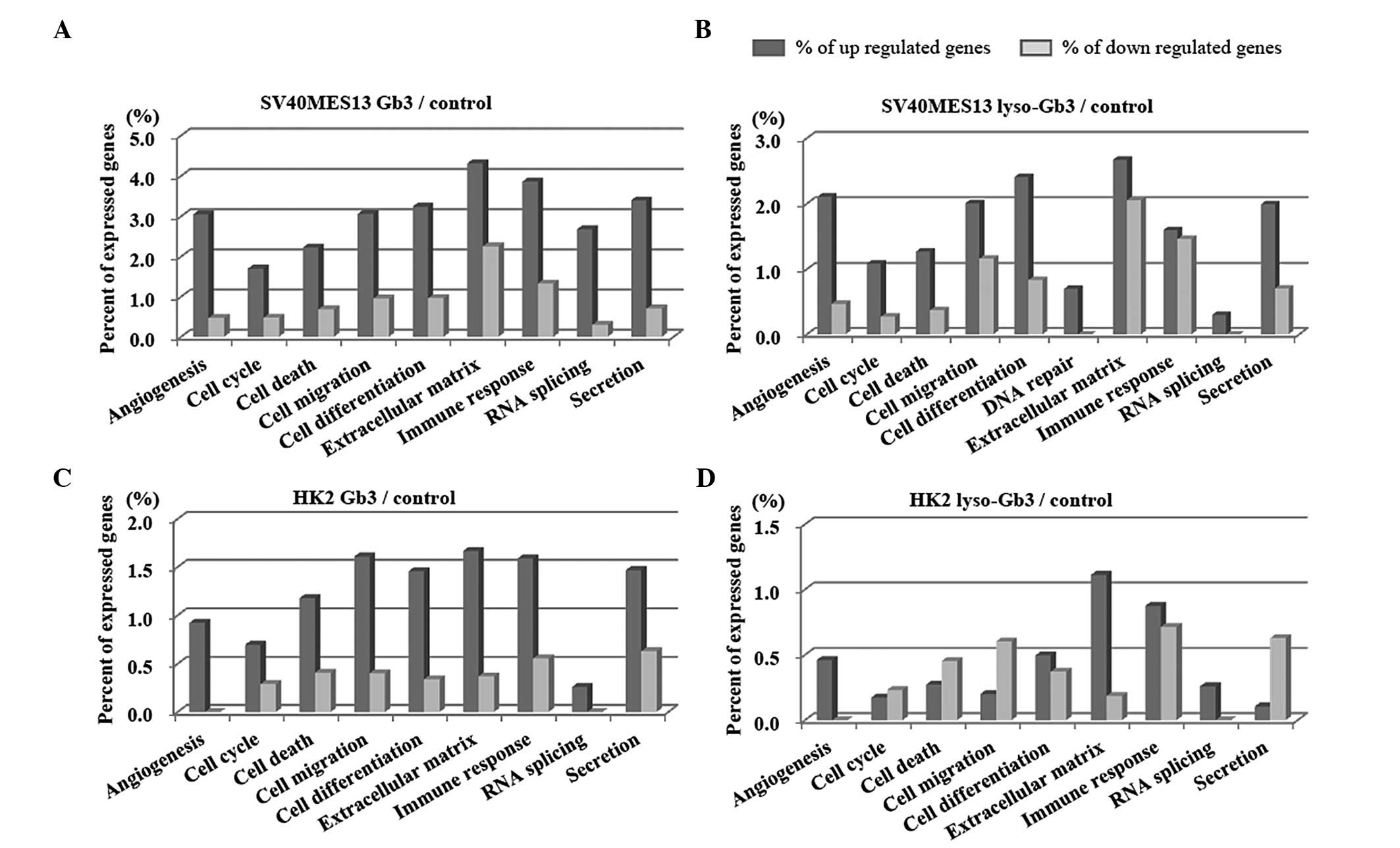
A total of 39,429 genes were detected in the SV40
MES 13 cells, of which 2,991 were significant. The majority of the
upregulated genes were associated with 'ECM', 'cell migration,'
'cell differentiation' or 'immune response'. The majority of the of
the downregulated genes were associated with 'ECM', 'immune
response' or 'cell migration' (Fig. 1A
and B).
HK-2 cells
A total of 34,127 genes were detected in the HK-1
cells, of which 1,141 were identified as either significantly
upregulated or downregulated. The majority of the upregulated genes
were associated with 'ECM', 'cell differentiation,' 'cell
migration' or 'immune response'. The majority of the down-regulated
genes were associated with 'cell migration', 'cell death', 'cell
differentiation' or 'immune response' (Fig. 1C and D).
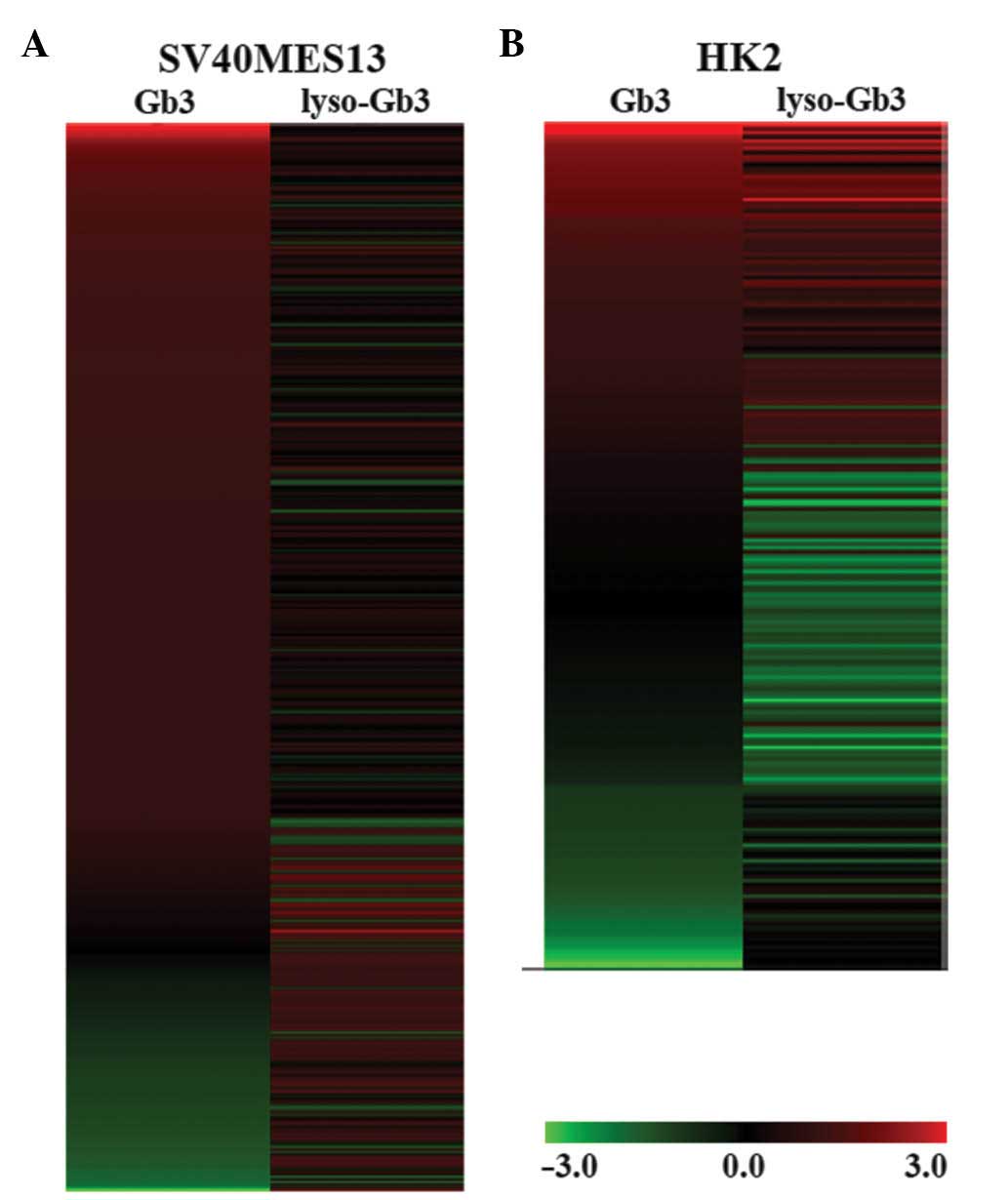
Hierarchical clustering analysis
Hierarchical clustering analysis revealed that,
following treatment of the SV40 MES 13 cells and HK-2 cells with
Gb3 or lyso-Gb3, the change in gene expression was dependant on the
cell type and substrate (Fig. 2).
In the SV40 MES 13 cells treated with Gb3, the majority of the
significantly altered genes were upregulated, however, their
corresponding gene expression patterns were different following
treatment with lyso-Gb3 (Fig. 2A).
In the HK-2 cells, the gene expression pattern was more stable,
compared with than that observed in the SV40 MES 13 cells (Fig. 2B), although the patterns differed
between treatments with Gb3 and lyso-Gb3. In contrast to the SV40
MES 13 cells treated with Gb3, the majorit of the significantly
altered genes in the HK-2 cells were upregulated following
treatment with lyso-Gb3.
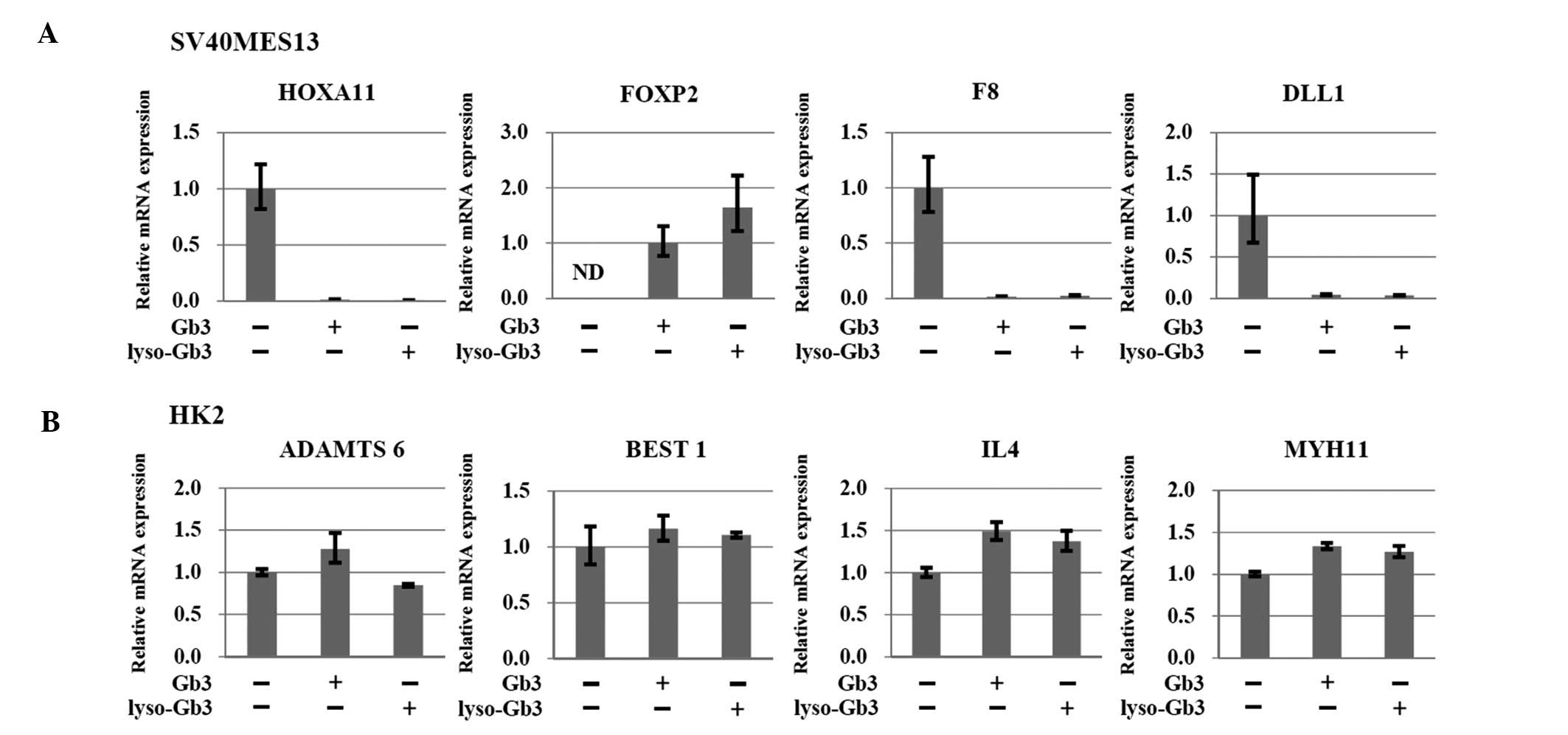
Validation of gene expression patterns in
the cell models using RT-qPCR
To confirm the microarray-predicted expression
patterns, the present study performed RT-qPCR to analyze the
candidate genes. The 'target' genes, which presented with
relatively consistent patterns in the micro-array and RT-qPCR,
including IL4, MYH11, ADAMTS6, BEST1,
FOXP2, F8, HOXA11, DLL1 and WT1
were identified. In the SV40 MES 13 mesangial cells treated with
Gb3 or lyso-Gb3, the expression of FOXP2 was upregulated and
the expression levels of HOXA11, F8 and DLL1
were downregulated, compared with the control cells (Fig. 3A). In the HK-2 epithelial cells,
the mRNA expression levels of ADAMTS6, BEST1,
IL4 and MYH11 were upregulated in the Gb3- and
lyso-Gb3-treated epithelial cells, compared with the control cells
(Fig. 3B).
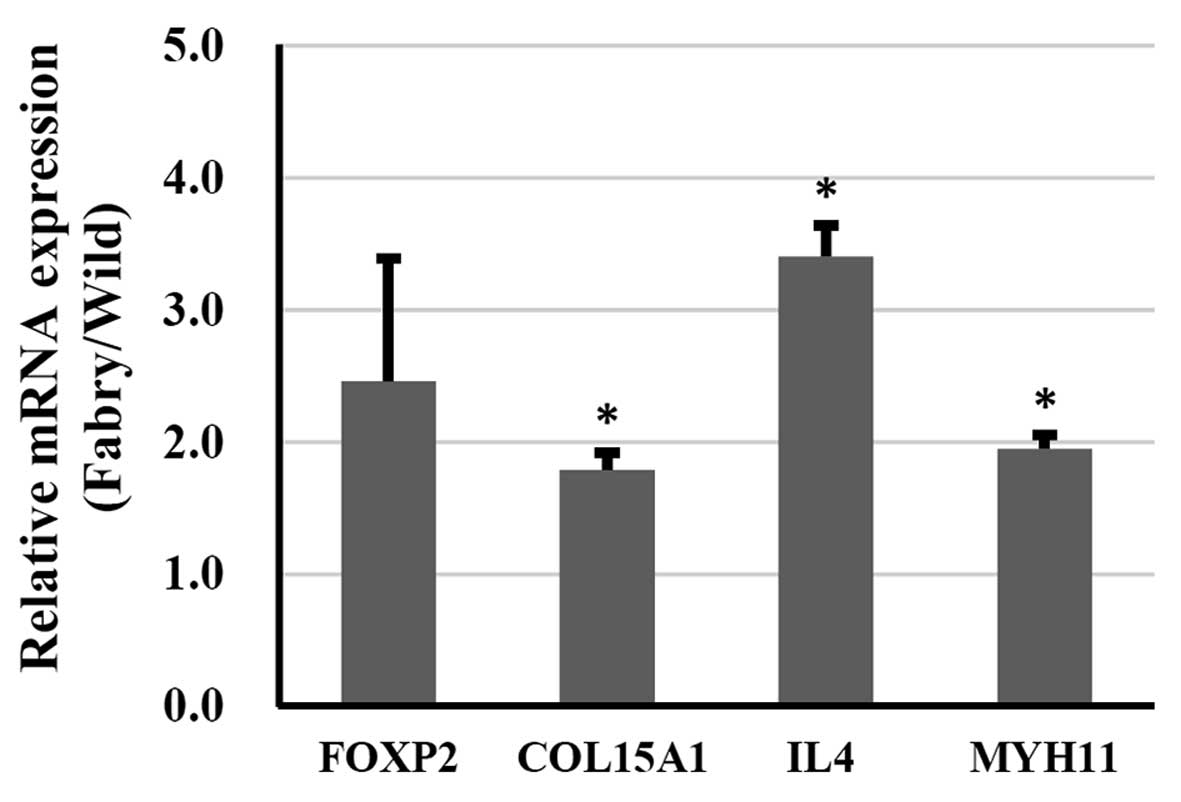
Validation of gene expression in kidney
tissues
The present study also examined the expression
levels of FOXP2, COL15A1, MYH11 and IL4
in kidney tissues from wild-type and Fabry mice (Fig. 4). Compared with the wild-type
tissues, the expression levels of COL15A1, IL4 and
MYH11 were significantly upregulated in the Fabry mouse
kidney tissues. No significant upregulation in the expression of
FOXP2 was observed in the Fabry mouse kidney (P=0.059).
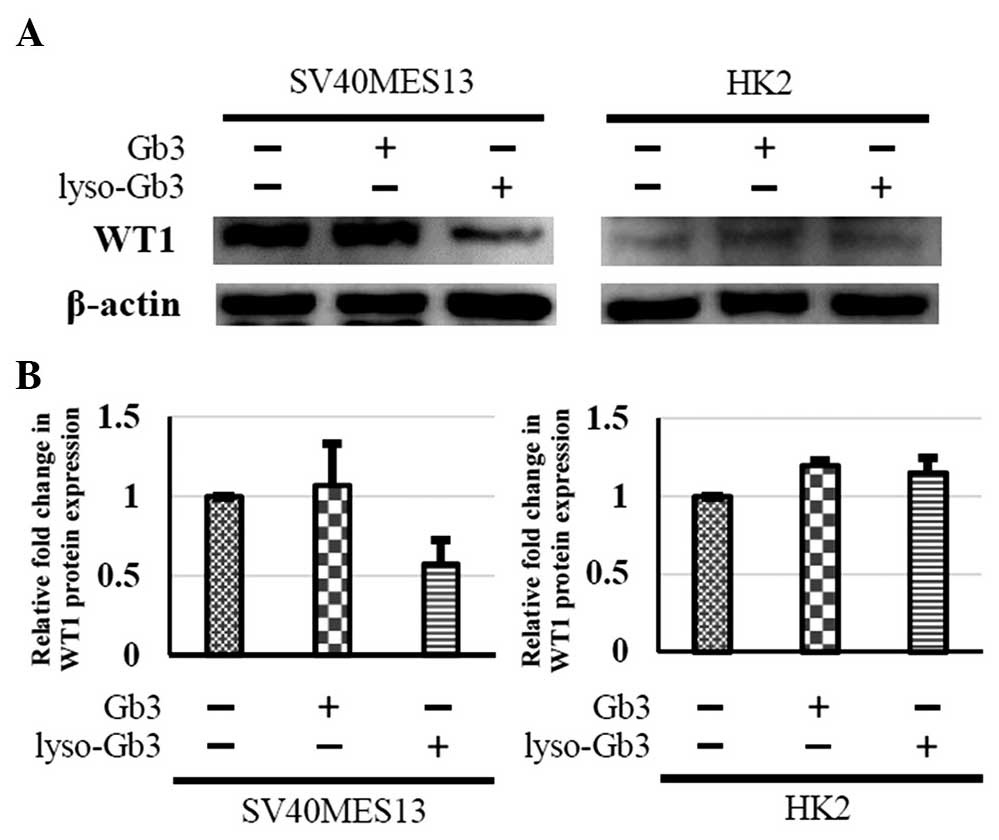
Expression of WT1 in the cell models
To further confirm the patterns of gene expression
identified in the present study, immunoblotting of the protein
encoded by the WT1 gene was performed. The protein
expression of WT1 was identified in the SV40 MES 13 cells and HK-2
cells following treatment with either Gb3 or lyso-Gb3 (Fig. 5). In agreement with the microarray
data, the expression of WT1 was significantly increased in the SV40
MES 13 cells treated with Gb3, compared with the control. However,
the expression of WT1 was decreased in SV40 MES 13 cells
treated with lyso-Gb3. In the HK-2 cells, the expression of
WT1 was increased marginally when treated with Gb3 or
lyso-Gb3, compared with the control.
Discussion
Fabry disease progresses to irreversible tissue
damage and organ dysfunction. Among the complications of Fabry
disease, renal failure significant contributes to rates of
morbidity and mortality. In Fabry nephropathy, glycolipids are
deposited in the glomerular cells, articularly in podocytes,
mesangial cells and endothelial cells, in tubular epithelial cells
of the distal nephron, and in vascular cells, including endothelial
cells of the capillaries, veins, arteries and vascular smooth
muscle cells (3). Early
progressive podocyte injury can lead to podocyte loss and fibrosis,
caused by epithelial cell segmental or global glomerulosclerosis,
which is a common finding in Fabry nephropathy (15,16),
with interstitial fibrosis and tubular atrophy usually observed
later.
In the past two decades, the contribution of the EMT
to fibrinogenesis has been noted. Therefore, the present study
examined whether the renal gene expression profiles in tissues with
Fabry disease are associated with the EMT, which is considered to
be involved in fibrosis of the kidney. The present study compared
the expression patterns of candidate genes in HK-2 tubular
epithelial and SV40 MES 13 mesangial cells in response to treatment
with Gb3 and lyso-Gb3. Gene ontology analysis revealed that the
most significantly altered genes were associated with 'ECM', 'cell
migration', or 'cell proliferation' in the two cell types. This
result indicated that Gb3 or lyso-Gb3 primarily regulated the
expression of genes associated with the EMT in these kidney
cells.
Following Gb3 or lyso-Gb3 treatment, BEST1,
ADAMTS6, MYH11 and IL4 were upregulated only
in the HK-2 cells, and ADAM18, COL4A3, PDE1C
and FOXP2 were upregulated only in the SV40 MES 13 cells.
The expression levels of DLL1, F8, and HOXA11
were downregulated in the SV40 MES 13 cells, but were unchanged in
the HK-2 cells. The gene expression patterns in the SV40 MES 13 and
HK-2 cells differed between the group treated with Gb3 and the
group treated with lyso-Gb3. The gene expression profile of the
SV40 MES 13 cells exhibited more marked changes following Gb3
treatment, suggesting that these cells are more sensitive to Gb3,
whereas the HK-2 cells appeared to be more sensitive to
lyso-Gb3.
In the present study, the expression levels of
FOXP2, COL15A1, IL4 and MYH11 were
upregulated in kidney tissues from Fabry mice. FOXP2 encodes
an evolutionarily conserved transcription factor, which is
expressed in fetal and adult brains and has been found to act as a
transcriptional repressor and be involved in multiple biological
processes, including development and immunoregulation (17). The expression of FOXP2 was
upregulated in the epithelial and mesangial cells, however, its
expression in the mesangial cells exceeded that in the epithelial
cells and was sensitive to lyso-Gb3 treatment. Upregulation of
COL15A1 was observed in the SV40 MES 13 and HK-2 cells, with
the exception of the HK-2 cells treated with lyso-Gb3. The
expression of COL15A1 is closed associated with the fibrotic
process in the kidney (18). Renal
interstitial type XV collagen staining in patients with kidney
fibrosis is pronounced due to these fibrotic changes (18).
Interleukin-4 (IL-4) is a ligand for the IL-4
receptor and mediates its numerous functions by binding to
receptors expressed on target cells. IL4 has been implicated
in the pathogenesis of experimental and human glomerulonephritis
(19). Aberrant multi-organ
expression of IL4, including in the kidney, causes
progressive glomerulosclerosis leading to end-stage renal failure.
Renal expression of IL-4 leads to increased ECM production
and alters the glomerular structure, possibly by promoting
immunoglobulin (Ig) deposition, and upregulation of the renal
expression of TGF-β1 following glomerular Ig deposition accelerates
sclerosis and exacerbates disease development (20). The present study observed that the
expression of IL4 was upregulated in epithelial cells and
was activated significantly by Gb3 treatment.
MYH11 encodes a smooth muscle myosin
belonging to the myosin heavy chain family, and mutation of
MYH11 has been reported to increase TGF-β signaling in
familial thoracic aortic aneurysm/dissection (21). In the present study, the expression
of MYH11 was upregulated in epithelial cells and
downregulated in mesangial cells. The expression of MYH11
was also upregulated more in the kidney tissues of Fabry mice,
compared with those from wild-type mice. WT1 acts as an activator
or a repressor of target genes, depending on the cell type and
target gene with which WT1 interacts. It is well known that WT1 is
critical during embryogenesis, despite the expression pattern of
WT1 being highly complex (22,23).
Following birth, terminally differentiated cells, including
epithelial tubular cells do not exhibit WT1 expression,
whereas cells with the potential for EMT, including podocytes,
continue to produce WT1 (23,24).
WT1 can be found outside podocytes, including mesangial cells, in
diabetic nephropathy or endothelial cells (25,26).
In the present study, the expression of WT1 in SV40 MES 13
mesangial cells was upregulated by Gb3, but was downregulated by
lyso-Gb3. By contrast, the expression of WT1 was weak in the
HK-2 epithelial cells and was marginally upregulated by either Gb3
or lyso-Gb3. This result suggested that WT1 may be involved
in the progression of fibrosis in kidney cells in Fabry
disease.
Lyso-Gb3 has been reported as a major bioactive
molecule in Fabry disease, and may promote the release of secondary
mediators of glomerular injury, including diabetic nephrop-athy and
disease manifestations. Lyso-Gb3 may activate target tissue cells,
including podocytes (4,6). Although intracellular Gb3
accumulation throughout the body has traditionally been ascribed as
a major cause of pathogenesis in Fabry disease, serum levels of Gb3
and Gb3 deposits do not necessarily correlate with clinical
manifestations (5). Lyso-Gb3 is
associated with vascular smooth muscle cell proliferation and
induces podocytes to produce mediators of glomerular injury,
including TGF-β1, a critical mediator of ECM production, fibrosis
and podocyte injury (4,5,27).
In conclusion, to identify biomarkers of the EMT,
which may be critical in renal fibrosis and end-stage renal
disease, the present study performed microarray analysis of
epithelial and mesangial cells, to compare the effects of Gb3 and
lyso-Gb3 treatment in models of Fabry disease. The microarray
results suggested that the gene expression patterns in kidney cells
are cell specific and substrate specific. Despite the relatively
high Gb3 and lyso-Gb3 concentrations in plasma and kidneys, these
levels did not reflect overt pathology or clinical manifestations,
which suggested that glycolipids are involved in determining the
target tissue or cell-specific pattern. The present study
hypothesized that Gb3 and lyso-Gb3 themselves are not specific
biomarkers for the diagnosis of Fabry disease, but each appears to
have a specific role and time course in relation to cell
activation, disease progression and clinical manifestations.
Further investigation is required to determine the precise
mechanisms through which Gb3 and lyso-Gb3 contribute to the
progression of fibrosis, and the potential roles of biomarkers of
the EMT in different cell types.
Acknowledgments
This study was supported by grants from the Health
Technology R&D project (grant. no. HI12C0598) of the Ministry
of Health and Welfare, Republic of Korea.
References
|
1
|
Brady RO, Gal AE, Bradley RM, Martensson
E, Warshaw AL and Laster L: Enzymatic defect in Fabry's disease.
Ceramidetrihexosidase deficiency. N Engl J Med. 276:1163–1167.
1967. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kint JA: The enzyme defect in Fabry's
disease. Nature. 227:11731970. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Densick RJ, Ioannou YA and Eng CM:
Alpha-galactosidase A deficiency: Fabry disease. The Metabolic and
Molecular Bases of Inherited Disease. 8th ed. Scriver CR, Beaudet
AL, Sly WS and Valle D: McGraw-Hill; New York: pp. 3733–3774.
2001
|
|
4
|
Weidemann F, Sanchez-Niño MD, Politei J,
Oliveira JP, Wanner C, Warnock DG and Ortiz A: Fibrosis: A key
feature of Fabry disease with potential therapeutic implications.
Orphanet J Rare Dis. 8:1162013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Aerts JM, Groener JE, Kuiper S,
Donker-Koopman WE, Strijland A, Ottenhoff R, van Roomen C, Mirzaian
M, Wijburg FA, Linthorst GE, et al: Elevated
globotriaosylsphingosine is a hallmark of Fabry disease. Proc Natl
Acad Sci USA. 105:2812–2817. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sanchez-Niño MD, Sanz AB, Carrasco S,
Saleem MA, Mathieson PW, Valdivielso JM, Ruiz-Ortega M, Egido J and
Ortiz A: Globotriaosylsphingosine actions on human glomerular
podocytes: Implications for Fabry nephropathy. Nephrol Dial
Transplant. 26:1797–1802. 2011. View Article : Google Scholar
|
|
7
|
Campanholle G, Ligresti G, Gharib SA and
Duffield JS: Cellular mechanisms of tissue fibrosis. 3. Novel
mechanisms of kidney fibrosis. Am J Physiol Cell Physiol.
304:C591–C603. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Iredale JP: Models of liver fibrosis:
Exploring the dynamic nature of inflammation and repair in a solid
organ. J Clin Invest. 117:539–548. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zeisberg M and Neilson EG: Mechanisms of
tubulointerstitial fibrosis. J Am Soc Nephrol. 21:1819–1834. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Iwano M, Plieth D, Danoff TM, Xue C, Okada
H and Neilson EG: Evidence that fibroblasts derive from epithelium
during tissue fibrosis. J Clin Invest. 110:341–350. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zeisberg EM, Potenta SE, Sugimoto H,
Zeisberg M and Kalluri R: Fibroblasts in kidney fibrosis emerge via
endothelial-to-mesenchymal transition. J Am Soc Nephrol.
19:2282–2287. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ohshima T, Murray GJ, Swaim WD,
Longenecker G, Quirk JM, Cardarelli CO, Sugimoto Y, Pastan I,
Gottesman MM, Brady RO, et al: α-Galactosidase A deficient mice: a
model of Fabry disease. Proc. Natl Acad Sci USA. 94:2540–2544.
1997. View Article : Google Scholar
|
|
13
|
National Research Council (US) Committee
for the Update of the Guide for the Care and Use of Laboratory
Animals: Guide for the Care and Use of Laboratory Animals. 8th
edition. Washington (DC): National Academies Press (US); 2011
|
|
14
|
Schefe JH, Lehmann KE, Buschmann IR, Unger
T and Funke-Kaiser H: Quantitative real-time RT-PCR data analysis:
Current concepts and the novel “gene expression's CT difference”
formula. J Mol Med. 84:901–910. 2006. View Article : Google Scholar
|
|
15
|
Gubler MC, Lenoir G, Grünfeld JP, Ulmann
A, Droz D and Habib R: Early renal changes in hemizygous and
heterozygous patients with Fabry's disease. Kidney Int. 13:223–235.
1978. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fogo AB, Bostad L, Svarstad E, Cook WJ,
Moll S, Barbey F, Geldenhuys L, West M, Ferluga D, Vujkovac B, et
al: Scoring system for renal pathology in Fabry disease: report of
the International Study Group of Fabry Nephropathy (ISGFN). Nephrol
Dial Transplant. 25:2168–2177. 2010. View Article : Google Scholar :
|
|
17
|
Konopka G, Bomar JM, Winden K, Coppola G,
Jonsson ZO, Gao F, Peng S, Preuss TM, Wohlschlegel JA and Geschwind
DH: Human-specific transcriptional regulation of CNS development
genes by FOXP2. Nature. 462:213–217. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hägg PM, Hägg PO, Peltonen S,
Autio-Harmainen H and Pihlajaniemi T: Location of type XV collagen
in human tissues and its accumulation in the interstitial matrix of
the fibrotic kidney. Am J Pathol. 150:2075–2086. 1997.PubMed/NCBI
|
|
19
|
Furusu A, Miyazaki M, Koji T, Abe K, Ozono
Y, Harada T, Nakane PK, Hara K and Kohno S: Involvement of IL-4 in
human glomerulonephritis: an in situ hybridization study of IL-4
mRNA and IL-4 receptor mRNA. J Am Soc Nephrol. 8:730–741.
1997.PubMed/NCBI
|
|
20
|
Rüger BM, Hasan Q, Erb KJ and Davis PF:
Progression of renal disease in interleukin-4 transgenic mice:
involvement of transforming growth factor-beta. Int J Exp Pathol.
80:113–123. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Renard M, Callewaert B, Baetens M, Campens
L, MacDermot K, Fryns JP, Bonduelle M, Dietz HC, Gaspar IM, Cavaco
D, et al: Novel MYH11 and ACTA2 mutations reveal a role for
enhanced TGFβ signaling in FTAAD. Int J Cardiol. 165:314–321. 2011.
View Article : Google Scholar
|
|
22
|
Pritchard-Jones K, Fleming S, Davidson D,
Bickmore W, Porteous D, Gosden C, Bard J, Buckler A, Pelletier J
and Housman D: The candidate Wilms' tumour gene is involved in
genitourinary development. Nature. 346:194–197. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Buckler AJ, Pelletier J, Haber DA, Glaser
T and Housman DE: Isolation, characterization and expression of the
murine Wilms' tumor gene (WT1) during kidney development. Mol Cell
Biol. 11:1707–1712. 1991.PubMed/NCBI
|
|
24
|
Grubb GR, Yun K, Williams BR, Eccles MR
and Reeve AE: Expression of WT1 protein in fetal kidneys and Wilms
tumors. Lab Invest. 71:472–479. 1994.PubMed/NCBI
|
|
25
|
Brunskill EW and Potter SS: Changes in the
gene expression programs of renal mesangial cells during diabetic
nephropathy. BMC Nephrol. 13:702012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mrowka C and Schedl A: Wilms' tumor
suppressor gene WT1: from structure to renal pathophysiologic
features. J Am Soc Nephrol. 11(Suppl 16): S106–S115.
2000.PubMed/NCBI
|
|
27
|
Hills CE and Squires PE: The role of TGF-β
and epithelial-to mesenchymal transition in diabetic nephropathy.
Cytokine Growth Factor Rev. 22:131–139. 2011.PubMed/NCBI
|