Introduction
The mammalian high temperature requirement factor A4
(HtrA4) is one of four members of the HtrA family of serine
proteases. First identified in bacteria as DegP (1), homologs of the HtrA family have been
identified in vertebrates, invertebrates and humans (2). All HtrAs share one or multiple highly
conserved C-terminal PDZ domains, which bind target proteins and
regulate protein-protein interactions, whereas the N-terminus is
variable and contains the signal and regulatory sequences (3). Members of the HtrA family are
involved in a variety of human diseases (4,5). The
most important previous study to date was a genome-wide association
study, which identified a marked genetic linkage between specific
overexpressed variants of the HtrA1 gene and the wet form of
age-associated macular degeneration (6). HtrA1, 2 and 3 have also been
implicated in osteoarthritis, cancer, Alzheimer's disease,
Parkinson's disease and muscular dystrophy (7–14).
Among the HtrA family members, HtrA4 has rarely been
investigated. The HtrA4 protein is predicted to be an oligomeric
chaperone protease, which degrades misfolded secretory proteins
(15). HtrA4 may exert its action
by cleaving within the β-actin sequence to inhibit its ability to
form insoluble fibers (16). The
transcripts of HtrA4 were upregulated in the placenta of patients
with severe pre-eclampsia (17).
The role of HtrA4 remains to be fully elucidated in mice. Mouse
HtrA4 is a protein of 483 amino acids, which shares 68.18%
similarity in amino acid sequence with its human homolog. The mouse
HtrA4 and HtrA1 proteins share a similar domain structure (4). The mouse HtrA4 gene is located
on chromosome 8 (at 13.70 cM), and consists of nine exons. Exon 1
codes for the insulin-growth-factor-binding and Kazal protease
inhibitor domains, exons 3–6 code for a trypsin-like catalytic
domain, and exons 7–9 code for the PDZ domains.
The present study aimed to further characterize the
cellular functions of HtrA4 in vivo, by generating an
HtrA4 conditional allele in mice, which allows the
inactivation of HtrA4 through Cre-mediated recombination in a
cell-type-specific manner. In addition, the phenotype of the
HtrA4 knockout mice was characterized.
Materials and methods
Animals
The mice with an HtrA4 floxed allele were
generated by Ozgene (Bentley, Australia). Briefly, a flippase
recognition target (FRT)-flanked phosphoglycerate kinase I
(PGK)-neomycin cassette was inserted downstream of exon 5 of the
HtrA4 gene. The loxP sites were inserted upstream of
exon 4, downstream of exon 6, and in between exon 5 and the
selection cassette. The construct was incorporated into the genomic
DNA of embryonic stem (ES) cells from C57BL/6J mice by
electroporation using GenePulser (Bio-rad Laboratories Inc.,
Hercules, CA, USA) at 230 V/500 µF and subsequent selection. The
targeted ES cells were injected into C57BL/6J blastocysts, and the
aggregates were transferred into the uterus of recipient mice to
produce chimeric mice. The chimeric mice were crossed with C57BL/6J
mice to generate HtrA4+/flox-neo mice, which were
bred with Rosa26-FlpE mice (provided by Ozgene) to remove
the FRT-flanked PGK-neo cassette. The HtrA4+/flox
mice were subsequently crossed with Rosa26-Cre mice to
create an HtrA4 null allele. All mice were maintained on a
C57BL/6J background. The animals (equal male to female ratio) were
housed in separate cages at 21±1°C with a 12 h light/dark cycle
with access to water and standard rodent pellets, in a
conventional, pathogen-free facility at Yale University School of
Medicine (New Haven, CT, USA). The present study and all
experiments were performed in accordance with the guidelines issued
by the Institutional Animal Care and Use Committee at Yale
University (New Haven, CT, USA). The genomic DNA was digested with
ScaI restriction enzyme (New England Biolabs, Ipswich, MA,
USA) and separated by electrophoresis in 0.8% agarose gel
(Calbiochem, San Diego, CA, USA) for 12 h prior to Southern blot
analyses. The following probes were used for Southern blot analysis
of the ES cell clones and HtrA4 alleles in mice: The 3′
probe, a 537 bp polymerase chain reaction (PCR) product targeting
the genome sequence beyond the 3′ end of the HtrA4 gene
(primers: Forward, 5′-TGG TGT CAC TGG GTC TCC TTA AAGC-3′ and
reverse, 5′-AGG GTT AGA TCG GGA TGT TTT AGGG-3′); the en probe, a
695 bp PCR product targeting genomic sequence on exon 8 (PCR
primers: Forward, 5′-AAC CAG ATG GGG AAG AGA TAG GCTC-3′ and
reverse, 5′-CAGATGCAGATGCAATACAAGCTGC-3′). Primers used for PCR
genotyping of the HtrA4 alleles as follows: Primer a, 5′-GCT
GGA GTT AGA GGT GGT TGT GGAC-3′ (forward); primer b, 5′-CAG GGA GAT
GGT TTA GAA CAG TGG-3′ (reverse); and primer c, 5′-TGC CTG TAA ATG
GAA AGC ATGA-3′ (reverse). All primers were obtained from Yale DNA
Analysis facility, New Haven, CT, USA).
Gene expression analysis
The relevant tissues were collected from age and
sex-matched mice of the wild-type and HtrA4−/−
genotypes following intra-cardiac perfusion with cold
phosphate-buffered saline (PBS). The tissues were snap frozen and
homogenized in liquid nitrogen using a mortar and pestle. The
homogenized tissues were incubated with TRIzol (Invitrogen; Thermo
Fisher Scientific, Inc., Grand Island, NY, USA) for 30 min on ice,
and total RNA was extracted using an RNeasy kit (Qiagen, Hilden,
Germany) with optional DNase I treatment (Qiagen) according to the
manufacturer's instructions. cDNA synthesis was conducted using the
RevertAid First strand cDNA Synthesis kit (Thermo Fisher
Scientific, Inc.) following the manufacturer's instruments. Reverse
transcription-quantitative (RT-q)PCR was performed by the qPCR
facility of Mt. Sinai School of Medicine (New York, NY, USA) with
the following thermocycling conditions: 50°C for 2 min and 95°C for
2 min, followed by 40 cycles of 95°C for 15 s, and 60°C for 1 min.
The RNA levels were normalized against the endogenous control, gene
mouse β-actin. The primers used were: mHtrA4, 5′-CTT GGA TGG
CGA CGT GAT TGGT-3′ and 5′-TCT GCA AAG GGG CCT TGC CTT-3′; β-actin,
5′-AGG TGA CAG CAT TGC TTC TG-3′ and 5′-GCT GCC TCA ACA CCT
CAAC-3′. All reactions were performed in triplicate and a
two-tailed t-test was used to determine statistical
significance.
Immunohistochemistry
The mouse tissues were snap-frozen and embedded in
optimal cutting temperature compound (Tissue-Tek; Miles, Inc.,
Elkhart, IN, USA), followed by serial sectioning at 6 µm. The
tissue sections were cut at 6 µm using a Leica CM1850 cryostat
(Leica Microsystems GmbH, Wetzlar, Germany) were air-dried and
fixed in cold acetone for 2 min at room temperature. Following
washing in PBS, the tissue sections were blocked for 15 min in 1%
BSA in PBS. The slides were incubated with polyclonal goat
anti-HtrA4 antibody (1:40; cat. no. sc-87773; Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA) overnight at 40°C. The
tissue sections were washed three times and were incubated for 30
min with Texas Red conjugated rabbit anti-goat immunoglobulin
(1:250; cat. no. TI-5000; Vector Laboratories, Burlingame, CA,
USA). Following washing, the slides were mounted in
VECTASHIELD® mounting medium, containing
4′,6-diamidino-2-phenylindole (Vector Laboratories) and were
visualized under a Zeiss Axio Imager M1 fluorescence microscope
(Zeiss, Jena, Germany). The tissue sections from
HtrA4−/− mice were used as a negative
control.
Echocardiography
Echocardiograms were obtained for lightly
anesthetized mice (1% isofluorane inhalation) using a
Micro-Ultrasounds instrument (Vevo 770 Imaging system; Visual
Sonics, Inc., Toronto, ON, Canada). Enlarged two-dimensional views
were used to determine a short-axis plane at the level of the
papillary muscles and the M-mode was obtained at this level.
Measurements were obtained using the Vevo 770 analysis
software.
Western blotting
Mouse tissues were harvested from 5-month-old
HtrA4−/− mice and their control littermates
following intra-cardiac perfusion with cold PBS. The tissues were
lysed in radioimmunoprecipitation assay buffer (EMD Millipore,
Billerica, MA, USA), supplemented with protease inhibitor cocktails
(Roche Diagnostics, Mannheim, Germany), and the protein
concentrations were measured by Bradford assay (Bio-Rad
Laboratories, Inc.) using a Bio-Rad 415 Visible Spectrophotometer
(Bio-Rad Laboratories, Inc.). The lysates were separated using a
12% Mini-protean TGX gel (Bio-Rad Laboratories, Inc.) and
electroblotted onto polyvinylidene fluoride membranes (EMD
Millipore). The membranes were blocked with 5% non-fat milk and
were incubated overnight at 4°C with goat anti-HtrA4 antibody
(1:500; Santa Cruz Biotechnology, Inc.). Following washing, the
membranes were probed with horseradish peroxidase-conjugated rabbit
anti-goat secondary antibody (1:2,500; cat. no. sc-2768; Santa Cruz
Biotechnology, Inc.). Western blotting signals were detected by
enhanced chemiluminescence (Thermo Fisher Scientific) and exposed
on X-ray films. The membranes were stripped using Mild Antibody
Stripping Solution (EMD Millipore) and reblotted with mouse
anti-β-actin antibody (1:10,000; Sigma-Aldrich, St. Louis, MO, USA)
as a loading control.
Statistical analysis
The data are expressed as the mean ± standard error
of the mean. The statistical difference between two groups was
evaluated using a two-tailed Student's t-test. All statistical
analyses were performed using SPSS 17.0 (SPSS Inc., Chicago, IL,
USA). P<0.05 was considered to indicate a statistically
significant result.
Results
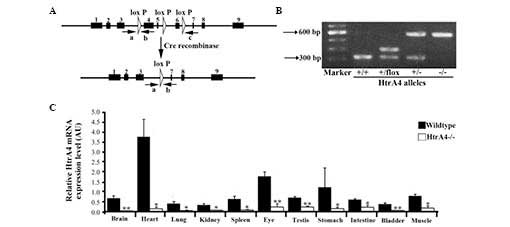
Generation of a conditional null allele
of the HtrA4 gene in mice
The targeting construct was generated by flanking
exons 4, 5 and 6 with loxP sites (Fig. 1A). Upon Cre excision, these three
exons were removed and translation was terminated. A PGK-neo
selection cassette, flanked by FRT sites, was inserted between
exons 5 and 6. Exons 1–3 were preserved, as their transcripts were
overlapping with a non-coding mRNA transcript of the Plekha2
gene. The targeting construct was linearized with AclI and
introduced into the Bruce4 ES cell line by electroporation.
Following neomycin selection and Southern blot analysis, two ES
cell clones with the correct integration were identified. These ES
cell clones were injected into blastocysts in order to develop
chimeras, and each cell exhibited a successful germline
transmission of the HtrA4 conditional allele, which is
termed HtrA4flox-neo. To produce the
HtrA4flox allele,
HtrA4flox−neo/+ mice were bred with
Flp deleter transgenic mice to remove the FRT-flanked PGK-neo
cassette. The presence of all alleles was confirmed by Southern
blotting (data not shown).
Confirmation of Cre-mediated excision of
the HtrA4 gene in mice
To assess the in vivo excision of the
HtrA4flox allele, the
HtrA4flox/+ mice were bred with
Rosa26-Cre transgenic mice to delete the floxed region
(18), generating the HtrA4
deletion mutants (Fig. 1A). The
complete deletion of exons 4–6 was demonstrated by RT-qPCR, thereby
confirming the effectiveness of the Cre excision (Fig. 1B). The
HtrA4flox/+;Rosa26-Cre mice were crossed with
C57/B6 mice to obtain HtrA4+/− mice lacking the
Rosa26-Cre transgene. HtrA4+/− mice were
intercrossed to obtain mice homozygotes of HtrA4 deletion
(HtrA4−/−; Fig.
1B). The HtrA4−/− mice exhibited normal
growth, without any obvious phenotypic differences. Among the
surviving pups of the interbreeding of HtrA4+/−
mice, the ratio of the identified HtrA4−/−
genotype was no less than the expected Mendelian ratio, suggesting
a lack of HtrA4 deletion-associated embryonic or neonatal
lethality. To examine the expression pattern of HtrA4 and to
confirm the loss of transcription of the HtrA4 gene in the
HtrA4−/− mice, the total mRNA was extracted from
11 organs (brain, heart, lung, kidney, spleen, bladder, eye,
testis, stomach, intestine and skeletal muscle) from the mice and
the level of transcription of the HtrA4 gene was determined
by RT-qPCR using primers, which were targeted to the floxed region.
In wild-type mice, HtrA4 was expressed at a low level in the
majority of organs, with the exception of the heart (Fig. 1C). HtrA4 transcription was
absent in tissues from the HtrA4−/− mice
(Fig. 1C).
Morphology and function of the
HtrA4−/− mouse heart
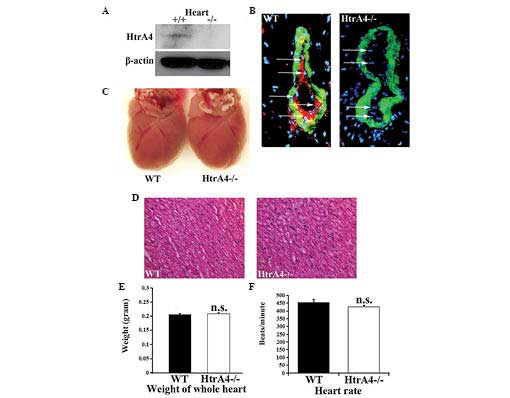
The proteins from mouse tissues were extracted for
western blot analysis. Among the 11 organs examined, the HtrA4
protein was only detected in the heart, which is consistent with
the results from the mRNA analysis (Fig. 2A). The HtrA4 protein expression
pattern was further assessed by immunofluorescence using an
antibody targeting the C-terminus of the HtrA protein, together
with an antibody against α-smooth muscle actin, the marker for
vascular smooth muscle cells. Consistent with the Western blotting
results, only the heart revealed a detectable level of protein
expression in the coronary vessels, which was absent in the hearts
of the HtrA4−/− mice (Fig. 2B). The expression of HtrA4
was restricted in the endothelial cells lining the inside layers of
the smooth muscle cells (Fig. 2B).
Therefore, the Cre-excised allele represented a null allele of
HtrA4. Upon the loss of HtrA4, the size, weight and tissue
morphology of the heart, and the heart rate (in beats/min) were
demonstrated to be unaffected (Fig.
2C–F). Subsequently, the pumping action of the mouse heart was
examined by echocardiography. The diameters of the left ventricle
were measured at the papillary muscle level (Fig. 3A). The left ventricular end
diastolic and end systolic diameters were similar in the wild-type
and HtrA4−/− mice (Fig. 3B). As shown in Fig. 3C, the fractional shortening and
ejection fraction, which are major indicators for heart function,
were not affected in the heart of the HtrA4−/−
mice. In addition, the mass of the left ventricles and cardiac
output of the HtrA4−/− mice were also similar to the
wild-type mice (Fig. 3D and E).
Therefore, the loss of HtrA4 failed to affect the morphology or
function of the heart.
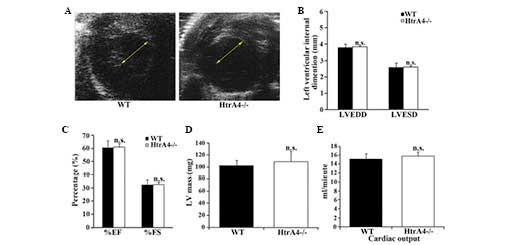 | Figure 3Echocardiographic analysis of the
cardiac function of
HtrA4−/− mice. (A)
Short-axis echocardiography demonstrated that the diameters of the
left ventricles of the WT and
HtrA4−/− mice were
similar. The arrows show the size of the intraventricular diameters
during diastole. (B) The LVEDD and LVESD of the left ventricles of
the WT and HtrA4−/−
mice were measured (n=6; P=0.73 and P=0.55, for LVEDD and LVESD,
respectively). (C) The EF and FS of the left ventricles of WT and
HtrA4−/− mice were
determined (n=6; P=0.39 and P=0.34, for %EF and %FS, respectively).
(D) The masses of the left ventricle from the WT and
HtrA4−/− mice were
measured and corrected by echocardiography (n=10; P=0.33). (E) The
cardiac output of the WT and
HtrA4−/− mice was
determined (n=9; P=0.61). The results of these experiments were
n.s. comparing WT with
HtrA4−/− mice. EF,
ejection fraction; FS, fractional shortening; LVEDD, left
ventricular end diastolic diameter; LVESD, left ventricular end
systolic diameter; HtrA4, high temperature requirement factor A4;
n.s., non-significant; WT, wild-type. |
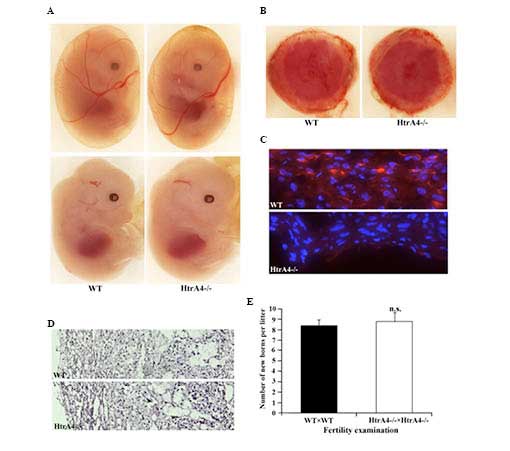
Morphology and function of the
HtrA4−/− mouse placenta
The embryonic development of the
HtrA4−/− mice was analyzed. At embryonic day
13.5, the HtrA4−/− embryos, including the yolk
sacs, exhibited a similar developmental pattern compared with their
wild-type littermates (Fig. 4A).
No HtrA4 expression was detectable by immunofluorescence at this
stage of embryonic development, including in the embryonic heart.
The HtrA4 transcript has been detected in human placenta
(GenBank accession no. BC057765), therefore, the placenta of
wild-type females with wild-type embryos and
HtrA4−/− females with HtrA4−/−
embryos were examined. The macroscopic morphology of the wild-type
and HtrA4−/− placentas was similar (Fig. 4B). A positive staining of HtrA4 was
observed only on the decidual layer of the placenta of wild-type
mice, and not on the HtrA4−/− placenta (Fig. 4C). Concerning the sections with
hematoxylin and eosin staining, HtrA4−/−
placentas demonstrated normal tissue structure and cell morphology
(Fig. 4D). In addition,
intercrossing of the HtrA4−/− mice produced a similar
number of offsprings from each litter as the intercrossing of
wild-type mice (Fig. 4E),
indicating a normal placental function.
Discussion
The HtrA family of proteins perform vital roles in
physiological processes, including apoptosis and cell signaling
(15,19). Previously, HtrA proteins 1, 2 and 3
were identified as potential drug targets for the treatment of
cancer and neurodegenerative disorders (4,5,19).
Although the protein structure of HtrA4 is similar to the other
three family members, HtrA4 has received little attention. In the
present study, a conditional null HtrA4 allele in mice was
successfully generated, and the phenotype of the HtrA4-deficient
mice was characterized.
Although the gene inactivation of
HtrA4−/− was successfully achieved, no obvious
differences in the phenotype between HtrA4−/−
adult mice and embryos was identified in the present study. The
function of HtrA4 under normal conditions may be redundant,
however, it is possible that HtrA4 may be activated in a
pathological situation. Similar to human HtrA4, mouse HtrA4 was
also expressed in the placenta. In the placenta of patients with
pre-eclampsia, the protein expression of HtrA4 was increased
(17). In addition, HtrA4
is a target gene of the placental transcription factor, glial cells
missing 1 (20). HtrA4 represses
the fusogenic activity of syncytin-1 and promotes trophoblast
invasion (20). It is possible
that HtrA4 works in association with the other HtrA family members.
HtrA1 was upregulated in the mouse model of arthritis, and
contributed to cartilage degradation (8). Another member of the HtrA family,
HtrA2, was increased in the rat testis in response to experimental
cryptorchidism (21). The mouse
model described in the present study may be useful in studying the
roles of HtrA4 in pathogenesis (17,19,20).
In the present study, it was observed that the mouse
HtrA4 protein is specifically expressed in the endothelial cells of
coronary vessels. However, a loss of HtrA4 failed to induce
abnormalities in the morphology and function of the heart. The
protein structure of HtrA4 is very similar to that of HtrA1, which
is highly expressed in endothelial cells, smooth muscle cells and
perivascular fibroblasts (22). In
the vascular system, HtrA1 regulates transforming growth factor-β
(TGF-β) signaling, and is involved in the pathogenesis of cerebral
autosomal recessive arteriopathy with subcortical infarcts and
leukoencephalopathy (23). The
N-termini of the HtrA1 and HtrA4 proteins contain predicted
cleavable signal peptides and a fragment of insulin growth
factor-binding protein (IGFBP)7 (15). IGFBP and kinase interaction domains
in HtrA1 and HtrA4 revealed a similarity to follistatin, which is a
potent antagonist of TGF-β signaling (24–26).
In addition, HtrA1 inhibited TGF-β signaling under a variety of
cellular conditions (27).
Therefore, HtrA4 may also interact with the TGF-β family of
proteins, although its function may be compensated by HtrA1.
Taken together, the conditional null HtrA4
allele in mice was generated and validated by effective Cre
excision, which led to the loss of transcription and protein
expression levels of HtrA4. This novel mouse line provides
opportunities to address the tissue and cell type-specific roles of
HtrA4 under different physiological and pathological
conditions.
Acknowledgments
The present study was funded by the Rosebay Medical
Foundation and Yale Medical School Dean's Research Fund, the
National Natural Science Foundation of China (grant no. 81370269),
and the Shandong Taishan Scholarship. The authors would like to
thank Professor Frank Giordano, Dr Yan Huang and Mr. Steve Viviano
of Yale University for their scientific input and technical
assistance.
References
|
1
|
Strauch KL and Beckwith J: An Escherichia
coli mutation preventing degradation of abnormal periplasmic
proteins. Proc Natl Acad Sci USA. 85:1576–1580. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pallen MJ and Wren BW: The HtrA family of
serine proteases. Mol Microbiol. 26:209–221. 1997. View Article : Google Scholar
|
|
3
|
Clausen T, Southan C and Ehrmann M: The
HtrA family of proteases: Implications for protein composition and
cell fate. Mol Cell. 10:443–455. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zurawa-Janicka D, Skorko-Glonek J and
Lipinska B: HtrA proteins as targets in therapy of cancer and other
diseases. Expert Opin Ther Targets. 14:665–679. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vande Walle L, Lamkanfi M and Vandenabeele
P: The mitochondrial serine protease HtrA2/Omi: An overview. Cell
Death Differ. 15:453–460. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dewan A, Liu M, Hartman S, Zhang SS, Liu
DT, Zhao C, Tam PO, Chan WM, Lam DS, Snyder M, et al: HTRA1
promoter polymorphism in wet age-related macular degeneration.
Science. 314:989–992. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Grau S, Richards PJ, Kerr B, Hughes C,
Caterson B, Williams AS, Junker U, Jones SA, Clausen T and Ehrmann
M: The role of human HtrA1 in arthritic disease. J Biol Chem.
281:6124–6129. 2006. View Article : Google Scholar
|
|
8
|
Tsuchiya A, Yano M, Tocharus J, Kojima H,
Fukumoto M, Kawaichi M and Oka C: Expression of mouse HtrA1 serine
protease in normal bone and cartilage and its upregulation in joint
cartilage damaged by experimental arthritis. Bone. 37:323–336.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chien J, Staub J, Hu SI, Erickson-Johnson
MR, Couch FJ, Smith DI, Crowl RM, Kaufmann SH and Shridhar V: A
candidate tumor suppressor HtrA1 is downregulated in ovarian
cancer. Oncogene. 23:1636–1644. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Baldi A, De Luca A, Morini M, Battista T,
Felsani A, Baldi F, Catricalà C, Amantea A, Noonan DM, Albini A, et
al: The HtrA1 serine protease is down-regulated during human
melanoma progression and represses growth of metastatic melanoma
cells. Oncogene. 21:6684–6688. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Park HJ, Seong YM, Choi JY, Kang S and
Rhim H: Alzheimer's disease-associated amyloid beta interacts with
the human serine protease HtrA2/Omi. Neurosci Lett. 357:63–67.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bakay M, Zhao P, Chen J and Hoffman EP: A
web-accessible complete transcriptome of normal human and DMD
muscle. Neuromuscul Disord. 12(Suppl 1): S125–S141. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ross OA, Soto AI, Vilariño-Güell C,
Heckman MG, Diehl NN, Hulihan MM, Aasly JO, Sando S, Gibson JM,
Lynch T, et al: Genetic variation of Omi/HtrA2 and Parkinson's
disease. Parkinsonism Relat Disord. 14:539–543. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhu F, Jin L, Luo TP, Luo GH, Tan Y and
Qin XH: Serine protease HtrA1 expression in human hepatocellular
carcinoma. Hepatobiliary Pancreat Dis Int. 9:508–512.
2010.PubMed/NCBI
|
|
15
|
Clausen T, Kaiser M, Huber R and Ehrmann
M: HTRA proteases: Regulated proteolysis in protein quality
control. Nat Rev Mol Cell Biol. 12:152–162. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ketteler R, Sun Z, Kovacs KF, He WW and
Seed B: A pathway sensor for genome-wide screens of intracellular
proteolytic cleavage. Genome Biol. 9:R642008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Inagaki A, Nishizawa H, Ota S, Suzuki M,
Inuzuka H, Miyamura H, Sekiya T, Kurahashi H and Udagawa Y:
Upregulation of HtrA4 in the placentas of patients with severe
pre-eclampsia. Placenta. 33:919–926. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Soriano P: Generalized lac Z expression
with the ROSA26 Cre reporter strain. Nat Genet. 21:70–71. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chien J, Campioni M, Shridhar V and Baldi
A: HtrA serine proteases as potential therapeutic targets in
cancer. Curr Cancer Drug Targets. 9:451–468. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang LJ, Cheong ML, Lee YS, Lee MT and
Chen H: High-temperature requirement protein A4 (HtrA4) suppresses
the fusogenic activity of syncytin-1 and promotes trophoblast
invasion. Mol Cell Biol. 32:3707–3717. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hayashi T, Yoshida S, Yoshinaga A, Ohno R,
Ishii N and Yamada T: HtrA2 is up-regulated in the rat testis after
experimental cryptorchidism. Int J Urol. 13:157–164. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu J, Dong F and Hoh J: Loss of
HtrA1-induced attenuation of TGF-β signaling in fibroblasts might
not be the main mechanism of CARASIL pathogenesis. Proc Natl Acad
Sci USA. 112:E16932015. View Article : Google Scholar
|
|
23
|
Hara K, Shiga A, Fukutake T, Nozaki H,
Miyashita A, Yokoseki A, Kawata H, Koyama A, Arima K, Takahashi T,
et al: Association of HTRA1 mutations and familial ischemic
cerebral small-vessel disease. N Engl J Med. 360:1729–1739. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kato MV: A secreted tumor-suppressor,
mac25, with activin-binding activity. Mol Med. 6:126–135.
2000.PubMed/NCBI
|
|
25
|
Nakamura T, Takio K, Eto Y, Shibai H,
Titani K and Sugino H: Activin-binding protein from rat ovary is
follistatin. Science. 247:836–838. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hu SI, Carozza M, Klein M, Nantermet P,
Luk D and Crowl RM: Human HtrA, an evolutionarily conserved serine
protease identified as a differentially expressed gene product in
osteoarthritic cartilage. J Biol Chem. 273:34406–34412. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Oka C, Tsujimoto R, Kajikawa M,
Koshiba-Takeuchi K, Ina J, Yano M, Tsuchiya A, Ueta Y, Soma A,
Kanda H, et al: HtrA1 serine protease inhibits signaling mediated
by Tgfbeta family proteins. Development. 131:1041–1053. 2004.
View Article : Google Scholar : PubMed/NCBI
|