Introduction
A glioma is a type of tumor that originates from
glial cells (astrocytes or oligodendrocytes), which are
non-neuronal supporting cells. Gliomas are the most fatal type of
brain tumor, and glioblastoma multiforme (GBM) is the most common
and aggressive subtype of glioma in humans (1). Although the etiology of the tumor
remains to be elucidated, GBM is hypothesized to occur as a result
of the interaction between multiple genetic and environmental
factors. Genetic factors have been reported to contribute to
gliomagenesis, as evidenced by familial aggregation studies
(2,3), as well as rare Mendelian syndromes,
including neurofibromatosis and Turcot syndrome (4). Associations have been established
between GBM susceptibility and single nucleotide polymorphisms
(SNPs) in genes that were independently identified to be candidate
genes involved in gliomagenesis in two genome-wide association
studies (5–7).
MicroRNAs (miRNAs/miRs) are a class of
~22-nucleotide-long non-protein coding RNAs, which are hypothesized
to regulate up to a third of all protein-coding genes via binding
to the 3′ untranslated region (UTR) of target gene messenger RNA
(mRNA), resulting in translational repression and/or degradation of
mRNA (8). miRNAs have previously
been reported to be involved in the regulation of various
biological processes, including the cell cycle, cell
differentiation, apoptosis and metastasis (8). Accumulating lines of evidence have
demonstrated that SNPs or mutations in the miRNA sequence may
influence cancer susceptibility via the alteration of miRNA
expression, maturation or miRNA-miRNA interactions (9,10).
An association between aberrant miRNA expression and oncogenesis
has been consistently reported (11,12),
and the dysregulated expression of miRNAs and their targets is
hypothesized to occur as a result of functional polymorphisms in
the miRNA sequence (13–17). The rs2910164 polymorphism in the
miR-146a precursor has previously been associated with various
types of cancer, including breast and ovarian cancer (18,19),
papillary thyroid cancer (11),
prostate cancer (20), esophageal
squamous cell carcinoma (21) and
gastric (22) and hepatocellular
cancer (23). Jazdzewski et
al (11) presented evidence
demonstrating that the C allele of rs2910164 may induce a reduction
in mature miR-146a levels and decreased inhibition of its target
genes, including tumor necrosis receptor-associated factor 6
(TRAF6) and interleukin-1 receptor-associated kinase 1
(IRAK1) by interfering with the processing of
pre-microRNA.
Notch signaling comprises a major pathway involved
in GBM development. Notch signaling has been observed to maintain
the proliferation of normal neural precursor cells and has been
defined as a survival marker in gliomas, as indicated by its
overexpression in glioma tissue and ability to promote glioma cell
migration and invasion by stimulation of β-catenin and nuclear
factor-κB (NF-κB) signaling via AKT activation (15,16).
Xu et al (21) revealed
that two major members of the Notch family, Notch1 and Notch2, have
opposite roles in regulating the proliferation of GBM cells.
Notably, by searching online microRNA databases and in
silico analysis, Notch1 and Notch2 were identified as potential
target genes of miR-146a. Additionally, Notch1 has been validated
as a target gene of miR-146a using a luciferase assay in GBM cells,
and it was induced as a negative-feedback regulator to suppress
tumor growth via the inhibition of Notch1 (17).
Based on the above evidence, the differential
inhibitory effect of miR-146a on Notch1 and Notch2 was evaluated at
various concentrations of the miRNA. Simultaneously, genotyping
analyses were performed for the miR-146a rs2910164 polymorphism and
their associations with the prognosis of GBM in 380 Chinese GBM
patients were evaluated.
Materials and methods
Patients
A total of 380 patients with histologically
confirmed GBM were recruited from the Department of Neurosurgery,
The First Affiliated Hospital of Dalian Medical University (Dalian,
China) between 2008 and 2012. Of this total, resected tumor
specimens were available from 138 patients. All participants were
ethnic Han Chinese individuals. The study was approved by
investigational review committees at Dalian Medical University.
Written informed consent was obtained from each participant.
Demographic data and information regarding known and potential risk
factors were obtained through interviewer administered
questionnaires.
Biospecimen collection
Genomic DNA samples for genotyping were isolated
from peripheral venous blood collected from all participants using
a DNA extraction kit obtained from Tiangen Biotech Co. Ltd.
(Beijing, China). A NanoPhotometer Ultraviolet/Vis
Spectrophotometer (Implen GmbH, Munich, Germany) and 0.6% agarose
electrophoresis were used to assess the concentration and purity of
the DNA prior to its storage at −20°C until future use.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was obtained using the TRIzol one-step RNA
isolation kit (Invitrogen Life Technologies, Carlsbad, CA, USA).
The miR-146a, Notch1, Notch2 and U6 (internal control)-specific
cDNA were synthesized from total RNA using gene-specific primers
according to the TaqMan MicroRNA assay protocol (Applied
Biosystems, Foster City, CA, USA). In a 96-well plate, 90 µl
master mix was added to 2 µg total RNA from each sample and
run under the following thermal cycling conditions: 25°C for 10
min, 48°C for 30 min and 95°C for 5 min. Relative quantification of
target miRNA expression was conducted using the ΔΔ cycle threshold
(ΔΔCt) method. Each sample was evaluated in triplicate and the raw
data are presented as the relative quantity of target miRNA,
normalized with respect to U6.
Genotyping of miR-146a rs2910164 SNP
Genotyping was performed as described previously
(11). Briefly, DNA specimens were
amplified using standard PCR protocols. The PCR products were
purified with the ExoSAP-IT purification kit (USB Corporation,
Cleveland, OH, USA) and were subsequently sent to the core
sequencing lab located in Dalian Medical University for sequencing
using the ABI sequencing system (PE Applied Biosystems, Foster
City, CA, USA). The sequencing results were analyzed using
Chromosome DNAstar software, version 12.1 (DNASTAR, Inc., Madison,
WI, USA). The PCR primers used for miR-146a sequencing were:
Forward, 5′-ATTTTACAGGGCTGGGACAG-3′ and reverse,
5′-TCTTCCAAGCTCTTCAGCAG-3′.
Western blot analysis
Total proteins were isolated from U251 glioma cells
(Type Culture Collection of the Chinese Academy of Sciences,
Shanghai, China) that were transfected with scramble, miR-146a
mimic or miR-146a inhibitor (Shanghai GenePharma Co., Ltd.,
Shanghai, China), respectively. The protein concentration was
quantitatively measured prior to being loaded onto 10%
SDS-polyacrylamide gel (Invitrogen Life Technologies) and
subsequently transferred onto a polyvinylidene difluoride membrane
(EMD Millipore, Billerica, MA, USA). The membrane was then
sequentially incubated with rabbit polyclonal IgG anti-Notch1
(sc-9170) and anti-Notch2 (sc-5545) primary antibodies (1:2,000; 2
h at room temperature) and horseradish peroxidase-conjugated
secondary antibody (sc-2371; 1:10,000; 1 h at room temperature)
according to manufacturer's instructions. All antibodies were
purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA)
and the signals were detected using an ECL chemifluorescence
detection kit (Pierce Biotechnology, Inc., Rockford, IL, USA). The
band density of specific proteins was densitometrically analyzed
following normalization to the density of β-actin.
MTT assay
The growth rate of control and transfected U251
cells was determined using an MTT assay. Briefly, 4×103
cells/well were plated onto a 96-well plate. Levels of cell growth
were measured each day for six consecutive days following plating,
by adding 20 µl (5 mg MTT/ml; Sigma-Aldrich, St. Louis, MO,
USA) to the well and incubating for an additional 4 h, prior to
termination of the assay by lysing the cells with 200 ml dimethyl
sulfoxide (Sigma-Aldrich) for 5 min. Optical density was measured
in triplicate at 570 nm using a xMark Microplate Absorbance
spectrophotometer (Bio-Rad Laboratories, Inc., Hercules, CA, USA)
and expressed as a percentage of the control.
Transwell assay
Transwell filters (Corning Costar, Inc., Corning,
NY, USA) were coated with Matrigel (3.9 mg/ml, 60–80 ml; BD
Biosciences Discovery Labware, Bedford, MA, USA) and incubated at
37°C for 30 min, prior to solidification of the Matrigel. Matrigel
is used as a surrogate for the major components of the
extracellular matrix in tumor cell invasion. Transfected and
control cells (1×105) suspended in 200 ml serum-free
Dulbecco's modified Eagle's medium (Invitrogen Life Technologies)
were added to the upper chamber and the conditioned medium
containing tumor cells was added into the lower chamber. Following
24 h of incubation at 37°C in 5% CO2, the medium was
removed from the upper chamber. The non-invaded cells on the upper
surface of the inserted filter were gently scraped off with a wet
cotton swab and the cells that had invaded the lower surface of the
filter were fixed in methanol and counted. The quantity was
expressed as a percentage of the control.
Statistical analysis
Fisher's exact χ2 test was used to
compare the frequency distribution of age, gender and smoking
status between high- and low-grade groups, if appropriate. The
Kaplan-Meier method was used to estimate overall survival (OS),
defined as the length of time between study registration and a
patient's mortality (of the patients who died). The OS periods were
compared using the log-rank test. Univariate and multivariate Cox
proportional hazard regression analyses were performed to estimate
the effect of miR-146a polymorphisms on survival rate and the
expression levels of miR-146 and Notch1 in the presence of known
prognostic factors, including age, gender and smoking status. Data
were analyzed using SPSS 19.0 software (IBM, Armonk, NY, USA) and
P<0.05 was considered to indicate a statistically significant
difference.
Results
Demographic and clinicopathological
characteristics of the participants
In the present study a total of 380 participants
(198 males and 82 females), with a mean age of 56.92 years, were
recruited. Information regarding the known risk factors of GBM
prognosis, including age, gender and smoking status are described
in Table I.
 | Table IClinicopathological and demographic
characteristics of the patients involved in the study. |
Table I
Clinicopathological and demographic
characteristics of the patients involved in the study.
| Characteristic | GG/GC (n=226) | CC (n=154) | P-value |
|---|
| Age (years ±
standard deviation) | 54.30±12.81 | 58.11±9.63 | 0.150 |
| Gender (male:
female) | 76:150 | 55:99 | 0.674 |
| Smoking status
(smoker: nonsmoker) | 101:125 | 63:91 | 0.465 |
Patients with the rs2910164 CC
polymorphism have a reduced survival rate
The genotype frequency of the SNP amongst the study
population fit the Hardy-Weinberg equilibrium. Initially, the
association between the SNP and the survival rate in the GBM
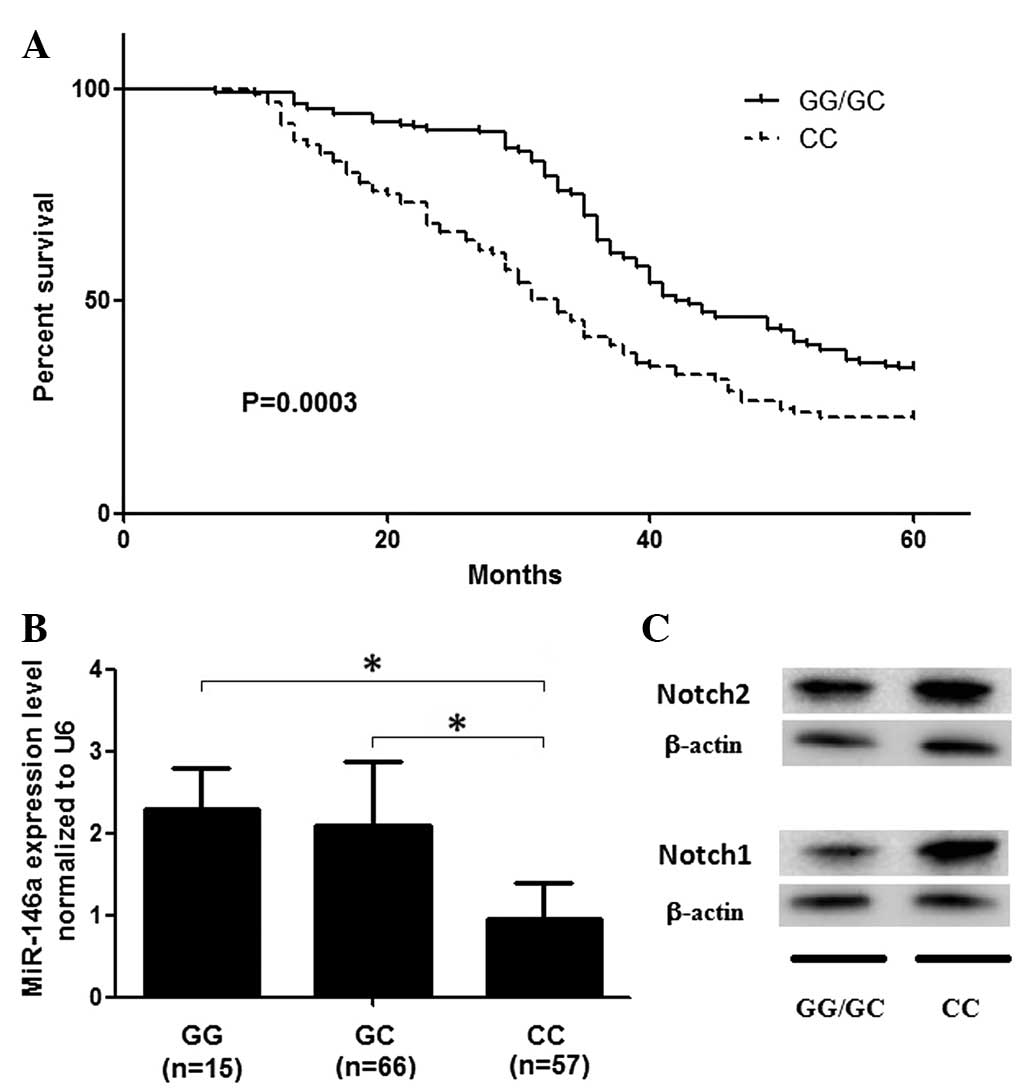
patients was evaluated. As shown in Fig. 1A, participants carrying the
rs2910164 CC genotype had a significantly decreased survival rate
compared with that of the GG and GC genotypes, following
adjustments for age, gender and smoking status (P=0.002). The
effect of the rs2910164 C allele on survival rate indicated a
recessive pattern in GBM patients (data not shown).
Patients carrying the rs2910164 CC
genotype exhibit increased miR-146a expression
The mRNA expression level of miR-146a was also
measured in the 138 patients whose tumor specimens were available
and its association with the genetic variant was evaluated. As
shown in Fig. 1B, the expression
levels of miR-146a in those who carried the rs2910164 CC genotype
were significantly higher than those of the GC and GG carriers
(P<0.001). The number of rs2910164 G alleles revealed a dominant
effect on the expression of miR-146a, as shown in Fig. 1. Furthermore, Notch1 and Notch2
were found to be differentially expressed in individuals with the
GG/GC and CC genotypes (Fig.
1C).
Notch1 and 2 are targets of miR-146a and
miR-146a expression levels affect the Notch1/2 ratio
The candidate target genes of miR-146a were further
computationally screened and identified using TargetScan Release
5.1 (http://www.targetscan.org). Notch1 and
Notch2 were identified as potential target genes of miR-146a, which
was subsequently confirmed using a luciferase assay, as shown in
Fig. 2. Considering the opposing
roles of Notch1 and Notch2 in gliomagenesis, the direct effect of
miR-146a on cell behavior, as well as the expression levels of
Notch1 and Notch2, were investigated using western blot analysis
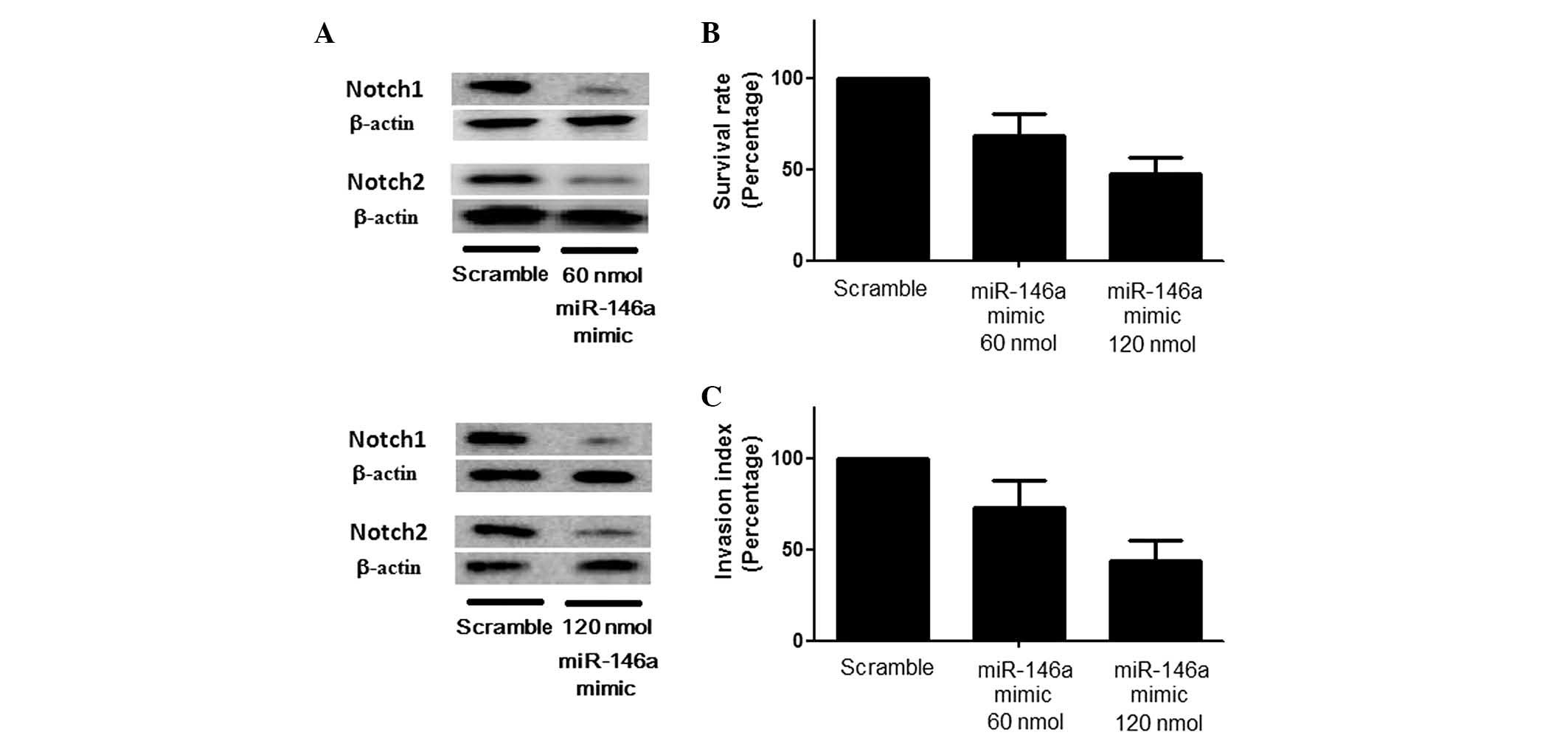
and the results revealed that transfection with miR-146a mimic
induced a significant downregulation of Notch1 and Notch2
expression to differing extents, presenting a stepwise reduction in
the Notch1/Notch2 ratio coupled with an ascending concentration of
the miR-146a mimic (Fig. 3A) and
suppressed the growth and invasion of U251 cells (Fig. 3B). Consistent with these results,
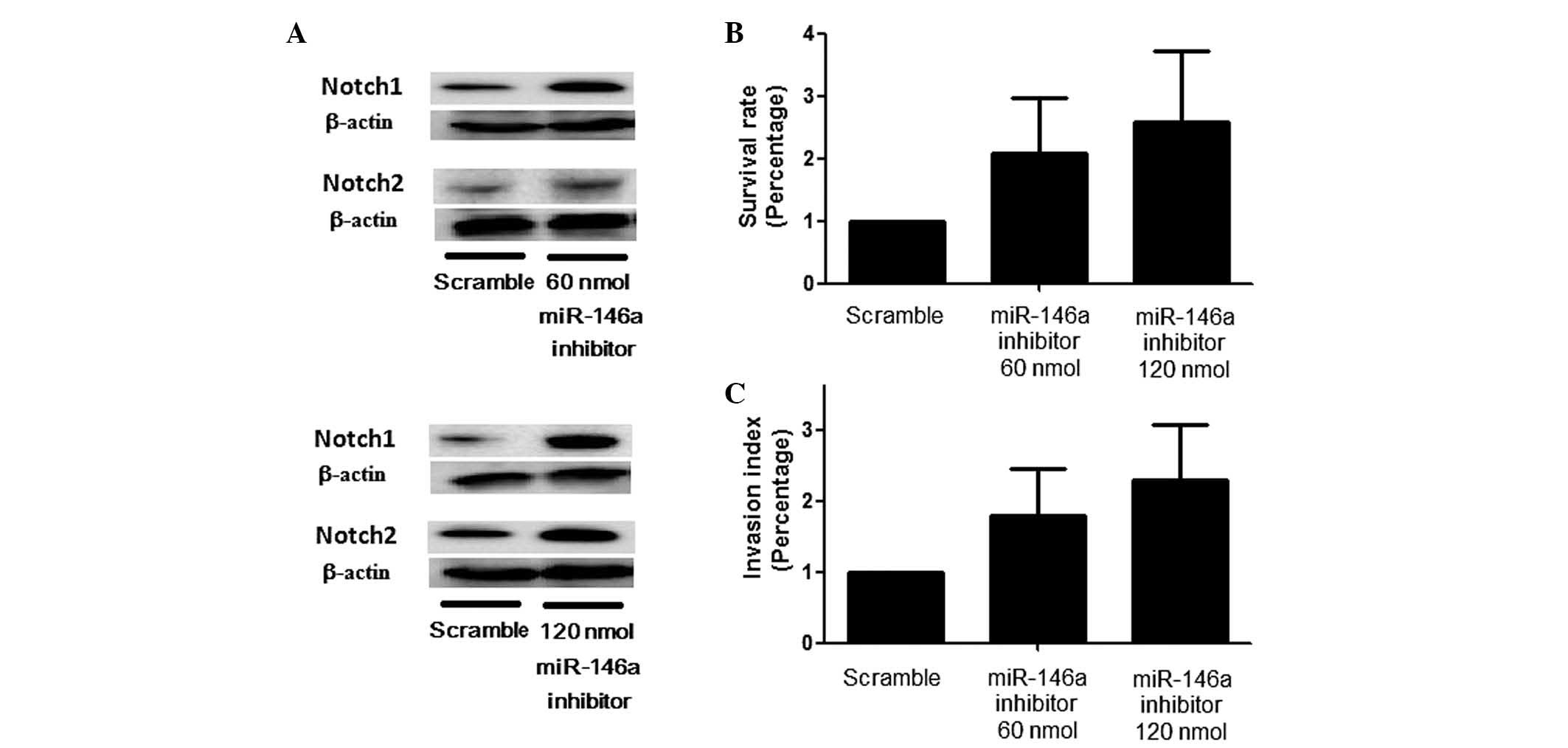
the introduction of an miR-146a inhibitor resulted in a marked
upregulation of Notch1 and Notch2 expression, with a corresponding
stepwise change in the Notch1/Notch2 ratio (Fig. 4A) and promoted cell proliferation
and invasion (Fig. 4B and C) with
various concentrations of the inhibitor.
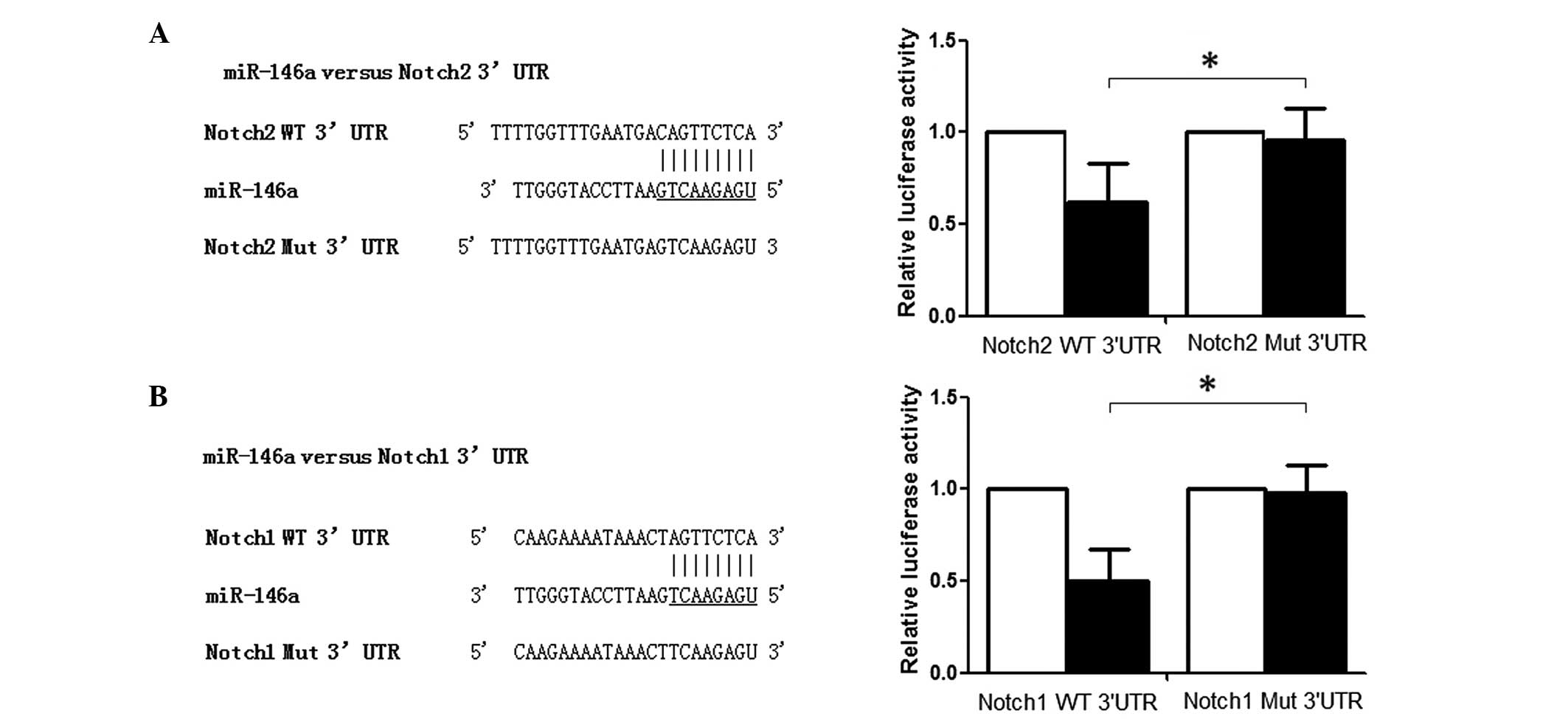 | Figure 2Identification of miR-146a target
genes. (A) Notch1 is a direct target of miR-146a in glioma cells.
Left, schematic representation of the miR-146a-binding sequence in
the 3′-UTR of Notch1 mRNA. Mutations were generated in the
miR-146a-binding sequence of the Notch1 3′-UTR as indicated; Right,
analysis of luciferase activity in U251 cells 48 h after
co-transfection with the control Renilla luciferase
expression construct pRL-TK and a Firefly luciferase reporter
plasmid containing wild-type or mutant Notch1 3′-UTR (indicated as
WT or MUT on the axis, respectively). Firefly luciferase activity
in each sample was normalized to Renilla activity, and
expressed relative to the normalized luciferase activity of control
cells (P<0.05). (B) Notch2 is a direct target of miR-146a in
glioma cells. Left, schematic representation of the
miR-146a-binding sequence in the 3′-UTR of Notch2 mRNA. Mutations
were generated in the miR-146a-binding sequence of the Notch2
3′-UTR as indicated; Right, analysis of luciferase activity in U251
cells 48 h after co-transfection with the control Renilla
luciferase expression construct pRL-TK and a Firefly luciferase
reporter plasmid containing either the wild-type or mutant Notch2
3′-UTR (indicated as WT or MUT on the axis, respectively). Firefly
luciferase activity in each sample was normalized to Renilla
activity, and expressed relative to the normalized luciferase
activity of control cells (P<0.05). miR, microRNA; UTR,
untranslated region; WT, wildtype; MUT, mutant.
*P<0.05. |
Tissue samples in the CC group exhibit
greater Notch1 and 2 expression compared with the GG/GC group
Tissue samples from 138 glioma patients were also
collected and the expression patterns of miR-146a, Notch1 and
Notch2 were determined using RT-qPCR and western blot analysis in
two genotypic groups. The expression levels of miR-146a were
higher, and Notch1 and Notch2 were lower in the GG/GC group than
those in the CC group, and the Notch1/Notch2 ratio was
significantly higher in the CC group than that in the GG/GC group
(data not shown).
Discussion
In the present study, the association between a
common miR-146a polymorphism (rs2910164) and glioma prognosis was
evaluated in a Chinese population of 380 GBM cases. It was
identified that the CC genotype was associated with a decreased
survival rate amongst patients with GBM.
Permuth-Wey et al (19) reported that the rs2910164 CC/GC
genotypes were associated with an increased risk of glioma,
particularly among older individuals, and that the C allele was
associated with a decreased survival rate among patients with
glioblastoma. Xu et al (20) demonstrated that the rs2910164 CC
genotype was associated with a decreased risk of prostate cancer.
Xu et al (22) observed
that the GG genotype was associated with an increased risk of
hepatocellular carcinoma. Jazdzewski et al (11) demonstrated that the GC genotype was
associated with an increased risk of papillary thyroid cancer
compared with that of the GC/CC genotypes. Guo et al
(23) identified an association
between the GG genotype and an increased risk of esophageal
squamous cell carcinoma, particularly among smokers. The C allele
carriers were reported to be more susceptible to gastric cancer in
a Japanese population (24), and
conversely the C allele was found to be protective against gastric
cancer in a Chinese population (25). Inconsistent results regarding the
association between the rs2910164 miR-146a genotype and cancer risk
may indicate a heterogeneous tumor etiology or differences in the
ethnicity of investigated populations.
In the present study, the rs2910164 CC genotype was
associated with the risk for high-grade glioma. However, unlike
previously reported age-specific effects for this SNP (25–27),
the association was comparable when stratified with age. Survival
analyses based on the rs2910164 CC genotype indicated a decreased
survival rate in the patients with high-grade glioma, with a
stepwise decrease in miR-146a expression levels. This trend was
also identified in low-grade gliomas, but the rs2910164 genotypes
did not influence the survival rate of this population.
Jazdzewski et al (11) demonstrated that the C allele of
rs2910164 in the pre-miR-146a sequence interfered with the correct
processing and expression of the miRNA and induced a 1.8-fold
reduction in mature miR-146a compared with that of the G allele
(27). The SNP was located at
position +60 relative to the first nucleotide on the passenger
strand of pre-miR-146a, with the C allele being hypothesized to
lead to mispairing within the mature hairpin (11). It has been suggested that the
rs2910164 genotype may contribute to carcinogenesis via mediation
of miR-146a interactions with key target genes (14). Jazdzewski et al (11) also demonstrated that the C allele
compromised the ability of miR-146a to bind with HeLa cell nuclear
proteins and resulted in inefficient inhibition of the miR-146a
target genes TRAF6 and IRAK1. Peng et al
(28) demonstrated that
TRAF6 was involved in the potentiation of growth,
proliferation, invasion and migration of glioma cells (U-87MG cell
line), and it exerted an inhibitory effect on the apoptosis of
glioma cells by activating NF-κB. Concurrently, Funakoshi et
al (29) observed that
overexpression of TRAF6 enhanced interleukin-1 mediated
NF-κB and activator protein 1 activation. TRAF6 is also able
to upregulate hypoxia inducible factor-1α expression and promote
tumor angiogenesis and growth (30). Additionally, TRAF6 has been
reported to have a significant oncogenic role in other types of
cancer, including esophageal squamous cell carcinoma, non-small
cell lung cancer and multiple myeloma, by either activating NF-κB
and Janus kinase/signal transducer and activator of transcription
pathways or via downregulation of activated caspase 3 and cleaved
poly ADP ribose polymerase, and upregulation of c-Jun, B cell
lymphoma 2 and c-Myc (31–33).
Notch signaling has been demonstrated to cumulate
oncogenic activities involved in glioma proliferation, apoptosis
inhibition and invasion, in addition to its functions in the
maintenance of non-neoplastic neural stem cells and
neovascularization (34). Notch1
is a validated target of miR-146a, and miR-146a has been observed
to be induced as a negative-feedback regulator to suppress glioma
growth via inhibition of Notch1 (17). Chen et al (15) demonstrated increased Notch1
expression levels in glioma and suggested that this increase may be
associated with tumor progression. Zhang et al (16) demonstrated that Notch activation
may stimulate the AKT pathway and subsequently activate β-catenin
and NF-κB signaling in glioma cells. By contrast, another major
member of Notch family, Notch2, was found to have a different role
in the control of glioma cell activities. Upregulation of Notch2
resulted in the induction of apoptosis, as well as the suppresion
of cell growth and invasion, similarly to the results of silencing
Notch1 (21). In addition, high
levels of Notch2 indicated a favorable prognosis in all subtypes of
glioma (35). In the present
study, it was verified that Notch1 and Notch2 were target genes of
miR-146a in glioma cells and that the introduction of miR-146a
suppressed Notch1 and Notch2 expression. It was therefore
hypothesized that Notch1 and Notch2 maintain a balance in
regulating cell behavior and the altered level of miR-146 induced
by the rs2910164 polymorphism alters the balance between the two
Notch family members, resulting in changes in cell behavior and
malignancy transformation. The hypothesis was investigated by
transfecting miR-146a mimic into U251 cells, and confirmed by the
observation that overexpression of miR-146a induced a significant
downregulation of Notch1 and Notch2 to differing extents,
presenting a stepwise decrease in Notch1/Notch2 ratio with
ascending concentration of the miR-146a mimic. Conversely, the
introduction of miR-146a inhibitor resulted in marked upregulation
of Notch1 and Notch2 expression with a corresponding stepwise
increase of Notch1/Notch2 ratio at various concentrations of the
inhibitor. Additionally, the expression pattern of miR-146a, Notch1
and Notch2 in the GG/GC group versus the CC group was consistently
in concordance with the aforementioned in vitro functional
studies.
In the present study, the rs2910164 C allele was
found to be associated with a decreased survival rate, indicating
that this variant may contribute to tumor progression by inducing
loss-of-function of miR-146a. Confirmation of the current
association in a larger population is required. Knowledge of the
inherited variation in miRNA-associated genes may aid the
identification of high-risk populations and the development of
diagnostic, prognostic and therapeutic strategies in order to
reduce the burden of gliomas and other malignancies.
References
|
1
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hemminki K and Li X: Familial risks in
nervous system tumors. Cancer Epidemiol Biomarkers Prev.
12:1137–1142. 2003.PubMed/NCBI
|
|
3
|
Scheurer ME, Etzel CJ, Liu M, El-Zein R,
Airewele GE, Malmer B, Aldape KD, Weinberg JS, Yung WK and Bondy
ML: Aggregation of cancer in first-degree relatives of patients
with glioma. Cancer Epidemiol Biomarkers Prev. 16:2491–2495. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bondy ML, Scheurer ME, Malmer B,
Barnholtz-Sloan JS, Davis FG, Il'yasova D, Kruchko C, McCarthy BJ,
Rajaraman P, Schwartzbaum JA, et al: Brain tumor epidemiology:
consensus from the brain tumor epidemiology consortium. Cancer.
113:1953–1968. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shete S, Hosking FJ, Robertson LB, Dobbins
SE, Sanson M, Malmer B, Simon M, Marie Y, Boisselier B, Delattre
JY, et al: Genome-wide association study identifies five
susceptibility loci for glioma. Nat Genet. 41:899–904. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wrensch M, Jenkins RB, Chang JS, Yeh RF,
Xiao Y, Decker PA, Ballman KV, Berger M, Buckner JC, Chang S, et
al: Variants in the CDKN2B and RTEL1 regions are associated with
high-grade glioma susceptibility. Nat Genet. 41:905–908. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Egan KM, Thompson RC, Nabors LB, Olson JJ,
Brat DJ, Larocca RV, Brem S, Moots PL, Madden MH, Browning JE and
Ann Chen Y: Cancer susceptibility variants and the risk of adult
glioma in a US case-control study. J Neurooncol. 104:535–542. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ryan BM, Robles AI and Harris CC: Genetic
variation in microRNA networks: the implications for cancer
research. Nat Rev Cancer. 10:389–402. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang Q, Jie Z, Ye S, Li Z, Han Z, et al:
Genetic variations in miR-27a gene decrease mature miR-27a level
and reduce gastric cancer susceptibility. Oncogene. 33:193–202.
2014. View Article : Google Scholar
|
|
11
|
Jazdzewski K, Murray EL, Franssila K,
Jarzab B, Schoenberg DR, et al: Common SNP in pre-miR-146a
decreases mature miR expression and predisposes to papillary
thyroid carcinoma. Proc Natl Acad Sci USA. 105:7269–7274. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Turner JD, Williamson R, Almefty KK,
Nakaji P, Porter R, Tse V and Kalani MY: The many roles of
microRNAs in brain tumor biology. Neurosurg Focus. 28:E32010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Duan R, Pak C and Jin P: Single nucleotide
polymorphism associated with mature miR-125a alters the processing
of pri-miRNA. Hum Mol Genet. 16:1124–1131. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chae YS, Kim JG, Lee SJ, Kang BW, Lee YJ,
Park JY, Jeon HS, Park JS and Choi GS: A miR-146a polymorphism
(rs2910164) predicts risk of and survival from colorectal cancer.
Anticancer Res. 33:3233–3239. 2013.PubMed/NCBI
|
|
15
|
Chen L, Zhang R, Li P, Liu Y, Qin K, Fa
ZQ, Liu YJ, Ke YQ and Jiang XD: P53-induced microRNA-107 inhibits
proliferation of glioma cells and down-regulates the expression of
CDK6 and Notch-2. Neurosci Lett. 534:327–332. 2013. View Article : Google Scholar
|
|
16
|
Zhang X, Chen T, Zhang J, Mao Q, Li S,
Xiong W, Qiu Y, Xie Q and Ge J: Notch1 promotes glioma cell
migration and invasion by stimulating β-catenin and NF-κB signaling
via AKT activation. Cancer Sci. 103:181–190. 2012. View Article : Google Scholar
|
|
17
|
Mei J, Bachoo R and Zhang CL:
MicroRNA-146a inhibits glioma development by targeting Notch1. Mol
Cell Biol. 31:3584–3592. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Treanor LM, Volanakis EJ, Zhou S, Lu T,
Sherr CJ and Sorrentino BP: Functional interactions between Lmo2,
the Arf tumor suppressor and Notch1 in murine T-cell malignancies.
Blood. 117:5453–5462. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Permuth-Wey J, Thompson RC, Burton Nabors
L, Olson JJ, Browning JE, Madden MH, Ann Chen Y and Egan KM: A
functional polymorphism in the pre-miR-146a gene is associated with
risk and prognosis in adult glioma. J Neurooncol. 105:639–646.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu B, Feng NH, Li PC, Tao J, Wu D, Zhang
ZD, Tong N, Wang JF, Song NH, Zhang W, Hua LX and Wu HF: A
functional polymorphism in Pre-miR-146a gene is associated with
prostate cancer risk and mature miR-146a expression in vivo.
Prostate. 70:467–472. 2010. View Article : Google Scholar
|
|
21
|
Xu P, Zhang A, Jiang R, Qiu M, Kang C, Jia
Z, Wang G, Han L, Fan X and Pu P: The different role of Notch1 and
Notch2 in astrocytic gliomas. PLoS One. 8:e536542013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xu T, Zhu Y, Wei QK, Yuan Y, Zhou F, Ge
YY, Yang JR, Su H and Zhuang SM: A functional polymorphism in the
miR-146a gene is associated with the risk for hepatocellular
carcinoma. Carcinogenesis. 29:2126–2131. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Guo H, Wang K, Xiong G, Hu H, Wang D, Xu
X, Guan X, Yang K and Bai Y: A functional varient in microRNA-146a
is associated with risk of esophageal squamous cell carcinoma in
Chinese Han. Fam Cancer. 9:559–603. 2010. View Article : Google Scholar
|
|
24
|
Okubo M, Tahara T, Shibata T, Yamashita H,
Nakamura M, Yoshioka D, Yonemura J, Kamiya Y, Ishizuka T, Nakagawa
Y, et al: Association study of common genetic variants in
pre-microRNAs in patients with ulcerative colitis. J Clin Immunol.
31:69–73. 2011. View Article : Google Scholar
|
|
25
|
Zeng Y, Sun QM, Liu NN, Dong GH, Chen J,
Yang L and Wang B: Correlation between premiR-146a C/G polymorphism
and gastric cancer risk in Chinese population. World J
Gastroenterol. 16:3578–3583. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shen J, Ambrosone CB, DiCioccio RA, Odunsi
K, Lele SB and Zhao H: A functional polymorphism in the miR-146a
gene and age of familial breast/ovarian cancer diagnosis.
Carcinogenesis. 29:1963–1966. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Iyer A, Zurolo E, Prabowo A, Fluiter K,
Spliet WGM, van Rijen PC, Gorter JA and Aronica E: MicroRNA-146a: a
key regulator of astrocyte-mediated inflammatory response. PLoS
One. 7:e447892012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Peng Z, Shuangzhu Y, Yongjie J, Xinjun Z
and Ying L: TNF receptor-associated factor 6 regulates
proliferation, apoptosis, and invasion of glioma cells. Mol Cell
Biochem. 377:87–96. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Funakoshi M, Sonoda Y, Tago K, Tominaga S
and Kasahara T: Differential involvement of p38 mitogen-activated
protein kinase and phosphatidyl inositol 3-kinase in the
IL-1-mediated NF-kappaB and AP-1 activation. Int Immunopharmacol.
1:595–604. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sun H, Li XB, Meng Y, Fan L, Li M and Fang
J: TRAF6 upregulates expression of HIF-1α and promotes tumor
angiogenesis. Cancer Res. 73:4950–4959. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yao F, Han Q, Zhong C and Zhao H: TRAF6
promoted the tumorigenicity of esophageal squamous cell carcinoma.
Tumour Biol. 34:3201–3207. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu H, Tamashiro S, Baritaki S, Penichet
M, Yu Y, Chen H, Berenson J and Bonavida B: TRAF6 activation in
multiple myeloma: a potential therapeutic target. Clin Lymphoma
Myeloma Leuk. 12:155–163. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu H, Zhang T, Ye J, Li H, Huang J, Li X,
Wu B, Huang X and Hou J: TNF receptor-associated factor 6 in
advanced non-small cell lung cancer: clinical and prognostic
implications. J Cancer Res Clin Oncol. 138:1853–1863. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hori K, Sen A and Artavanis-Tsakonas S:
Notch signaling at a glance. J Cell Sci. 126:2135–2140. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Boulay JL, Miserez AR, Zweifel C,
Sivasankaran B, Kana V, Ghaffari A, Luyken C, Sabel M, Zerrouqi A,
Wasner M, et al: Loss of NOTCH2 positively predicts survival in
subgroups of human glial brain tumors. PLoS One. 2:e5762007.
View Article : Google Scholar : PubMed/NCBI
|