Introduction
Endometrial cancer is one of the most common types
of gynecologic malignancy, increasing each year (1). Based on a pathological view,
endometrial cancer can be divided into two subtypes (2). Type 1 endometrial cancer includes
highly differentiated endometrioid adenocarcinoma, which is
characterized by stepwise carcinogenesis through endometrial
hyperplasia to endometrial cancer. Type 2 endometrial cancer,
including poorly differentiated, serous and clear cell
adenocarcinoma, is reported to occur alongside de novo
mutation of TP53 (3). The
principal cause of type 1 endometrial cancer is considered to be
the prolonged exposure to estrogens without antagonistic effect of
progesterone, and this pathophysiology is closely associated with
first grade amenorrhea, polycystic ovary syndrome, obesity and
hormonal supplementation therapy (3). Estrogen replacement therapy is
utilized to control menopausal symptoms, however, it is known to
increase the risk of developing endometrial cancer between 2- and
20-fold for females possessing uteri (4), and the increment of risk is well
correlated with the duration of its use. In order to reduce the
risk of endometrial cancer, it is recommended that postmenopausal
females possessing uteri use progestin together with estrogens
(4). A representative progestin,
medroxyprogesterone acetate (MPA) is used for fertility-sparing
treatment in type 1 endometrial cancer (5). The way in which MPA maintains
quiescence of the endometrium remains to be elucidated, although
MPA is known to possess detrimental effects on breast tissue, and
MPA marginally increases the risk of developing breast cancer
(6). Therefore, analysis of the
physiological role of estrogens for the prevention of endometrial
cancer is urgently required.
HAND1 and HAND2 constitute the HAND subclass of the
basic helix-loop-helix (bHLH) family, and were independently
identified during analyses to identify candidate E-box binding
transcription factors in yeast two-hybrid screens (7). HAND2 is known as a critical regulator
of morphogenesis in a variety of tissues, as HAND2 is expressed in
the heart and neural crest-derived tissues, and is essential for
the formation of the brachial arch, cardiovasculature and limbs
(8). It has been reported that
HAND2 interacts with GATA4, Nkx2.5, MEF2C, Phox2 and Mash1
(9–12). GATA4, Nkx2.5 and MEF2C are
associated with cardiogenesis, whereas Phox2 and Mash1 are
associated with the development of the autonomic nervous system
(9–12). It has been demonstrated that HAND2
forms homo- or heterodimers with other bHLH proteins, and activates
transcription by binding to the E-box elements (13,14).
However, the downstream factors of HAND2 and the associations
between HAND2 and nuclear receptors remain to be fully elucidated.
A previous study revealed that HAND2 is localized exclusively in
the uteri of stromal tissue, and progesterone-induced expression of
HAND2 in the murine uterine stroma suppresses the production of
fibroblast growth factors (FGFs), which act as paracrine mediators
of the mitogenic effects of estrogen on the uterine epithelium
(15). Whether HAND2 affects the
transcriptional activation function of ERα as a transcriptional
factor remains to be elucidated.
Considering previous data, demonstrating that the
expression of HAND2 is impaired in endometrial cancer, compared
with normal endometrium and endometrial hyperplasia, as determined
using DNA methylation analysis (16), the present study aimed to
investigate the interaction between HAND2 and ERα, and aimed to
identify the physiological function of HAND2, particularly
associated with endometrial cancer. The results of this
investigation may prove to be useful in identifying novel
molecular-targeted therapies for the treatment of endometrial
cancer.
Materials and methods
Chemicals and antibodies
The MG132 proteasome inhibitor,, 17β-estradiol
(E2) and anti-Flag M2 agarose were purchased from
Sigma-Aldrich (St. Louis, MO, USA). The ERα selective ligand,
propylpyrazole triol (PPT), was obtained from Tocris Bioscience
(Ellisville, MO, USA). Mouse monoclonal antibodies used were
anti-ERα (D-12; Santa Cruz Biotechnology, Inc., Santa Cruz, CA,
USA), anti-β-Actin (cat no. sc-47778; Santa Cruz Biotechnology,
Inc.) and anti-HA (cat. no. 12CA5; Roche Applied Science, Basel,
Switzerland). Rabbit polyclonal antibodies included anti-ERα
(MC-20; Santa Cruz Biotechnology, Inc.), anti-ERβ (H-150; Santa
Cruz Biotechnology, Inc.), anti-DYKDDDDK Tag (cat. no. #2368 Cell
Signaling Technology, Inc., Danvers, MA, USA), and anti-HAND2 (cat.
no. PAB4702; Abnova, Taipei, Taiwan).
Cell culture
The ERα-positive MCF-7 (cat. no. HTB-22) and ERβ
positive MDA-MB-231 (cat. no. HTB-26) human breast cancer cell
lines, and the AN3CA human endometrial cancer cell line (cat. no.
HTB-111) were purchased from American Type Culture Collection
(Manassas, VA, USA). These cells were maintained in Dulbecco's
modified Eagle's medium (Wako Pure Chemical Industries, Ltd.,
Osaka, Japan) supplemented with 10% fetal bovine serum
(Sigma-Aldrich) at 37°C in a humidified 5% CO2
incubator.
Expression vectors
Human ERα vectors and the ERE-tk-Luc and
17M8-AdMLP-Luc reporter constructs were used, as described
previously (17,18).
Immunoprecipitation, western blot
analysis and ubiqutination assay
The formation of endogenous HAND2-ERα and HAND2-ERβ
complexes were analysed by co-immunoprecipitation using specific
antibodies raised against human ERα and ERβ, followed by
immunoblotting using anti-human HAND2 antibody, as described
previously (17,18).
For evaluating the HAND2-mediated degradation of
ERα, 2×105 AN3CA cells were transfected with 0.2
µg/ml pcDNA FLAG ERα and/or pcDNA Myc HAND2 in 6 cm dishes.
The cells were treated with or without MG132 (10−5 M),
and were harvested 24 h following the addition of MG132.
For evaluating the HAND2-mediated degradation of
ERα, 0.2 µg/ml HA-tagged ubiquitin (HA-Ub), pcDNA FLAG ERα
and pcDNA Myc HAND2 were transfected into 4×104 HEK293T
cells, in the presence or absence of E2 (10−8
M). The cell lysates were subjected to anti-Flag M2 agarose (1:100;
Sigma-Aldrich) and the level of Ub-bound ERα protein was evaluated
using western blotting, as previously described (17,18).
The antibodies used for western blotting were as
follows: Anti-ERα (1:1,000; mouse monoclonal and rabbit
polyclonal), anti-β-actin (1:10,000), anti-HA (1:1,000), anti-ERβ
(1:1,000), anti-DYKDDDDK Tag (1:1,000) and anti-HAND2 (1:1,000).
These primary antibodies were incubated overnight at 4°C and the
results were visualized by ImageQuant™ LAS-3000 (GE Healthcare Life
Sciences, Chalfont, UK).
In vitro glutathione S-transferase
(GST)-pull down assay
The GST fusion proteins, GST-ERα activation function
(AF)-1/AF-2, or GST alone were expressed in Escherichia coli
(Takara Bio, Inc., Otsu, Japan) and bound to glutathione-sepharose
4B beads (GE Healthcare Life Sciences). The expression levels of
GST-ERα AF-1 and AF-2 were confirmed using Coomassie Brilliant Blue
(Thermo Fisher Scientific, Waltham, MA, USA) staining (17,18).
The immobilized GST-ERα AF-2 fusion proteins were preincubated for
30 min in GST binding buffer, containing 20 mM Tris-HCl (pH 7.5),
200 mM NaCl and 1 mM EDTA, with or without E2
(10−6 M). The GST proteins were incubated at 4°C with
the indicated [35S] methionine-labeled proteins. After 1
h incubation, unbound proteins were removed by washing the beads in
GST binding buffer containing 0.5% Nonidet P-40 (Wako Pure Chemical
Industries, Ltd.) and protease inhibitor cocktail (Roche Applied
Science). The specifically-bound proteins were eluted by boiling in
SDS sample buffer and analysed using 10% SDS polyacrylamide gel
electrophoresis and autoradiography (ImageQuant™ LAS-3000; GE
Healthcare Life Sciences).
Luciferase reporter assay
For the luciferase assay, 4×104 HEK293T
cells were transfected with 0.2 µg/ml pcDNA, pcDNA FLAG ERα,
pcDNA, pM ERα AF-2 and 0.2–0.6 µg/ml pcDNA Myc HAND2 vectors
using Effectene reagent (Qiagen, Hilden, Germany), according to the
manufacturer's instructions. As an internal control to equalize the
transfection efficiency, a phRL CMV-Luc vector (Promega
Corporation, Madison, WI, USA) was also transfected in all the
experiments. Individual transfections, each consisting of
triplicate wells, were repeated at least three times (17,18).
Reverse transcription-semi-quantitative
polymerase chain reaction (RT-qPCR)
The AN3CA cells were transfected with either pcDNA3
(control; Invitrogen, Carlsbad, CA, USA) or pcDNA Myc HAND2. These
cells were then treated with vehicle, E2 or PPT for 24
h. Total RNA was extracted from the cells using an RNeasy Mini kit
(Qiagen), and cDNA was synthesized using ReverTra Ace (Toyboyo, Co,
Ltd., Tokyo, Japan). The expression of each mRNA was normalised for
RNA loading in each sample using glyceraldehyde 3-phosphate
dehydrogenase (GAPDH). The primers and conditions for the
amplification of GAPDH were as described previously (19). The PCR primers for GAPDH VEGF and
ERα were as follows: GAPDH, forward 5′-TGC ACC ACC AAC TGC TTA
GC-3′ and reverse 5′-GGC ATG GAC TGT GGT CAT GAG-3; VEGF, forward
5′-CCA GCA GAA AGA GGA AAG AGG TAG-3′ and reverse 5′-CCC CAA AAG
CAG GTC ACT CA C-3; and ERα, forward 5′-TGT GCA ATG ACT ATG CTT
CA-3′ and reverse 5′-GCT CTT CCT CCT GTT TTT A-3′. Firstly, 250 ng
cDNA, 0.1 µl Ex Taq Polymerase (Takara Bio, Inc.) and 0.2
µM primers were mixed. Thereafter each PCR regimen involved
a 2 min initial denaturation step (94°C), which was followed by
15-30 cycles at 94°C for 30 sec, then at 55°C for 30 sec and
finally at 72°C for 30 sec using a Thermal Cycler Gene Atlas
instrument (ASTEC Co., Ltd., Kasuya, Japan).
Statistical analysis
Data are presented as the mean ± standard error of
the mean from at least three independent determinations. Multiple
comparisons between more than two groups were analysed using
one-way analysis of variance and post-hoc tests using GraphPad
Prism version 6.0 with the Bonferoni/Dunn post hoc tests (GraphPad
Software, San Diego, CA, USA). P<0.05 were considered to
indicate a statistically significant difference.
Results
HAND2 directly binds to the AF-1 region
of ERα in a ligand-independent manner
To assess the hypothesis that ERα interacts with
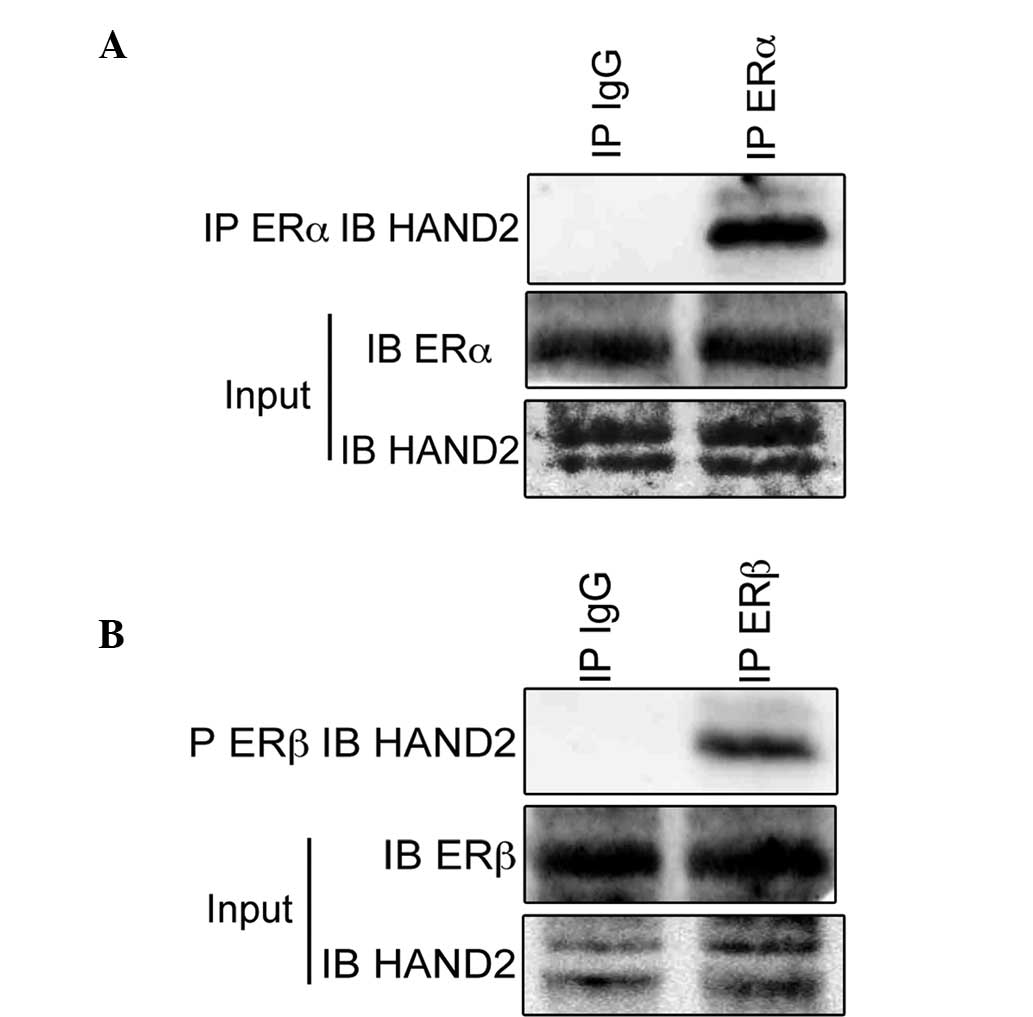
HAND2 protein, the present study performed immunoprecipitation
assays using antibodies raised against ERα. The immunoblotting
revealed the existence of HAND2 protein in the cell lysates of the
ERα-proficient MCF-7 cells (Fig.
1A). The present study also performed immunoprecipitation
assays using antibodies raised against ERβ, and the immunoblotting
revealed the existence of HAND2 protein in the cell lysates of the
ERβ-proficient MDA-MB-231 cell (Fig.
1B), which indicated that HAND2 was physically associated with
ERα and ERβ. Subsequently, the present study examined the function
of HAND2 in association with ERα to determine the physiological
function of HAND2 in endometrial cancer.
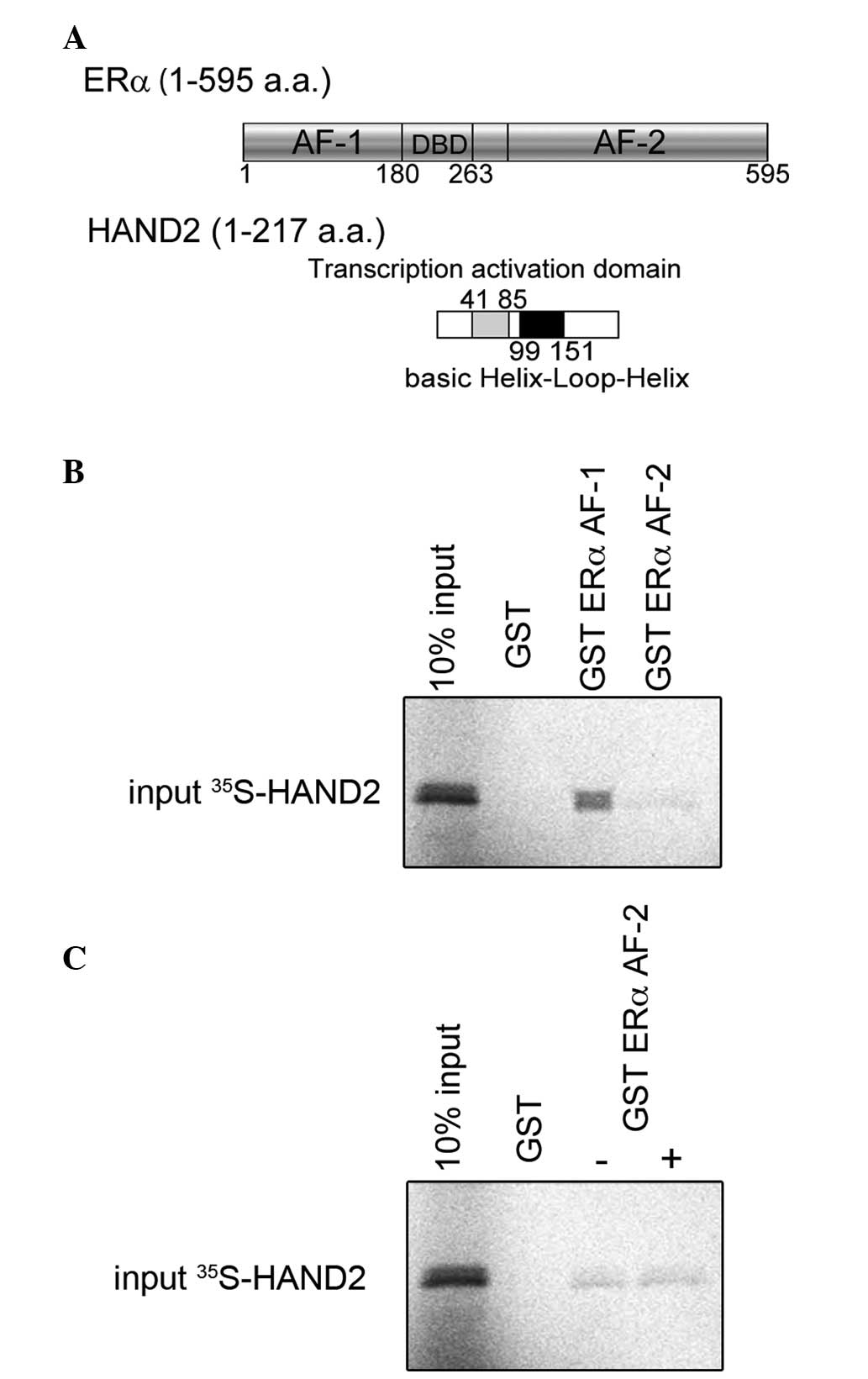
The results, described above, were further confirmed
using in vitro pull-down assays to demonstrate the
functional importance of the HAND2-ERα interaction. To map the
region of ERα that interacts with HAND2 as a transcription factor,
GST-fused ERα activation function (AF)-1 or AF-2 (Fig. 2A) and [35S]
methionine-labelled HAND2 were incubated and their interactions
were assessed. As shown in Fig.
2B, the GST-fused ERα AF-1 protein possessed the ability to
retain HAND2 on the column. The GST ERα AF-2 column exhibited weak
interaction with HAND2, compared with the GST ERα AF-1 column and
the interaction between HAND2, and GST ERα AF-2 was unchanged,
regardless of the presence of E2 (Fig. 2C). These data indicated that HAND2
interacted directly with ERα AF-1 in a ligand-independent
manner.
HAND2 represses the transcriptional
activation function of ERα
To examine the cofactor activity of HAND2 in the
transcriptional activation function of ERα, the present study
performed transient transfection assays using a luciferase reporter
plasmid, driven by a thymidine kinase promoter containing three
tandem repeats of the canonical estrogen responsive element
(AGGTCAnnnTGACCT). Although ERα exhibited a ligand-dependent
transactivation function in the HEK293T cells (Fig. 3A, lane 4), the transient expression
of HAND2 led to a significant decrease in luciferase activity of
ERα, and this downregulation increased as the quantity of HAND2
expression vector increased (Fig.
3A, lanes 5–7).
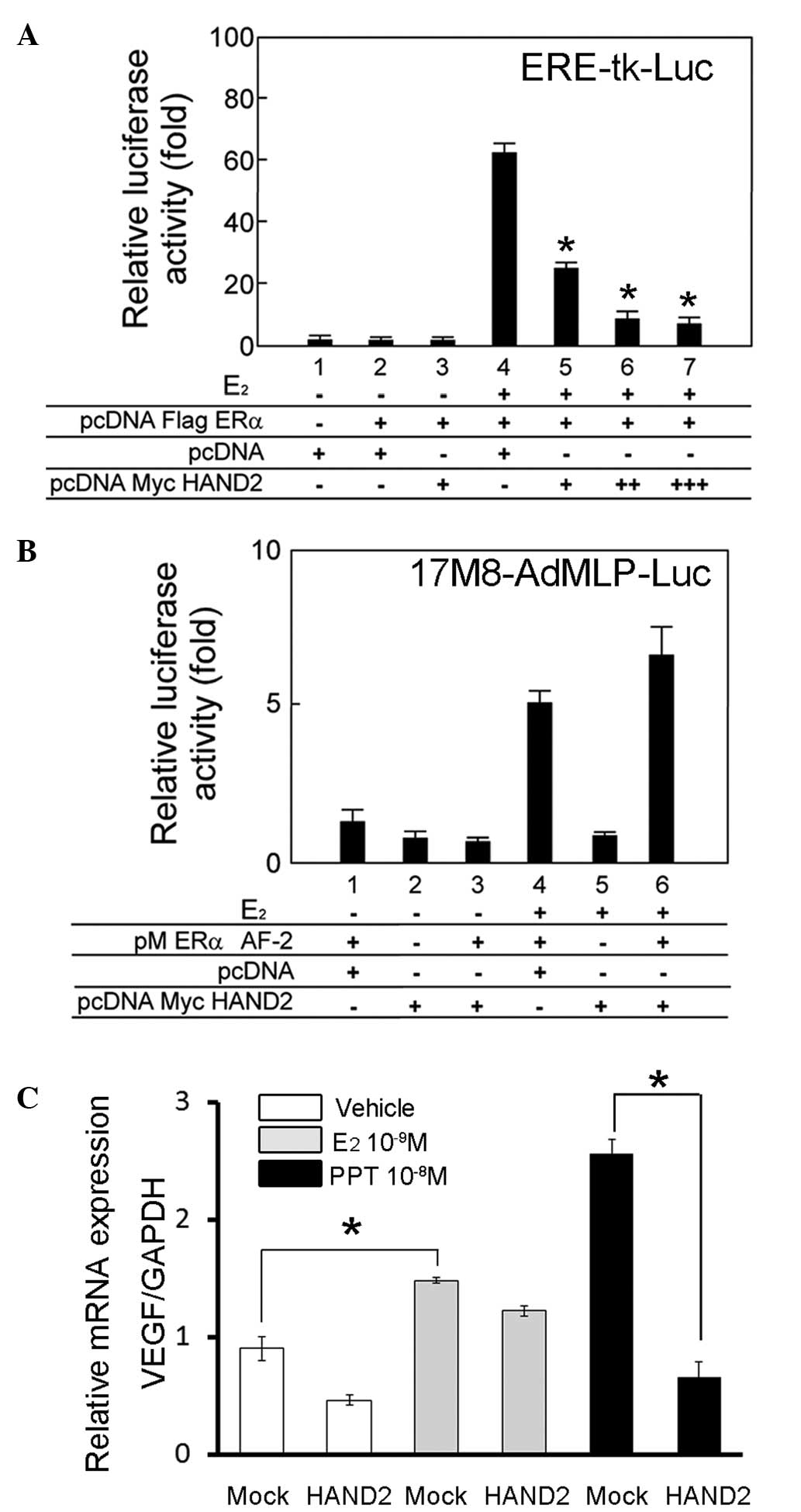 | Figure 3HAND2 attenuates the
ligand-independent transcriptional activation function of ERα. (A)
Transient transfection assays were performed to examine the
activity of HAND2 in the transcriptional activation function of
ERα. Indicated plasmids were cotransfected into the HEK293T cells
in the presence or absence of E2 (10−8 M).
The cells were harvested 24 h after transfection with the
expression vectors and reporter constructs (ERE-tk-Luc), and the
transfected whole cell lysates were assayed for luciferase
activity, produced from the reporter plasmid. HAND2 exhibited
specific repression of the ligand-dependent transactivation
function of ERα in the HEK293T cells, in a dose-dependent manner.
Individual transfections, each consisting of triplicate wells, were
repeated at least three times. *P<0.05 vs. lane 4.
(B) Transient transfection assays were performed to examine the
activity of HAND2 in the transcriptional activation function of ERα
AF-2. The cells were harvested 24 h after transfection with the
expression vectors and reporter constructs (17M8-AdMLP-Luc), and
the transfected whole cell lysates were assayed for luciferase
activity, produced from the reporter plasmid. HAND2 exhibited no
significant effect on the ligand-dependent transactivation function
of ERα AF-2 in the HEK293T cells. Individual transfections, each
consisting of triplicate wells, were repeated at least three times.
(C) mRNA expression of VEGF was analysed as the representative
downstream gene of ERα. The exogenous expression of HAND2 in the
AN3CA cells decreased the mRNA level of VEGF, particuarly in the
presence of PPT. mRNA levels were normalized by GAPDH.
*P<0.05. Mock denotes transfection of empty (pcDNA)
vector, and vehicle denotes solvent. ERα, estrogen receptor α; AF,
activation function; PPT, propylpyrazole triol; VEGF, vascular
endothelial growth factor; GAPDH, glyceraldehyde 3-phosphate
dehydrogenase. |
The present study subsequently aimed to determine
the role of HAND2 in the transactivation function of GAL4-fused ERα
AF-2. For this purpose, transient transfection assays were
performed in HEK293T cells using a 17M8-AdMLP-luc luciferase
reporter plasmid (20). The
ligand-induced trans-activation function of ERα AF-2 (Fig. 3B, lane 4) was not significantly
affected by the exogenous expression of HAND2 (Fig. 3B, lane 6).
To evaluate the effect of HAND2 on endogenous gene
expression, the mRNA expression of VEGF was examined in AN3CA
endometrial cancer cells, as expression of the VEGF gene is driven
by estrogen (21). As expected,
treatment of the AN3CA cells with E2 led to a
significant increase in the mRNA expression of VEGF, and
overexpression of HAND2 suppressed the mRNA expression of VEGF, in
the presence or absence of ERα agonists. However, suppression was
most prominent in the presence of PPT, an ERα specific agonist
(Fig. 3C). Therefore, these
results indicated that HAND2 suppressed the transcriptional
activation function of ERα via its AF-1 domain, and the suppression
was specific for ERα.
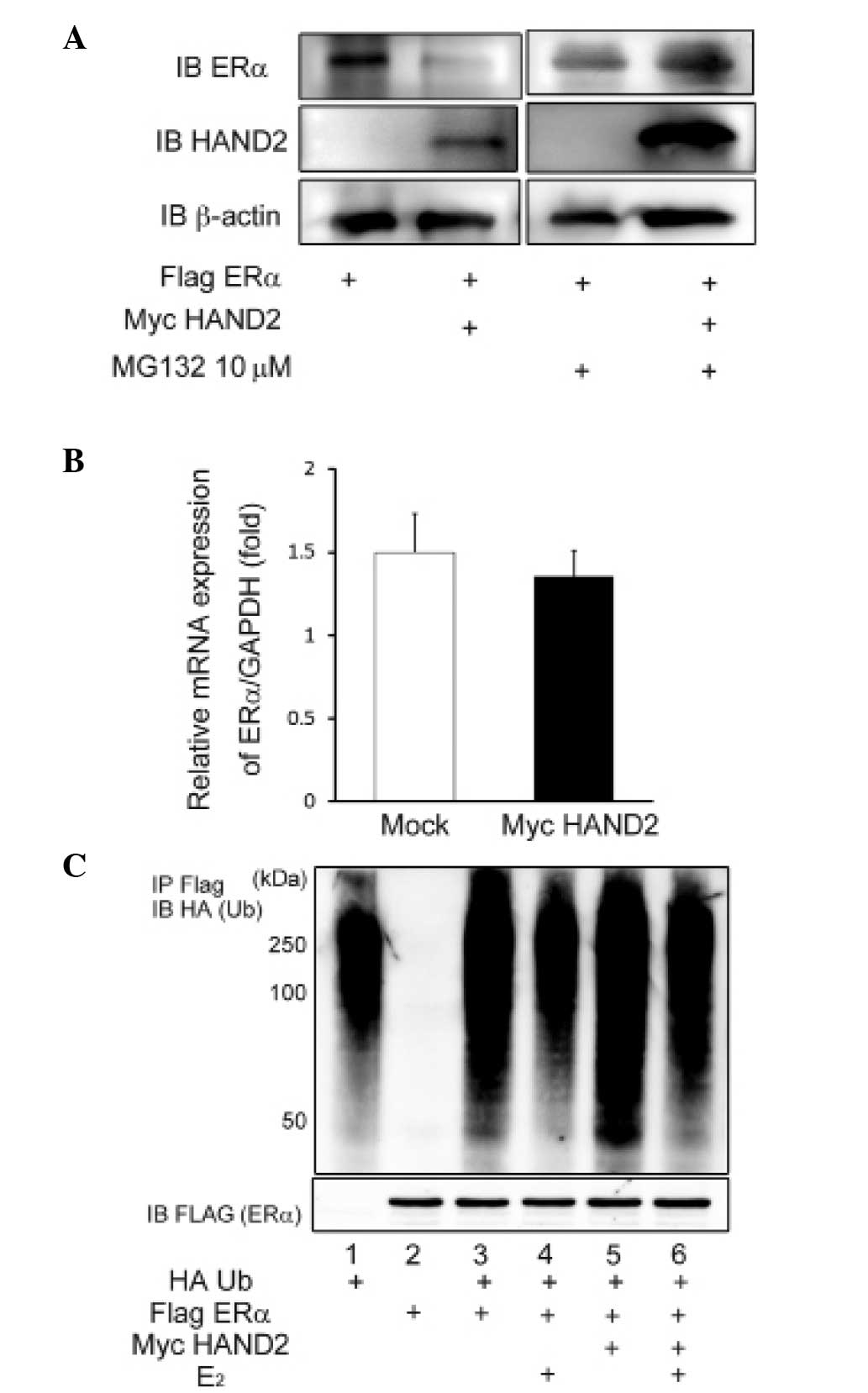
Stimulation of proteasomal degradation by
HAND2 decreases the expression level of ERα
To elucidate the mechanism underlying the
HAND2-induced decrease in the ligand-dependent transcriptional
activation function of ERα, the present study investigated the
possibility of post-transcriptional modification of ERα by HAND2.
The protein expression of ERα was significantly reduced by
exogenous expression of HAND2 in the AN3CA cells (Fig. 4A), although the mRNA level of ERα
was unaffected by forced expression of HAND2 (Fig. 4B). In addition, the decreased
protein expression of ERα was reversed by the addition of the MG132
proteasome inhibitor (Fig. 4A).
Thus, it was hypothesized that HAND2 protein may stimulate
degradation of the ERα protein, and this degradation of ERα protein
results in the downregulation of transcriptional activation of ERα.
To confirm this hypothesis, HA-Ub and Flag-tagged ERα were
transfected into the HEK293T cells, and the protein level of ERα
and ubiqutination status of ERα were determined using western
blotting with anti-Flag M2 agarose and anti-HA antibody. The
ubiquitination assays demonstrated the polyubiquitinated status of
ERα (Fig. 4C), indicating that ERα
was degraded via the ubiquitin-proteasome pathway, irrespective of
the presence or absence of E2.
Discussion
Although it has been reported that HAND2 belongs to
the bHLH transcription factor, which binds to the E-box domain, its
transcriptional function on ERα remained to be fully elucidated.
The present study demonstrated that HAND2 functioned as a modifier
of ERα. Representative transcription factors that contain the bHLH
domain include hypoxia-inducible factor (HIF), Myc, aryl
hydrocarbon receptor (AhR), aryl hydrocarbon receptor nuclear
translocator (ARNT) and TWIST1/2 (22), and the characterization of the bHLH
family as a factor that affects the nuclear receptor superfamily
has been investigated substantially. Originally, the interaction
between AhR and ERα was identified through the investigation of
2,3,7,8-tetrachlorodibenzo-[p]-dioxin (TCDD) (23), and AhR was identified as a
ligand-dependent ubiquitin E3 ligase (24,25).
AhR also belongs to the nuclear receptor super-family, and the
AhR/ARNT heterodimer inhibits ERα activity by binding to the AF-1
lesion of ERα (26), similar to
HAND2 in the present study (Fig.
2B). In the present study, exogenous expression of HAND2
repressed the transcriptional activity of ERα (Fig. 3A and B). This mechanism is similar
to that of AhR. Therefore, it is not surprising that HAND2
repressed the ligand-dependent transcriptional activation function
of ERα.
It has been accepted that the function of ERα is
regulated at transcriptional and post-transcriptional levels. The
latter includes phosphorylation, acetylation, sumoylation,
methylation, palmitoylation, modulation by microRNA and
ubiquitination (27). For
recognition by ubiquitin ligases, a substrate protein requires
phosphorylation or methylation. The sequential administration of
ubiquitin activating enzyme (E1), ubiquitin conjugating enzyme (E2)
and ubiquitin ligase (E3) is followed by 26S proteasomal
degradation (27). Several
ubiquitin proteasome pathway components, including E6AP, MDM2,
BRCA1 and SCFSKP2 are considered to be ERα cofactors
(28). Among these, BRCA1, a
breast and ovarian cancer-susceptible gene product, is known to
form a complex with BARD1, and this complex decreases the
ligand-dependent transcriptional activation function of ERα
(29). The observation that HAND2
functioned as a co-repressor of ERα in the present study resembled
the function of BRCA1, and this inhibition leads to the continuous
suppression of transcriptional activity of ERα, regardless of the
presence of ligands.
Although the expression of HAND2 is regulated by
progesterone (15), a previous
report suggested that increased calcineurin/Nfat signalling and
decreased expression of miR-25 integrated to re-express HAND2
(30), and others have reported
that the expression of HAND2 is silenced by methylation of the
promoter lesion of HAND2 (16).
The present study investigated whether the expression of HAND2 was
manipulated by a DNA de-methylation chemical using
5-Aza-2′-deoxycytidine (AZA). However, the protein level of HAND2
remained unchanged following treatment of the AN3CA cells with AZA
(data not shown). Therefore, another possible mechanism requires
consideration to fully elucidate how the expression of HAND2 may be
modulated. Although it has been demonstrated that the expression of
HAND2 correlates with the ubiquitin fusion degradation 1L (UFD1L)
ubiquitin-conjugating protein (31), the effects of HAND2 on the
expression of UFD1 L were not examined in the present study.
Clarification of the mechanism underlying how HAND2 recruits
ubiquitin-proteasome machinery is required, as HAND2 itself is not
an ubiquitin-conjugating enzyme.
The findings of the present study suggested that
HAND2 may contribute to the suppression of tumorigenesis, as ERα
generally contributes to tumorigenic function by stimulating
cellular proliferation (32) and
E2 may have a principal role in homeostasis of the
uterine endometria, as continuous and unopposed stimulation of the
uterine epithelia by E2 results in the increased
frequency of endometrial cancer (33). Consistent with this hypothesis, the
expression of HAND2 was attenuated in the epithelia of endometrial
cancer, compared with those in the normal endometrium and in
endometrial hyperplasia (16), and
the expression of VEGF was abrogated by concomitant overexpression
of HAND2 particularly in the presence of the ERα-specific ligand,
PPT (Fig. 3C). It was suggested
that HAND2 is involved in maintaining the quiescence of uterine
endometria, however, determining the detailed mechanism underlying
the effects on ERα by HAND2 requires further investigation. The
present study also revealed the interaction between HAND2 and ERβ.
VEGF is a downstream factor of ERβ, as well as ERα, and
E2 stimulates ERα and ERβ (34), however, the role of ERβ in
endometrial cancer remains to be fully elucidated. The present
study suggested that HAND2, a transcription factor for
morphogenesis, may have a function in suppressing
estrogen-dependent cancer. Breast cancer is known to be an
estrogen-associated cancer, and the role of HAND2 in breast cancer
remains to be elucidated. HAND2 may provide an important molecular
target for these hormone-dependent types of cancer, and
identification of the regulating mechanism of HAND2 may improve the
control of these types of cancer.
In conclusion, the present study demonstrated the
role of HAND2 as a negative transcriptional regulator of ERα. HAND2
was involved in the inactivation of ERα by associating with the
amino-terminus of ERα, and exerted degradation of ERα by
stimulating the ubiquitin-proteasome pathway. Therefore, in
addition to inhibition of FGF signalling in the uterine tissue,
HAND2 directly affected the mitogenic effects of ERα, and these
results suggested that inactivation of HAND2 may be detrimental in
the regulation of cellular proliferation. Consistent with this
hypothesis, hypermethylation and silencing of HAND2 is commonly
found in endometrial cancer (16).
Taken together, it can be hypothesized that HAND2 may serve as a
possible tumor suppressor, particularly in uterine tissue. However,
further investigations are required to confirm the physiological
implications of HAND2 in the uterus.
Acknowledgments
The authors would like to thank Professor Murakami
(Tokyo University of Science, Tokyo, Japan) for providing the HAND2
expression vector (pcDNA3.1 Myc-His B HAND2). This study was
supported by a grant from the Grant-in-Aid for Scientific Research
from the Ministry of Education, Science and Culture (grant no.
24592505).
Abbreviations:
|
AF
|
activation function
|
|
AhR
|
aryl hydrocarbon receptor
|
|
ARNT
|
aryl hydrocarbon receptor nuclear
translocator
|
|
AZA
|
Aza-2′-deoxycytidine
|
|
bHLH
|
basic helix-loop-helix
|
|
E2
|
17β-estradiol
|
|
ER
|
estrogen receptor
|
|
FGF
|
fibroblast growth factor
|
|
GAPDH
|
glyceraldehyde 3-phosphate
dehydrogenase
|
|
MPA
|
medroxyprogesterone acetate
|
|
PPT
|
propylpyrazole triol
|
|
Ub
|
ubiquitin
|
|
UFD1L
|
ubiquitin fusion degradation 1 L
|
|
VEGF
|
vascular endothelial growth factor
|
References
|
1
|
SGO Clinical Practice Endometrial Cancer
Working Group; Burke WM, Orr J, Leitao M, Salom E, Gehrig P,
Olawaiye AB, Brewer M, Boruta D, Villella J, et al: Endometrial
cancer: A review and current management strategies: Part I. Gynecol
Oncol. 134:385–392. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bokhman JV: Two pathogenetic types of
endometrial carcinoma. Gynecol Oncol. 15:10–17. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hecht JL and Mutter GL: Molecular and
pathologic aspects of endometrial carcinogenesis. J Clin Oncol.
24:4783–4791. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Henderson BE, Casagrande JT, Pike MC, Mack
T, Rosario I and Duke A: The epidemiology of endometrial cancer in
young women. Br J Cancer. 47:749–756. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Koskas M, Uzan J, Luton D, Rouzier R and
Daraï E: Prognostic factors of oncologic and reproductive outcomes
in fertility-sparing management of endometrial atypical hyperplasia
and adenocarcinoma: Systematic review and meta-analysis. Fertil
Steril. 101:785–794. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Manson JE, Chlebowski RT, Stefanick ML,
Aragaki AK, Rossouw JE, Prentice RL, Anderson G, Howard BV, Thomson
CA, LaCroix AZ, et al: Menopausal hormone therapy and health
outcomes during the intervention and extended post-stopping phases
of the Women's Health Initiative randomized trials. JAMA.
310:1353–1368. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hollenberg SM, Sternglanz R, Cheng PF and
Weintraub H: Identification of a new family of tissue-specific
basic helix-loop-helix proteins with a two-hybrid system. Mol Cell
Biol. 15:3813–3822. 1995.PubMed/NCBI
|
|
8
|
Firulli AB: A HANDful of questions: The
molecular biology of the heart and neural crest derivatives
(HAND)-subclass of basic helix-loop-helix transcription factors.
Gene. 312:27–40. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dai YS, Cserjesi P, Markham BE and
Molkentin JD: The transcription factors GATA4 and dHAND physically
interact to synergistically activate cardiac gene expression
through a p300-dependent mechanism. J Biol Chem. 277:24390–24398.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yamagishi H, Yamagishi C, Nakagawa O,
Harvey RP, Olson EN and Srivastava D: The combinatorial activities
of Nkx2.5 and dHAND are essential for cardiac ventricle formation.
Dev Biol. 239:190–203. 2001. View Article : Google Scholar
|
|
11
|
Rychlik JL, Gerbasi V and Lewis EJ: The
interaction between dHAND and Arix at the dopamine beta-hydroxylase
promoter region is independent of direct dHAND binding to DNA. J
Biol Chem. 278:49652–49660. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Morikawa Y, Dai YS, Hao J, Bonin C, Hwang
S and Cserjesi P: The basic helix-loop-helix factor Hand 2
regulates autonomic nervous system development. Dev Dyn.
234:613–621. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Firulli BA, Hadzic DB, McDaid JR and
Firulli AB: The basic helix-loop-helix transcription factors dHAND
and eHAND exhibit dimerization characteristics that suggest complex
regulation of function. J Biol Chem. 275:33567–33573. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dai YS and Cserjesi P: The basic
helix-loop-helix factor, HAND2, functions as a transcriptional
activator by binding to E-boxes as a heterodimer. J Biol Chem.
277:12604–12612. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li Q, Kannan A, DeMayo FJ, Lydon JP, Cooke
PS, Yamagishi H, Srivastava D, Bagchi MK, Bagchi IC, et al: The
antiproliferative action of progesterone in uterine epithelium is
mediated by Hand2. Science. 331:912–916. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jones A, Teschendorff AE, Li Q, Hayward
JD, Kannan A, Mould T, West J, Zikan M, Cibula D, Fiegl H, et al:
Role of DNA methylation and epigenetic silencing of HAND2 in
endometrial cancer development. PLoS Med. 10:e10015512013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wada-Hiraike O, Yano T, Nei T, Matsumoto
Y, Nagasaka K, Takizawa S, Oishi H, Arimoto T, Nakagawa S, Yasugi
T, et al: The DNA mismatch repair gene hMSH2 is a potent
coactivator of oestrogen receptor alpha. Br J Cancer. 92:2286–2291.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Koyama S, Wada-Hiraike O, Nakagawa S,
Tanikawa M, Hiraike H, Miyamoto Y, Sone K, Oda K, Fukuhara H,
Nakagawa K, et al: Repression of estrogen receptor beta function by
putative tumor suppressor DBC1. Biochem Biophys Res Commun.
392:357–362. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fu H, Wada-Hiraike O, Hirano M, Kawamura
Y, Sakurabashi A, Shirane A, Morita Y, Isono W, Oishi H, Koga K, et
al: SIRT3 positively regulates the expression of folliculogenesis-
and luteinization-related genes and progesterone secretion by
manipulating oxidative stress in human luteinized granulosa cells.
Endocrinology. 155:3079–3087. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hiraike H, Wada-Hiraike O, Nakagawa S,
Koyama S, Miyamoto Y, Sone K, Tanikawa M, Tsuruga T, Nagasaka K,
Matsumoto Y, et al: Identification of DBC1 as a transcriptional
repressor for BRCA1. Br J Cancer. 102:1061–1067. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cullinan-Bove K and Koos RD: Vascular
endothelial growth factor/vascular permeability factor expression
in the rat uterus: Rapid stimulation by estrogen correlates with
estrogen-induced increases in uterine capillary permeability and
growth. Endocrinology. 133:829–837. 1993.PubMed/NCBI
|
|
22
|
Bersten DC, Sullivan AE, Peet DJ and
Whitelaw ML: bHLH-PAS proteins in cancer. Nat Rev Cancer.
13:827–841. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
White TE and Gasiewicz TA: The human
estrogen receptor structural gene contains a DNA sequence that
binds activated mouse and human Ah receptors: A possible mechanism
of estrogen receptor regulation by
2,3,7,8-tetrachlorodibenzo-p-dioxin. Biochem Biophys Res Commun.
193:956–962. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ohtake F, Baba A, Takada I, Okada M,
Iwasaki K, Miki H, Takahashi S, Kouzmenko A, Nohara K, Chiba T, et
al: Dioxin receptor is a ligand-dependent E3 ubiquitin ligase.
Nature. 446:562–566. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Swedenborg E and Pongratz I: AhR and ARNT
modulate ER signaling. Toxicology. 268:132–138. 2010. View Article : Google Scholar
|
|
26
|
Ohtake F, Takeyama K, Matsumoto T,
Kitagawa H, Yamamoto Y, Nohara K, Tohyama C, Krust A, Mimura J,
Chambon P, et al: Modulation of oestrogen receptor signalling by
association with the activated dioxin receptor. Nature.
423:545–550. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Welboren WJ, Sweep FC, Span PN and
Stunnenberg HG: Genomic actions of estrogen receptor alpha: What
are the targets and how are they regulated? Endocr Relat Cancer.
16:1073–1089. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou W and Slingerland JM: Links between
oestrogen receptor activation and proteolysis: Relevance to
hormone-regulated cancer therapy. Nat Rev Cancer. 14:26–38. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fan S, Wang J, Yuan R, Ma Y, Meng Q, Erdos
MR, Pestell RG, Yuan F, Auborn KJ, Goldberg ID, et al: BRCA1
inhibition of estrogen receptor signaling in transfected cells.
Science. 284:1354–1356. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dirkx E, Gladka MM, Philippen LE, Armand
AS, Kinet V, Leptidis S, El Azzouzi H, Salic K, Bourajjaj M, da
Silva GJ, et al: Nfat and miR-25 cooperate to reactivate the
transcription factor Hand2 in heart failure. Nat Cell Biol.
15:1282–1293. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yamagishi H, Garg V, Matsuoka R, Thomas T
and Srivastava D: A molecular pathway revealing a genetic basis for
human cardiac and craniofacial defects. Science. 283:1158–1161.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bondesson M, Hao R, Lin CY, Williams C and
Gustafsson JA: Estrogen receptor signaling during vertebrate
development. Biochim Biophys Acta. 1849:142–151. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Grady D, Rubin SM, Petitti DB, Fox CS,
Black D, Ettinger B, Ernster VL and Cummings SR: Hormone therapy to
prevent disease and prolong life in postmenopausal women. Ann
Intern Med. 117:1016–1037. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mueller MD, Vigne JL, Minchenko A, Lebovic
DI, Leitman DC and Taylor RN: Regulation of vascular endothelial
growth factor (VEGF) gene transcription by estrogen receptors alpha
and beta. Proc Natl Acad Sci USA. 97:10972–10977. 2000. View Article : Google Scholar : PubMed/NCBI
|