Introduction
Mitochondrial (mt) nucleoids as functional units are
composed of circular DNA coding 13 genes involved in the electron
transport chain. The DNA is maintained by several nuclear-encoded
proteins. Topically, it is attached to the inner mitochondrial
membrane (1). The basic
nucleoid-composing proteins are responsible for transcription,
replication and repair. A detailed overview of their function,
composition and structure, which exceeds the capacities of the
present study, was given by Bogenhagen (2). Mitochondrial transcription factor A
(TFAM) is responsible for mtDNA binding and space conformation
(3), stabilization of the
mitochondrial genome (4) and
transcription (5). TFAM regulates
the mt genome copy number (6). In
certain cases, its overexpression rescues this copy number and
restores the activity of respiratory chain-associated molecules and
ATP synthesis, resulting in restoration of insulin secretion in
Pdx1-defective INS1 cells (7) or
protection of 3T3-L1 adipocytes from NYGGF4 (PID1)
overexpression-induced insulin resistance (8). mtSSB protein is expressed in the
nucleoids of mammalian mitochondria (9) and is responsible for replication and
repair of mtDNA and also for mtDNA maintenance (10). It acts via covering single-stranded
mtDNA (11), and by enhancing
mtDNA polymerase (12) and
helicase (13) activity. Twinkle
is an mtDNA helicase which is, in a complex with mtDNA polymerase
and mtSSB, responsible for unwinding mtDNA (14). Transcription and replication is
then performed by specific mitochondrial RNA or DNA polymerases
(15,16). Distribution and nucleoid
interrelation during physiological and pathophysiological
conditions of fission/fusion processes have also been studied
(17). Disturbance in nucleoid
components and mutations in mtDNA were identified to be significant
in various diseases (18,19), including carcinogenesis (20).
Photoactivable proteins, including green, yellow and
cyan fluorescence protein as well as Keima tagged to nucleoid
protein components are widely used for studying these
mtDNA-containing units (21). The
properties of green-to-red photoconvertible fluorescent protein Eos
(EosFP) are utilized in a novel method of superresolution
fluorescence photo-activation localization microscopy (fPALM)
(22). However, oligomer
aggregation is a natural effect of the wild-type (WT) protein
variant (23). The aim of the
present study was to follow up mitochondrial nucleoid behavior
under natural conditions of living cells transfected with mtSSB
wild-type EosFP-tagged protein, also with regard to the contents of
other nucleoid components (mtDNA, TFAM).
Materials and methods
DNA vectors and lentiviral (LTV) particle
production
The ORF of mtSSB was purchased from Invitrogen Life
Technologies (Carlsbad, CA 92008, USA), amplified by polymerase
chain reaction (PCR) with attb primers and sub-cloned into modified
pLenti 6.3/V5-DEST (Invitrogen Life Technologies) vector with
dimeric EosFP enabling N-terminal fusion after LR recombination.
Lentiviral particles based on mtSSB-EosFP plasmids were multiplied
in the 239LTV cell line using common calcium phosphate transfection
utilizing the packaging plasmids LP1, LP2 and VSV-G (Invitrogen
Life Technologies). The lentiviral stock was filtered and
concentrated by PEG-it Virus Precipitation Solution (System
Biosciences, Mountain View, CA, USA).
Cell cultures
The human hepatocellular cancer HEPG2 cell line
(European Collection of Cell Cultures, Salisbury, UK; 85011430) was
maintained as described previously (17). The human neuroblastoma SH-SY5Y cell
line (American Type Culture Collection, Manassas, VA, USA;
CRL-2266) was cultivated at 37°C in humidified air with 5%
CO2 in glucose-free DMEM medium (Invitrogen Life
Technologies) supplemented with 2 mM glutamine (Sigma-Aldrich, St.
Louis, MO, USA), 0.5 mM sodium pyruvate (Sigma-Aldrich), 15% (v/v)
fetal calf serum (Biochrom GmbH, Berlin, Germany), 10 mM
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Sigma-Aldrich),
100 IU/ml penicillin (Sigma-Aldrich), 100 μg/ml streptomycin
(Sigma-Aldrich) and 11 mM glucose (Sigma-Aldrich). For microscopy,
HEPG2 and SH-SY5Y cells were cultured for 3–5 days or 2–4 days,
respectively, on glass coverslips coated with poly-L-lysine.
Primary polyclonal antibodies
Rabbit anti-human anti-TFAM antibody was kindly
provided by Professor D.F. Bogenhagen (Department of
Pharmacological Sciences, University at Stony Brook, Stony Brook,
NY, USA). Rabbit anti-human anti-mtSSB was purchased from
Sigma-Aldrich (cat. no. HPA002866), mouse anti-human anti-DNA was
obtained from Progen Biotechnik GmbH (Heidelberg, Germany; cat. no.
61014) and mouse anti-human anti-TIM23 was from BD Biosciences
(Franklin Lakes, NJ, USA; cat. no. 611222).
Immunocytochemistry and confocal
microscopy
Samples were fixed with cooled (4°C) 4%
paraformaldehyde in phosphate-buffered saline (PBS) overnight and
washed three times for 10 min with PBS. The samples were then
permeabilized for 1 h with PBS + 0.1% (w/v) Triton X-100 0.1% (w/v)
Tween 20 and 0.3 M glycine (all from Sigma-Aldrich). Coverslips
were blocked with 5% bovine serum (Sigma-Aldrich) for 1 h and
incubated overnight at 4°C with the appropriate primary antibody.
Immunostaining with primary anti-TFAM or anti-DNA was followed with
secondary Alexa-647- or Alexa-568-conjugated antibodies (Invitrogen
Life Technologies).
EosFP was excited at 488 nm and Alexa-568-conjugated
antibody was excited at 543 nm using an Olympus IX81 fluorescent
microscope (Olympus, Tokyo, Japan). An inverted confocal
fluorescent Leica TCS SP2 AOBS microscope (Leica Microsystems Inc.,
Wetzlar, Germany) was used with a PL APO 100×/1.40–0.70 oil
immersion objective (a pinhole of 1 Airy unit). EosFP was excited
at 488 nm with a 20-mW argon laser, whereas Alexa-647-conjugated
antibodies were excited at 630 nm with a 1.2-mW HeNe laser.
Biplane fPALM and direct stochastic
optical reconstruction microscopy (dSTORM) superresolution
microscopy
Samples were visualized with a Biplane fPALM
instrument, a prototype of Vutara (SR-200; Vutara Inc., Salt Lake
City, UT, USA) with settings described previously (24). The composition of the dSTORM
imaging buffer was 10% glucose, 169 units glucose oxidase, 1.4
units catalase and 50 mM Tris-HCl (pH 8.0) (all from
Sigma-Aldrich), all in 10 mM NaCl (Lach-Ner, Neratovice, Czech
Republic).
mtDNA copy number
The number of mtDNA copies per cell was determined
by qPCR as described previously (25) with human primer sequences as
follows: Nuclear forward, 5′GGCAGCTTTGAAGAACGGGAC3′ and reverse,
5′CACAGGGTTAGGAGGCAGCAA3′; mitochondrial forward,
5′CAGTCTGCGCCCTTACACAAAA3′ and reverse, 5′TGGACCCGGAGCACATAAATA3′.
The SYBR Green (Promega Corp., Madison, WI, USA) PCR detection was
performed on a LightCycler® 480 Instrument, with
LightCycler software version 1.5 (Roche Diagnostics Ltd., Basel,
Switzerland).
Western blot analysis
Immunoblot analyses were performed using respective
cell lysates; proteins were separated by 12% SDS-PAGE and
transferred onto a polyvinylidene difluoride membrane (Bio-Rad
Laboratories, Hercules, CA, USA). The total quantity of protein was
measured using a Bicinchoninic Acid Assay kit (Sigma-Aldrich) and
30 μg total protein was loaded into each lane. Blots were
blocked with 5% bovine serum, incubated with a primary antibody
overnight at 4°C, washed again, and the membrane was incubated with
horseradish peroxidase-conjugated secondary antibody for 1 h at
room temperature. Proteins were visualized using Amersham Enhanced
Chemiluminescence Western Blotting Detection Reagent (GE Healthcare
Life Sciences, Little Chalfont, UK) and a LAS-1000 (Fujifilm Life
Science, Tokyo, Japan).
Statistical analysis
T-tests were conducted using SigmaPlot software,
version 9 (Systat Software, Inc., San Jose, CA, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
Transcription efficacy and construct
confirmation
Between the two cell types, a difference in the time
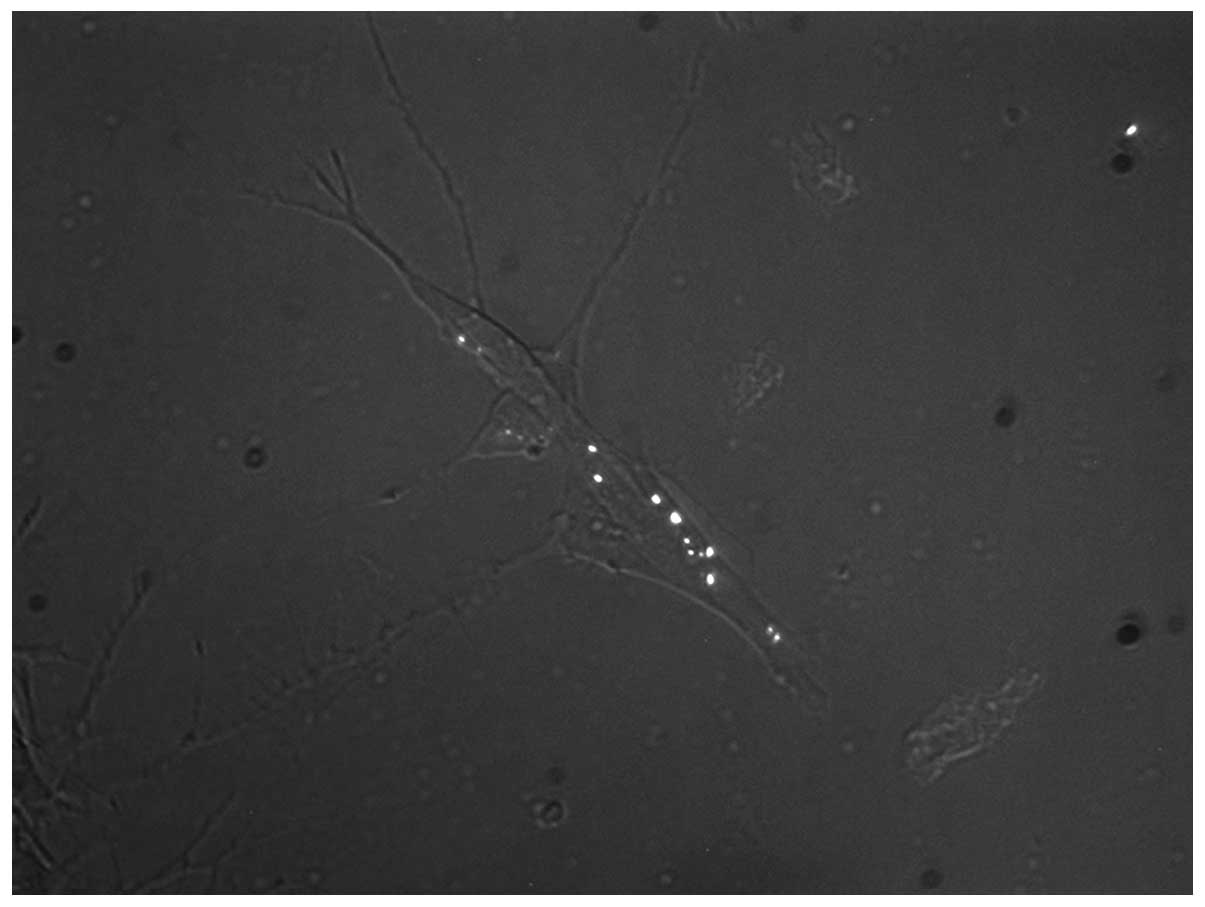
course of mtSSB-EosFP appearance was noted: While in SH-SY5Y cells,
the first fluorescent dots were already observed on day 1 (within
the first 48 h; Fig. 1), the first
fluorescent dots in HEPG2 cells were visible late on day 3 after
lentiviral transduction. Furthermore, a difference in lentiviral
transduction efficacy was observed between the two cell lines: In
SH-SY5Y, ~40–50% of cells were fluorescent on day 4, while in HEPG2
cells, only 5–10% were fluorescent on day 4 and/or day 5. Western
blot analysis confirmed an additional mtSSB-positive band
corresponding to the molecular weight of this protein (17 kDa) plus
26 kDa of EosFP (data not shown). The results did not show any
significant difference in the natural mtSSB content between
transduced and control samples of the two cell lines.
Microscopic observations
Early mtSSB-EosFP nucleoids in SH-SY5Y
and HEPG2 cells
Depending on the time course of lentiviral
transduction in the individual cell line, the first coupled
mtSSB-EosFP dots were observed in the two cell lines on day 1 and
day 3, respectively, following transduction (day 0). The
observation of mtSSB-EosFP nucleoid coupling was confirmed by
common fluorescent microscopy and fPALM superresolution
microscopy.
Later mtSSB-EosFP nucleoids and
immunocytochemistry in SH-SY5Y and HEPG2 cells
Generally, coupling in later stages following
lentiviral treansduction was recorded at high rates, but not in
absolutely all nucleoid figures in 100% of all cells; however,
equal twins of variable size were clearly distinguished.
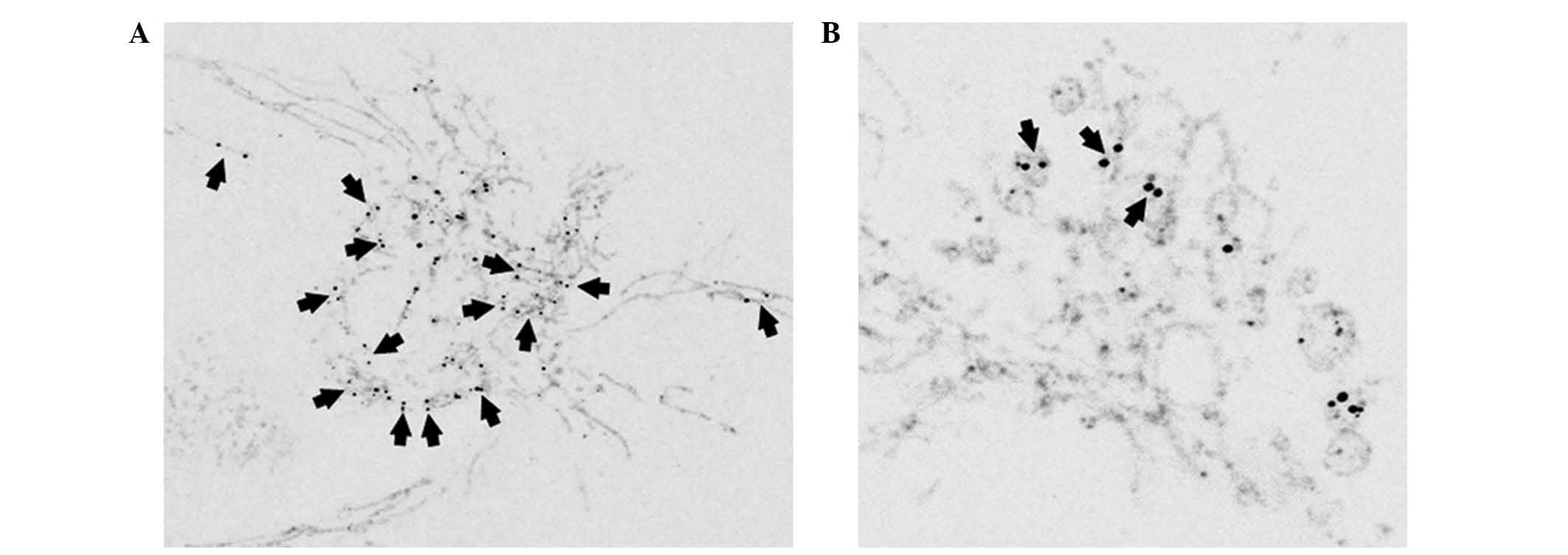
TIM23
As verified by confocal and superresolution
microscopy approaches, mtSSB-Eos-coupled nucleoids (late as well as
early) were located in the mitochondria as confirmed by the
anti-TIM23 antibody (Figs. 2 and
3A). In later stages (days 4 and
5) in HEPG2 cells, the size of nucleoid couples differed among
cells and, depending on mitochondrial morphology, were located
either in tubular mitochondria or mitochondrial bulbs (Fig. 2).
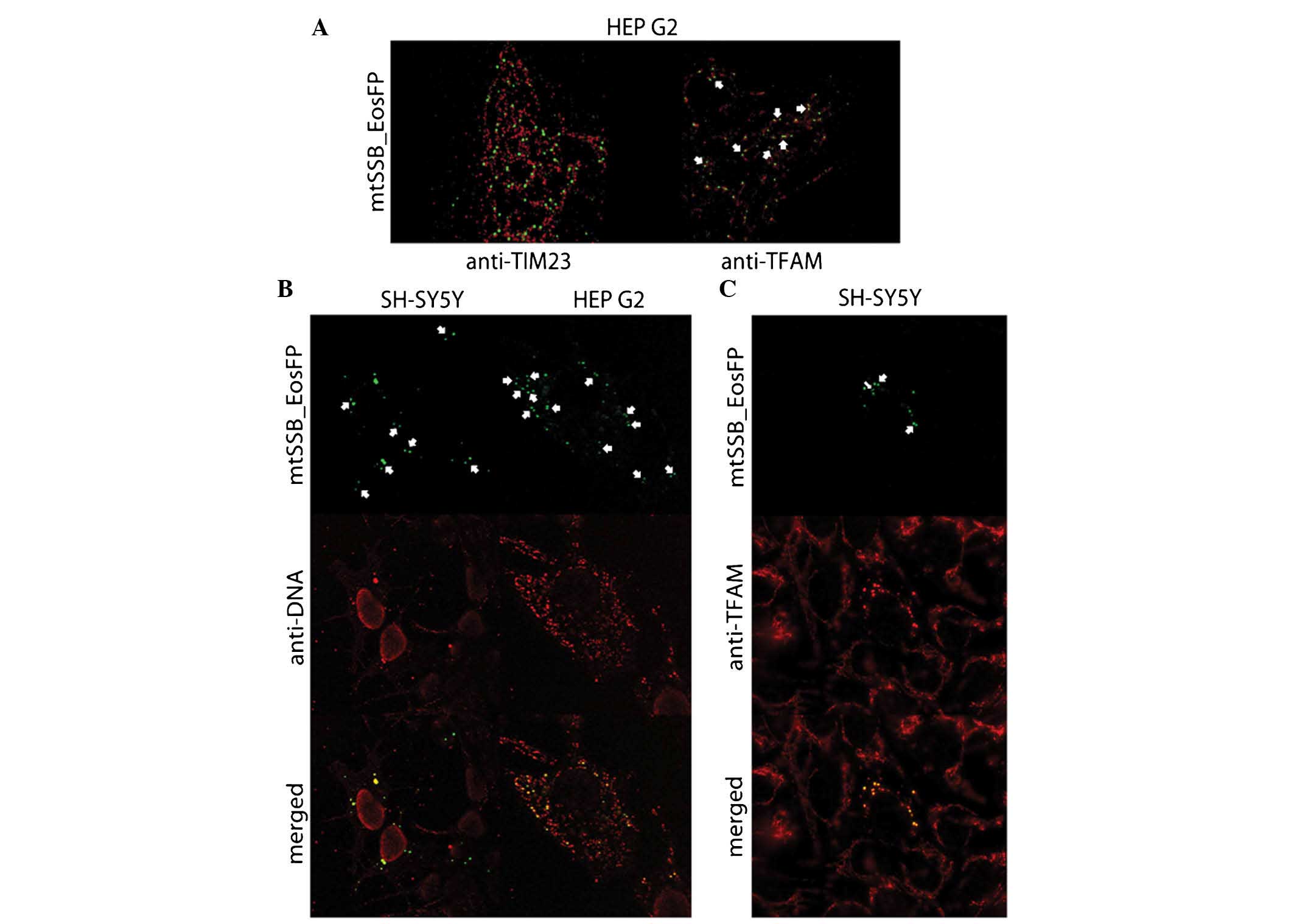 | Figure 3(A) Superresolution microscopy
visualizing early coupled mtSSB-EosFP nucleoids (green) with TIM23
counterstain (red) in HEP G2 cells and coupled mtSSB-EosFP
nucleoids (green) surrounded by TFAM (red) in HEP G2 cells on day 3
after lentiviral transduction. Clerarly visible are couples in
mitochondria couterstained with antibody against TIM23. The
coupling counterstained with TFAM is indicated by arrows. (B)
mtSSB-EosFP nucleoids (green) in SH-SY5Y cells on day 4 after
lentiviral transduction. Counterstaining with antibody against DNA
(red) proved co-localization of mtSSB-EosFP protein construct with
mtDNA. Eos-mediated aggregation of mtSSB is accompanied by the
aggregation of mtDNA in transducted cells. This mtDNA aggregation
is not observed in cells without the presence of EosFP. However,
couples without counterstaining with mtDNA are also observed.
Preservation of coupling (arrows) in later stages of aggregation
together with the presence of larger mtSSB-EosFP-coupled masses in
central parts of cells also suggests certain levels of hierarchy in
nucleoid organization. Confocal microscopy; original magnification,
×400. However, in HEPG2 cells, different mtSSB-EosFP nucleoids
(green) were observed after lentiviral transduction.
Counterstaining with antibody against DNA (red) proved
co-localization of mtSSB-EosFP protein construct with mtDNA.
Accumulations of mtDNA were present together with coupled
accumulations of mtSSB-EosFP without topical predilection; however,
in these cells, not all mtDNA was attracted to mtSSB-EosFP and a
dominant proportion of common wild-type mtDNA nucleoids remained
unaffected. However, the mass of mtDNA co-expressed with
mtSSB-EosFP was also increased. Similarly to SH-SY5Y,
mtSSB-EosFP-mediated coupling (arrows) was preserved until later
stages on day 5. Confocal microscopy; original magnification,
×1,000. (C) mtSSB-EosFP-coupled nucleoid aggregation (green) in
SH-SY5Y cells on Day 4 after lentiviral transduction (arrows).
Counterstaining with antibody against human TFAM (red) proved
co-localization of mtSSB-EosFP protein construct with TFAM.
Similarly to the observation with anti-DNA counterstaining,
Eos-mediated aggregation of mtSSB was accompanied by the
aggregation of TFAM in transducted cells. This TFAM aggregation was
not observed in the other cells without the presence of EosFP, and
the preservation of coupling was also recorded. mtSSB-EosFP,
mitochondrial single-strand DNA-binding protein/Eos fluorescent
protein; TIM, translocase of the inner membrane; TFAM,
transcription factor A, mitochondrial. |
mtDNA
Using confocal microscopy, co-localization of
mtSSB-EosFP nucleoid couples with mtDNA was proven. In the two cell
types, co-localization of mtSSB-EosFP together with mtDNA was
observed. In SH-SY5Y cells on day 4, almost all mtDNA was attached
to a limited number of accumulated and enlarged mtSSB-EosFP
aggregates close to the nucleus (Fig.
3B) in which, however, coupling was preserved as described
above. However, in HEPG2 cells on day 4 and/or day 5, the presence
of accumulations of copied mtDNA coupled to accumulations of
mtSSB-EosFP without topical predilection; however, not all mtDNA
was attached and a dominant proportion of common wild-type mtDNA
nucleoids remained unaffected (Fig.
3B). However, accumulations of mtDNA co-localization with
mtSSB-EosFP were also enlarged. In compliance with TIM23
co-localization, mtSSB-EosFP couples were observed to either be
coupled in mtDNA chains, suggesting tubular mitochondria, but also
in mtDNA clusters, suggesting the presence of mitochondrial
bulbs.
TFAM
mtSSB-EosFP and TFAM co-localization was observed
since the early stages of lentiviral transduction in the two cell
lines (Fig. 3A). In later stages
of lentiviral transduction, aggregation of mtSSB-EosFP and TFAM,
similar to that of mtDNA accumulations, was observed mostly in the
SH-SY5Y cell line (Fig. 3C). Cells
without the presence of mtSSB-EosFP did not show any TFAM
aggregation.
mtDNA copy number
The mtDNA copy number per cell decreased from 100%
[standard deviation (SD)=6.44%] in the control SH-SY5Y cells to
79.63% (SD=7.79%) with statistically significant difference between
the groups (P=0.007). In HEPG2 cells, the level decreased from 100%
(SD=2.80%) to 82.06% (SD=4.48%) with statistically significant
difference between the groups (P=0.007).
Discussion
The present study reported the cell-by-cell
observation of a relatively high rate of nucleoid coupling and
subsequent aggregation mediated by mtSSB tagged with wild-type
EosFP. This effect was accompanied by aggregation of mtDNA and
TFAM, suggesting an important role of mtSSB in nucleoid/mtDNA
distribution within the mitochondrial network. These aggre gates
are fully located to the mitochondria, co-localizing with TIM23
protein.
It is also known that mtSSB itself influences
mitochondrial biogenesis (26).
Overexpression of this nucleoid component caused the
re-distribution of the mitochondria to a perinuclear location,
implying its specialized function in cellular metabolism. mtSSB
overexpression also led to increased mitochondrial fragmentation
(27). However, co-localization of
mtDNA with dynamin-related protein (DRP)1, the membrane component
responsible for mitochondrial fission, in the context of the
immediate transmembrane neighborhood of mtDNA and cytoplasmatic
cytosceletal components, including tubulin and/or the associated
transporter kinesin (28),
suggests clear evidence of nucleoid-organized mitochondrial
biogenesis and branching. DRP1 knockdown caused nucleoid clustering
and aggregation in HeLa cells, suggesting prevention of this effect
and possible maintenance of mtDNA quality by mitochondrial fission
(29). According to the
observations of the present study, aggregation of mtSSB-EosFP
nucleoids governing the distribution of either mtDNA or TFAM in
parallel suggests the possible leading role of natural mtSSB in
mtDNA/nucleoid distribution within the mitochondrial network.
Aggregation of mtSSB-EosFP with TFAM or mtDNA in a couple suggested
a key role of mtSSB as an organizational core for nucleoid location
within the network, in parallel utilizing tetramerization as a
natural effect of mtSSB together with the ability of its C-terminal
to interact with other proteins (30,31).
However, a decrease of the mtDNA copy number in SH-SY5Y and HEPG2
cells suggested improper nucleoid replication in mtSSB-EosFP
lentivirus-transduced cells.
In spite of this, mtSSB-EosFP overexpression
visualized clear coupling of nucleoids in HEPG2 and SH-SY5Y cells.
Nucleoid couples observed in the present study most probably arose
from equal mtSSB binding capacity of neighboring nucleoids,
resulting either from equal mtDNA content and/or
transcription/replication activity and may indicate an important
harmonization of their activity. Preservation of this nucleoid
coupling in stages of aggregates, as described above, suggested a
certain topical affiliation, and together with the observation in
SH-SY5Y cells that larger couples are located perinuclearly, also a
certain level of hierarchical organization. In HEPG2 cells,
mtSSB-EosFP aggregation and coupling was observed without topic
perinuclear predilection, but was randomly dispersed within the
mitochondrial network. This organization is, however, dependent on
the mode of mitochondrial branching, mitochondrial
activity/metabolism and possibly also on the different levels of
mitophagy in individual cell types. It has already been proven that
Twinkle-dependent mtSSB co-localizes with only a subset of
nucleoids (32). In the present
study, this observation was confirmed for HEPG2, but not in SH-SY5Y
cells, where the two investigated nucleoid components (mtDNA and
TFAM) were fully attracted to mtSSB-EosFP aggreagates. An
interesting observation was made in HEPG2 cell, where couples were
identified either in nucleoids in tubular mitochondria, but also in
nucleoid clusters located in mitochondrial bulbs that are even
physiologically present in this cell type.
The differential susceptibility to lentiviral
transduction between SH-SY5Y and HEPG2 cells cannot be easily and
precisely explained; however, their differential mitochondrial
metabolism, mtDNA replication and/or transcription activity,
number, composition and behavior of natural mtDNA nucloids,
mitophagy and numerous others must be taken into account.
The present study pointed out possible essential
mechanisms of synchronized activity of nucleoids visualized by
artificial mtSSB-EosFP coupling. Subsequent to mtSSB-EosFP-mediated
aggregation of these neighboring mtDNA-organizing units containing
other components, including mtDNA and TFAM, suggests a leading role
of mtSSBs in their distribution within the mitochondrial
network.
Acknowledgments
This work was supported by The Centre of Biomedical
Research (CZ.1.07/2.3.00/30.0025) and by the Grant Agency of the
Czech Republic (grant nos. 13-02033 and P302/10/0346). This project
was co-funded by the European Social Fund and the state budget of
the Czech Republic.
References
|
1
|
Brown TA, Tkachuk AN, Shtengel G, Kopek
BG, Bogenhagen DF, Hess HF and Clayton DA: Superresolution
fluorescence imaging of mitochondrial nucleoids reveals thein
spatial range, limits and membrane interaction. Mol Cell Biol.
31:4994–5010. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bogenhagen DF: Mitochondrial DNA nucleoid
structure. Biochim Biophys Acta. 1819:914–920. 2012. View Article : Google Scholar
|
|
3
|
Ngo HB, Kaiser JT and Chan DC: The
mitochondrial transcription and packaging factor Tfam imposes a
U-turn on mitochondrial DNA. Nat Struct Mol Biol. 18:1290–1296.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Scarpulla RC: Transcriptional paradigms in
mammalian mitochondrial biogenesis and function. Physiol Rev.
88:611–638. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Falkenberg M, Larsson NG and Gustafsson
CM: DNA replication and transcription in mammalian mitochondria.
Annu Rev Biochem. 76:679–699. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Campbell CT, Kolesar JE and Kaufman BA:
Mitochondrial transcription factor A regulates mitochondrial
transcription initiation, DNA packaging and genome copy number.
Biochim Biophys Acta. 1819:921–929. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gauthier BR, Wiederkehr A, Baquié M, Dai
C, Powers AC, Kerr-Conte J, Pattou F, MacDonald RJ, Ferrer J and
Wollheim CB: PDX1 deficiency causes mitochondrial dysfunction and
defective insulin secretion through TFAM suppression. Cell Metab.
10:110–118. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shi CM, Xu GF, Yang L, Fu ZY, Chen L, Fu
HL, Shen YH, Zhu L, Ji CB and Guo XR: Overexpression of TFAM
protects 3T3-L1 adipocytes from NYGGF4 (PID1)
overexpression-induced insulin resistance and mitochondrial
dysfunction. Cell Biochem Biophys. 66:489–497. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kaguni LS: DNA polymerase gamma, the
mitochondrial replicase. Annu Rev Biochem. 73:293–320. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ruhanen H, Borrie S, Szabadkai G,
Tyynismaa H, Jones AW, Kang D, Taanman JW and Yasukawa T:
Mitochondrial single-stranded DNA binding protein is required for
maintenance of mitochondrial DNA and 7S DNA but is not required for
mitochondrial nucleoid organisation. Biochim Biophys Acta.
1803:931–939. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Van Tuyle GC and Pavco PA: The rat liver
mitochondrial DNA-protein complex: Displaced single strands of
replicative intermediates are protein coated. J Cell Biol.
100:251–257. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hoke GD, Pavco PA, Ledwith BJ and Van
Tuyle GC: Structural and functional studies of the rat
mitochondrial single strand DNA binding protein P16. Arch Biochem
Biophys. 282:116–124. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Korhonen JA, Gaspari M and Falkenberg M:
TWINKLE Has 5′->3′ DNA helicase activity and is specifically
stimulated by mitochondrial single-stranded DNA-binding protein. J
Biol Chem. 278:48627–48632. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Milenkovic D, Matic S, Kühl I, Ruzzenente
B, Freyer C, Jemt E, Park CB, Falkenberg M and Larsson NG: TWINKLE
is an essential mitochondrial helicase required for synthesis of
nascent D-loop strands and complete mtDNA replication. Hum Mol
Genet. 22:1983–1993. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Blomain ES and McMahon SB: Dynamic
regulation of mitochondrial transcription as a mechanism of
cellular adaptation. Biochim Biophys Acta. 1819:1075–1079. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Asin-Cayuela J and Gustafsson CM:
Mitochondrial transcription and its regulation in mammalian cells.
Trends Biochem Sci. 32:111–117. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tauber J, Dlasková A, Santorová J,
Smolková K, Alán L, Spaček T, Plecitá-Hlavatá L, Jabůrek M and
Ježek P: Distribution of mitochondrial nucleoids upon mitochondrial
network fragmentation and network reintegration in HEPG2 cells. Int
J Biochem Cell Biol. 45:593–603. 2013. View Article : Google Scholar
|
|
18
|
Li Z, Shen J, Chen Y, Pan J, Zeng H, Fang
H, Ye Z, Zeng C, Zhang R and Cai D: Mitochondrial genome sequencing
of chondrocytes in osteoarthritis by human mitochondria RT2
Profiler™ PCR array. Mol Med Rep. 6:39–44. 2012.PubMed/NCBI
|
|
19
|
Piao L, Han Y and Li D: Correlation study
on adiponectin gene SNP45 and long-term oxidative stress in
patients with diabetes and carotid atherosclerosis. Exp Ther Med.
8:707–712. 2014.PubMed/NCBI
|
|
20
|
Grzybowska-Szatkowska L and Slaska B:
Mitochondrial DNA and carcinogenesis (review). Mol Med Rep.
6:923–930. 2012.PubMed/NCBI
|
|
21
|
Li WH and Zheng G: Photoactivatable
fluorophores and techniques for biological imaging applications.
Photochem Photobiol Sci. 11:460–471. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hedde PN and Nienhaus GU: Super-resolution
localization microscopy with photoactivatable fluorescent marker
proteins. Protoplasma. 251:349–362. 2014. View Article : Google Scholar
|
|
23
|
Nienhaus GU, Nienhaus K, Hölzle A,
Ivanchenko S, Renzi F, Oswald F, Wolff M, Schmitt F, Röcker C,
Vallone B, et al: Photoconvertible fluorescent protein EosFP:
Biophysical properties and cell biology applications. Photochem
Photobiol. 82:351–358. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mlodzianoski MJ, Schreiner JM, Callahan
SP, Smolková K, Dlasková A, Santorová J, Ježek P and Bewersdorf J:
Sample drift correction in 3D fluorescence photoactivation
localization microscopy. Opt Express. 19:15009–15019. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Alán L, Špaček T, Zelenka J, Tauber J,
Berková Z, Zacharovová K, Saudek F and Ježek P: Assessment of
mitochondrial DNA as an indicator of islet quality: An example in
Goto Kakizaki rats. Transplant Proc. 43:3281–3284. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Korr H, Thorsten Rohde H, Benders J,
Dafotakis M, Grolms N and Schmitz C: Neuron loss during early
adulthood following prenatal low-dose X-irradiation in the mouse
brain. Int J Radiat Biol. 77:567–580. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Arakaki N, Nishihama T, Kohda A, Owaki H,
Kuramoto Y, Abe R, Kita T, Suenaga M, Himeda T, Kuwajima M, et al:
Regulation of mitochondrial morphology and cell survival by
Mitogenin I and mitochondrial single-stranded DNA binding protein.
Biochim Biophys Acta. 1760:1364–1372. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Iborra FJ, Kimura H and Cook PR: The
functional organization of mitochondrial genomes in human cells.
BMC Biol. 2:92004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ban-Ishihara R, Ishihara T, Sasaki N,
Mihara K and Ishihara N: Dynamics of nucleoid structure regulated
by mitochondrial fission contributes to cristae reformation and
release of cytochrome c. Proc Natl Acad Sci USA. 110:11863–11868.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang C, Curth U, Urbanke C and Kang C:
Crystal structure of human mitochondrial single-stranded DNA
binding protein at 2.4 A resolution. Nat Struct Biol. 4:153–157.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Antony E, Weiland E, Yuan Q, Manhart CM,
Nguyen B, Kozlov AG, McHenry CS and Lohman TM: Multiple C-terminal
tails within a single E. coli SSB homotetramer coordinate DNA
replication and repair. J Mol Biol. 425:4802–4819. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rajala N, Gerhold JM, Martinsson P, Klymov
A and Spelbrink JN: Replication factors transiently associate with
mtDNA at the mitochondrial inner membrane to facilitate
replication. Nucleic Acids Res. 42:952–967. 2014. View Article : Google Scholar :
|