Introduction
Primates and humans possess intrinsic immunity to
inhibit viral replication immediately and directly (1,2). The
sterile α motif and HD domain-containing protein 1 (SAMHD1) is a
newly identified anti-viral factor in this immunity (3). SAMHD1 is a deoxynucleoside
triphosphate triphosphohydrolase (dNTPase) that depletes the
intracellular pool of deoxynucleoside triphosphates (dNTPs) and
restricts the replication of human immunodeficiency virus type 1
(HIV-1) in non-cycling myeloid cells (4–8).
Most recently, it was discovered that SAMHD1 prevented HIV-1
infection by directly degrading HIV-1 RNA through its ribonuclease
activity (9).
Besides HIV-1, SAMHD1 has the ability to restrict
other retroviruses, including simian immunodeficiency virus, feline
immunodeficiency virus, bovine immunodeficiency virus, equine
infectious anemia virus, murine leukemia virus (MLV), Mason Pfizer
monkey virus, Rous sarcoma virus and human T-cell leukemia virus
type 1 (10–12). In addition, it has been discovered
that SAMHD1 restricts two DNA viruses, herpes simplex virus type 1
(HSV-1) and vaccinia virus, in non-dividing myeloid cells (13,14).
A previous study by our group showed that SAMHD1 also restrained
the replication of another DNA virus, hepatitis B virus (HBV)
(15). Most recently, porcine
SAMHD1 was demonstrated to block the replication of porcine
reproductive and respiratory syndrome virus, a positive-stranded
RNA virus, in MARC-145 cells (16). Thus, SAMHD1 is a relatively
broad-spectrum anti-viral factor against numerous types of
virus.
Interferons (IFNs) often strongly induce the
expression of restriction factors during the anti-viral state
(17). It is well known that type
I IFNs may induce the expression of IFN-stimulated genes (ISGs)
through the canonical and non-canonical signaling pathway (18,19).
In the canonical pathway, the binding of IFN-α to the IFN-α
receptor results in the activation of Janus kinase (JAK) members
Tyk2 and JAK1, which phosphorylate signal transducer and activator
of tran-scription 1 (STAT1) and STAT2. Phosphorylated STAT1 and
STAT2 dimerize and further assemble with IFN-regulatory factor 9
(IRF9) to form a transcription factor complex called IFN-stimulated
gene factor 3 (ISGF3). ISGF3 binds to IFN-stimulated response
elements (ISRE) and directly activates the transcription of ISGs.
Accumulating evidence has shown that non-canonical IFN-α signaling
pathways exist and function beyond ISGF3 (19).
Apolipoprotein B mRNA-editing enzyme catalytic
polypeptide-like 3G protein is a well-studied anti-viral factor
that may be induced by IFN-α in liver cells through a novel
STAT1-independent signaling pathway (20). A previous study by our group
reported that IFN-α induced SAMHD1 expression in liver cells
(15). However, the mechanism of
how SAMHD1 expression is upregulated by IFN-α in liver cells has
remained elusive. The present study found that ISGF3 complex was
required for the induction of SAMHD1 expression by IFN-α in
SMMC-7721 cells, suggesting that IFN-α induced SAMHD1 expression in
liver cells through the canonical IFN-α signaling pathway.
Materials and methods
Cell culture, IFN-α stimulation and
transfection
SMMC-7721 cells (Type Culture Collection of the
Chinese Academy of Sciences, Shanghai, China) were cultured in
Dulbecco's modified Eagle's medium supplemented with 10% (v/v)
fetal calf serum (Zhejiang Tianhang Biological Technology Co.,
Ltd., Hangzhou, China), in a 37°C incubator containing 5%
CO2. SMMC-7721 cells were plated in six-well plates
(4×105 cells/well) and grown to 80–90% confluency. Then
cells were treated with 1,000 IU/ml IFN-α (Anhui Anke Biotechnology
Co., Ltd., Hefei, China) for 0, 2, 4, 6, 8, 10 or 24 h,
respectively, and were then harvested for reverse-transcription
quantitative polymerase chain reaction (RT-qPCR) analysis.
SMMC-7721 cells were plated in 12-well plates (2×105
cells/well) and transfected with small interfering (si)RNAs
(negative control siRNA, STAT1 siRNA, STAT2 siRNA or IRF9 siRNA)
using Lipofectamine™ 2000 (Life Technologies, Grand Island, NY,
USA) according to the manufacturer's instructions. 48 h
post-transfection, cells were further treated with 1,000 IU/ml
IFN-α for 10 h. Cells were harvested for western blot and RT-qPCR
analyses.
RNA interference
RNA interference against STAT1, STAT2 and IRF9 was
performed using known siRNAs which had been used in previous
studies (21–23). The siRNAs had the following
sequences: STAT1 sense, 5′-r(CAC GAG ACC AAU GGU GUGG)d(TT)-3′ and
anti-sense, 5′-r(CCA CAC CAU UGG UCU CGUG)d(TT)-3′; STAT2 sense,
5′-GGACUG AGUUGCCUGGUUAUU-3′ and anti-sense, 5′-(P)UAA
CCAGGCAACUCAGUCCUU-3′; IRF9 sense, 5′-GCAGAG ACUUGGUCAGGUAUU-3′ and
anti-sense, 5′-(P)UACCUG ACCAAGUCUCUGCUU-3′; negative control
sense, 5′-UUC UCCGAACGUGUCACGUTT-3′ and anti-sense, 5′-ACG
UGACACGUUCGGAGAATT-3′. All of these siRNAs were synthesized by
Shanghai GenePharma Co., Ltd. (Shanghai, China). SMMC-7721 cells
were transfected with the respective siRNAs and incubated for 48 h.
After further stimulation with IFN-α, cells were harvested for
western blot and RT-qPCR.
Western blot analysis
After RNA interference and IFN-α treatment,
SMMC-7721 cells were lysed in radioimmunoprecipitation assay lysis
buffer (Beyotime Institute of Biotechnology, Haimen, China). The
protein concentration of the supernatants was determined using a
bicinchoninic acid kit (cat. no. P00125; Beyotime Institute of
Biotechnology) after centrifugation at 12,000 g for 5 min, 20
µg protein was loaded onto each lane and separated by 10%
SDS-PAGE. Then proteins were transferred onto Immuno-Blot
polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA).
Following blocking, the membranes were incubated with rabbit
anti-STAT1 polyclonal antibody (cat. no. 10144-2-AP; 1:1,000),
rabbit anti-STAT2 polyclonal antibody (cat. no. 16674-1-AP;
1:1,000) or rabbit anti-IRF9 polyclonal antibody (cat. no.
14167-1-AP; 1:1,000) at 4°C overnight, respectively. These primary
antibodies were purchased from Proteintech Group (Wuhan, China).
β-actin was used as a loading control and mouse anti-β-actin
monoclonal antibody (cat. no. TA-09; 1:500) was obtained from
Beijing ZSGB-Biotechnology Co., Ltd. (Beijing, China) and incubated
at 4°C overnight. The following secondary antibodies were used in
the present study: Horseradish peroxidase-conjugated goat
anti-rabbit immunoglobulin IgG (cat. no. BL003A; 1:10,000),
incubated at room temperature for 1 h, and goat anti-mouse IgG
(cat. no. BL001A; 1:10,000), incubated at room temperature for 1 h
(Biosharp Co., Hefei, China). Immunoreactive proteins were
visualized using the Super Signal West Femto kit (cat. no. 34094;
Thermo Fisher Scientific, Waltham, MA, USA) and images were
captured using the digital gel image analysis system (4500SF; Tanon
Science & Technology Co., Ltd., Shanghai, China).
RT-qPCR analysis
Total RNA was isolated from SMMC-7721 cells after
RNA interference and IFN-α treatment using TRIzol®
reagent (Life Technologies). cDNAs were then prepared by reverse
transcription from total RNA using M-MLV Reverse Transcriptase
(Life Technologies). Real-time qPCR experiments were performed in
0.2 ml 96-well PCR plates using TaqMan® Gene Expression
Master Mix (Life Technologies). Each reaction well in the 96-well
PCR plates contained a total volume of 20 µl: 1 µl
20xTaqMan® Gene Expression Assay, 10 µl 2X
TaqMan® Gene Expression Master Mix, 4 µl cDNA
template (500 ng) and 5 µl RNase-free water. The following
primer/probe sets were utilized in the present study: SAMHD1
(Hs00210019_m1), ISG15 (Hs00192713_m1) and GAPDH (Hs99999905_m1)
(Life Technologies). Real-time PCR reactions were performed using
StepOnePlus™ Real-time PCR System (Life Technologies). Thermal
cycling conditions were as follows: 2 min at 50°C and 10 min at
95°C, followed by 40 cycles of 15 sec at 95°C and 1 min at 60°C.
Data analysis and quantification were performed using the
2−ΔΔCT comparative method (24).
Statistical analysis
Values are expressed as the mean ± standard
deviation. Statistical significance of differences between STAT1,
STAT2 or IRF9 siRNA-transfected groups with IFN-α-treatment and the
control siRNA-transfected group with IFN-α treatment were analyzed
by Student's t test using GraphPad Prism 5 (GraphPad Inc.,
La Jolla, CA, USA). P<0.05 was considered to indicate a
statistically significant difference between values.
Results
IFN-α treatment increases SAMHD1 mRNA
levels in SMMC-7721 cells in a time-dependent manner
A previous study by our group identified that IFN-α
induced SAMHD1 expression at the protein level in liver cells by
using western blot analysis (15).
The present study further assessed whether IFN-α induced SAMHD1
expression at the mRNA level using RT-qPCR analysis. The results
showed that IFN-α treatment increased the mRNA levels of SAMHD1 in
SMMC-7721 cells in a time-dependent manner (Fig. 1A). The fold induction of SAMHD1
mRNA expression by IFN-α was ~25-fold following 10 h of incubation,
while at 24 h, it was reduced to ~two-fold of the levels at the
beginning of the experiment. As the positive control, ISG15, a the
well-defined IFN-α-responsive gene, was used. ISG15 mRNA levels in
SMMC-7721 cells were markedly upregulated by IFN-α following 10 h
of incubation (Fig. 1B). Together
with the results of the previous study by our group (15), the results of the present study
demonstrated that IFN-α induced SAMHD1 expression in SMMC-7721
cells at the mRNA as well as the protein level in a time-dependent
manner.
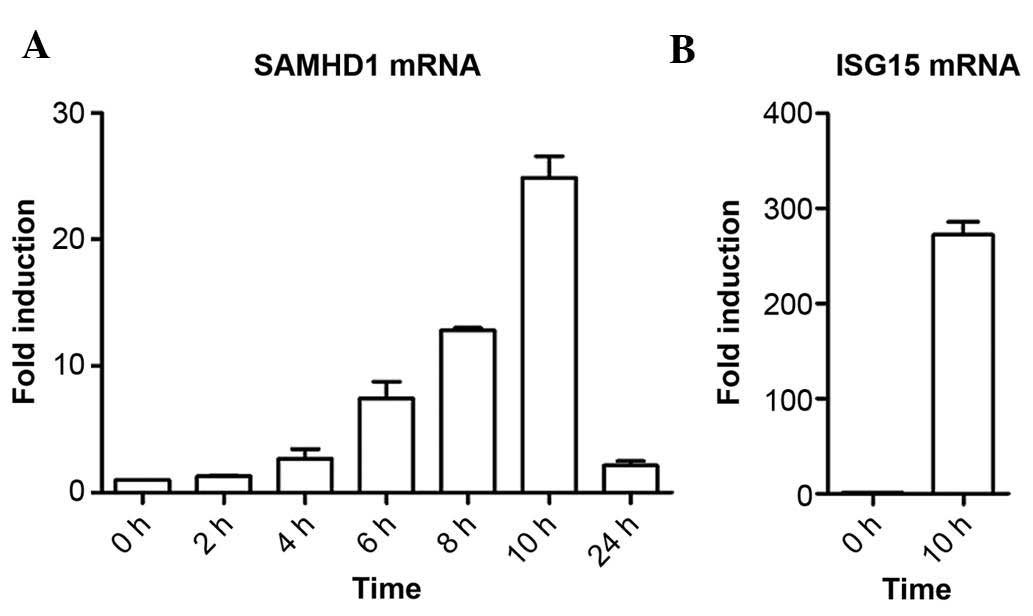 | Figure 1IFN-α induces SAMHD1 expression at the
mRNA level in SMMC-7721 cells in a time-dependent manner. (A)
SMMC-7721 cells were treated with 1,000 IU/ml IFN-α and harvested
following incubation for 0, 2, 4, 6, 8, 10 or 24 h for assessment
of SAMHD1 mRNA levels using RT-qPCR. (B) SMMC-7721 cells were
stimulated with 1,000 IU/ml IFN-α for 10 h and the mRNA levels of
the known IFN-α-responsive ISG15 were detected using RT-qPCR
method. Results are representative of at least three independent
experiments with triplicate samples, and are expressed as the mean
± standard deviation. IFN, interferon; SAMHD1, sterile α motif and
HD domain-containing protein 1; ISG15, interferon-stimulated gene
factor 3; RT-qPCR, reverse-transcription quantitative polymerase
chain reaction. |
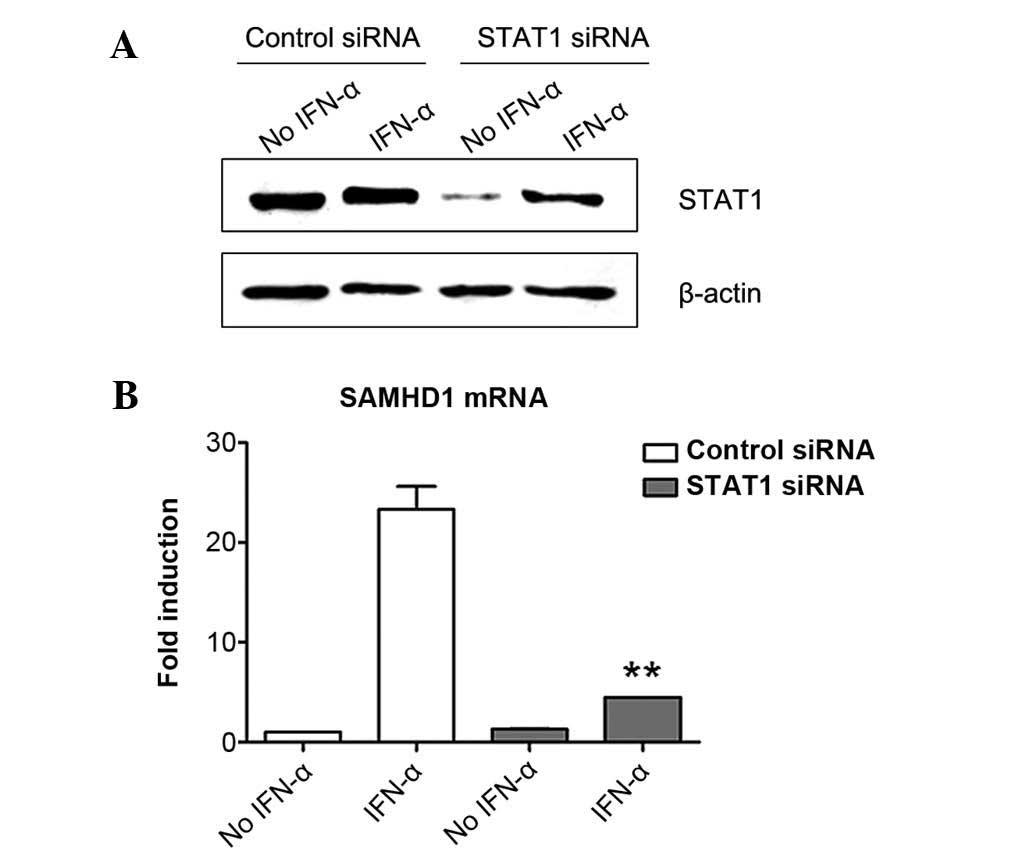
Induction of SAMHD1 expression in
SMMC-7721 cells by IFN-α is inhibited by STAT1 knockdown
ISGF3 transcription factor complex consists of
STAT1, STAT2 and IRF9, which are involved in the canonical type I
interferon signaling pathway (19). To explore the role of ISGF3 in the
induction of SAMHD1 expression by IFN-α, the present study firstly
downregulated the expression of STAT1 in SMMC-7721 cells by RNA
interference and then evaluated its influence in the induction of
SAMHD1 expression by IFN-α using RT-qPCR. The results showed that
STAT1 was expressed in SMMC7-7721 cells and that IFN-α treatment
induced its expression (Fig. 2A).
This phenomenon was similar to that reported by a previous study
(25). STAT1-specific siRNA
efficiently silenced STAT1 expression in SMMC-7721 cells, although
IFN-α treatment partially abrogated the effects of STAT1 siRNA,
suggesting that STAT1-knockdown was efficient (Fig. 2A). Of note, STAT1 siRNA treatment
significantly reduced the induction of SAMHD1 expression by IFN-α
in SMMC-7721 cells, while control siRNA had no effect on
IFN-α-induced SAMHD1 expression (Fig.
2B). These results indicated that STAT1 was required for the
induction of SAMHD1 expression by IFN-α in SMMC-7721 cells.
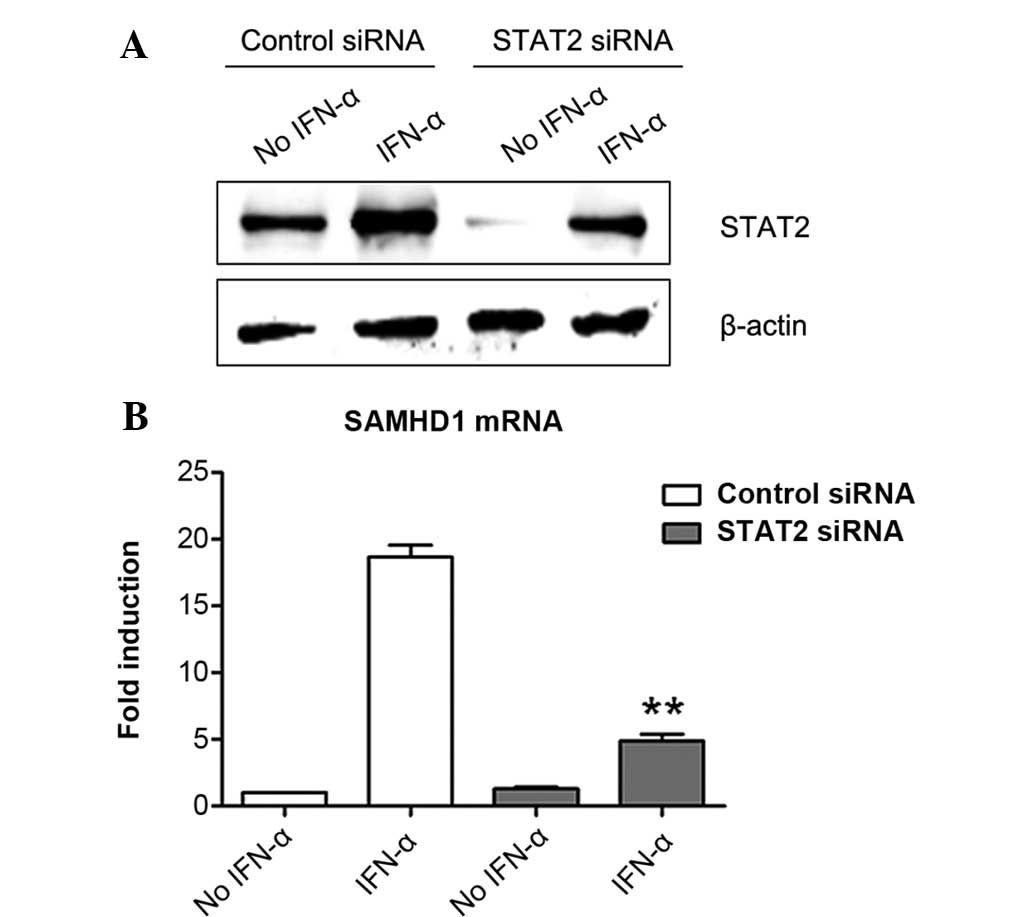
STAT2 silencing suppresses the induction
of SAMHD1 expression by IFN-α in SMMC-7721 cells
Next, the present study determined the role of STAT2
in the induction of SAMHD1 expression by IFN-α in SMMC-7721 cells.
Similarly to the results on STAT1, it was shown that STAT2 was also
expressed in SMMC-7721 cells and that IFN-α treatment induced its
expression (Fig. 3A). STAT2
expression was almost completely silenced by STAT2-specific siRNA
in the group without IFN-α treatment. With IFN-α treatment, STAT2
protein levels in the STAT2 siRNA-transfected cells was obviously
lower than those in control siRNA-transfected cells (Fig. 3A). These results demonstrated that
STAT2 siRNA successfully inhibited STAT2 expression in SMMC-7721
cells. Of note, the results showed that silencing of STAT2
expression markedly reduced the induction of SAMHD1 expression by
IFN-α in SMMC-7721 cells (Fig.
3B), indicating that STAT2 was involved in the signaling
pathway of IFN-α-induced SAMHD1 expression.
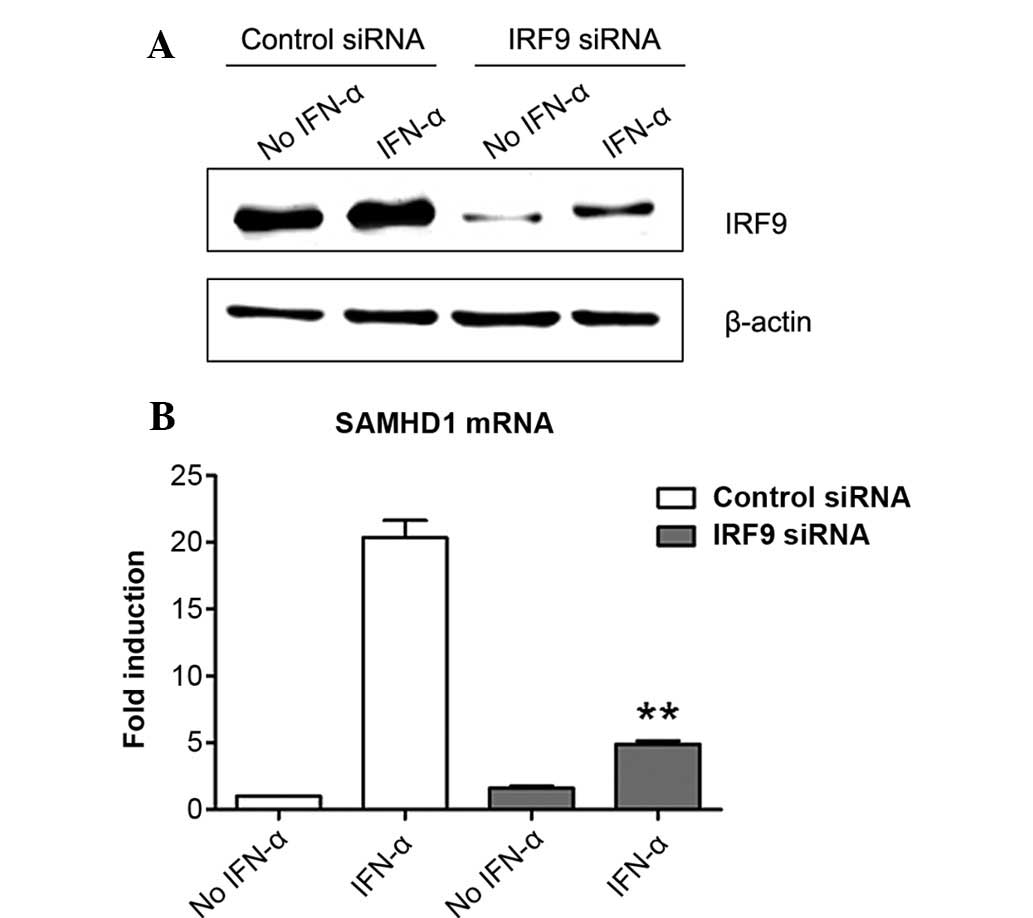
Induction of SAMHD1 expression by IFN-α
is IRF9-dependent in SMMC-7721 cells
The present study further tested whether IRF9 was
also involved in the induction of the expression of SAMHD1 by IFN-α
in SMMC-7721 cells. The results showed that IRF9 was expressed in
SMMC-7721 cells and that IFN-α treatment markedly enhanced its
expression (Fig. 4A). Of note,
IRF9 siRNA markedly inhibited IRF9 expression in the presence or
absence of IFN-α (Fig. 4A). More
importantly, siRNA-mediated knockdown of IRF9 largely suppressed
the induction of SAMHD1 expression by IFN-α in SMMC-7721 cells.
These results revealed that IRF9 was required for the induction of
SAMHD1 expression by IFN-α in SMMC-7721 cells.
Discussion
It has been demonstrated that IFN-α induces SAMHD1
expression in monocytic cells (26), U87-MG cells (27), human embryonic 293T cells and HeLa
cells (28). A previous study
showed that SAMHD1 expression was obviously induced by IFN-α at the
protein level in SMMC-7721 and BEL-7402 hepatoma cell lines
(15). However, the signaling
pathway via which the induction of SAMHD1 expression by IFN-α is
mediated has remained elusive. The formation of ISGF3 transcription
factor complex comprising STAT1, STAT2 and IRF9 is a hallmark of
the canonical type I IFN signaling pathway (19). The present study revealed that
silencing of STAT1, STAT2 and IRF9 by their specific siRNAs blocked
IFN-α-induced SAMHD1 expression in SMMC-7721 cells, indicating that
the ISGF3 complex was required for the induction of SAMHD1
expression by IFN-α in SMMC-7721 cells.
Previous studies showed that STAT1, STAT2 and IRF9
mRNA as well as protein levels were upregulated by IFN-α treatment
in human hepatoma HepG2 cells (29), human peripheral blood mononuclear
cells and macrophages (30).
Consistent with these results, the present study indicated that
IFN-α upregulated STAT1, STAT2 and IRF9 in human hepatoma SMMC-7721
cells. However, STAT1, STAT2 and IRF9 expression levels in the
specific siRNA-transfected groups were markedly lower than those in
the control siRNA-transfected groups even after IFN-α stimulation,
indicating that these siRNAs were efficient regardless of the
presence or absence of IFN-α.
In the classical type I IFN signaling pathway, ISGF3
binds to the consensus ISRE DNA sequence and then activates gene
expression. Whether ISRE is located upstream of the SAMHD1 gene
requires further investigation. It has been reported that STAT1 may
be phosphorylated by inhibitor of nuclear factor κB kinase ε (IKKε)
and certain type I IFN-stimulated genes remain inactive in the
absence of IKKε, indicated by ISGF3 not binding to the promoter
elements of these genes (31).
Further studies are required to explore whether IKKε or JAKs are
engaged in the formation of ISGF3 in IFN-α-induced SAMHD1
expression in SMMC-7721 cells.
Zhang et al (29) reported that the ISGF3 complex has a
key role in IFN-α-mediated anti-HBV responses in human hepatoma
cells. The present study demonstrated that IFN-α induced SAMHD1
expression through the ISGF3 complex in SMMC-7721 cells. Together
with the results of a previous study by our group (15), it may be concluded that SAMHD1 is
induced by IFN-α in liver cells through the canonical IFN-α
signaling pathway and inhibits HBV replication during IFN-α
treatment of HBV-infected patients.
Acknowledgments
The present study was supported by grants from the
Natural Science Foundation of Anhui Province (no. 1208085MH134 to
C.H. and no. 158085MH158 to S.Y) and the Research Fund of Anhui
Medical University (no. 0116025101 to S.Y.).
References
|
1
|
Yan N and Chen ZJ: Intrinsic antiviral
immunity. Nat Immunol. 13:214–222. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kawamura T, Ogawa Y, Aoki R and Shimada S:
Innate and intrinsic antiviral immunity in skin. J Dermatol Sci.
75:159–166. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen Z, Zhang L and Ying S: SAMHD1: A
novel antiviral factor in intrinsic immunity. Future Microbiol.
7:1117–1126. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Laguette N, Sobhian B, Casartelli N,
Ringeard M, Chable-Bessia C, Ségéral E, Yatim A, Emiliani S,
Schwartz O and Benkirane M: SAMHD1 is the dendritic- and
myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx.
Nature. 474:654–657. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hrecka K, Hao C, Gierszewska M, Swanson
SK, Kesik-Brodacka M, Srivastava S, Florens L, Washburn MP and
Skowronski J: Vpx relieves inhibition of HIV-1 infection of
macrophages mediated by the SAMHD1 protein. Nature. 474:658–661.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Goldstone DC, Ennis-Adeniran V, Hedden JJ,
Groom HC, Rice GI, Christodoulou E, Walker PA, Kelly G, Haire LF,
Yap MW, et al: HIV-1 restriction factor SAMHD1 is a deoxynucleoside
triphosphate triphosphohydrolase. Nature. 480:379–382. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Powell RD, Holland PJ, Hollis T and
Perrino FW: Aicardi-Goutieres syndrome gene and HIV-1 restriction
factor SAMHD1 is a dGTP-regulated deoxynucleotide
triphosphohydrolase. J Biol Chem. 286:43596–43600. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lahouassa H, Daddacha W, Hofmann H, Ayinde
D, Logue EC, Dragin L, Bloch N, Maudet C, Bertrand M, Gramberg T,
et al: SAMHD1 restricts the replication of human immunodeficiency
virus type 1 by depleting the intracellular pool of deoxynucleoside
triphosphates. Nat Immunol. 13:223–228. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ryoo J, Choi J, Oh C, Kim S, Seo M, Kim
SY, Seo D, Kim J, White TE, Brandariz-Nuñez A, et al: The
ribonuclease activity of SAMHD1 is required for HIV-1 restriction.
Nat Med. 20:936–941. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
White TE, Brandariz-Nuñez A, Valle-Casuso
JC, Amie S, Nguyen L, Kim B, Brojatsch J and Diaz-Griffero F:
Contribution of SAM and HD domains to retroviral restriction
mediated by human SAMHD1. Virology. 436:81–90. 2013. View Article : Google Scholar :
|
|
11
|
Gramberg T, Kahle T, Bloch N, Wittmann S,
Müllers E, Daddacha W, Hofmann H, Kim B, Lindemann D and Landau NR:
Restriction of diverse retroviruses by SAMHD1. Retrovirology.
10:262013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sze A, Belgnaoui SM, Olagnier D, Lin R,
Hiscott J and van Grevenynghe J: Host restriction factor SAMHD1
limits human T cell leukemia virus type 1 infection of monocytes
via STING-mediated apoptosis. Cell Host Microbe. 14:422–434. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hollenbaugh JA, Gee P, Baker J, Daly MB,
Amie SM, Tate J, Kasai N, Kanemura Y, Kim DH, Ward BM, et al: Host
factor SAMHD1 restricts DNA viruses in non-dividing myeloid cells.
PLoS Pathog. 9:e10034812013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim ET, White TE, Brandariz-Núñez A,
Diaz-Griffero F and Weitzman MD: SAMHD1 restricts herpes simplex
virus 1 in macrophages by limiting DNA replication. J Virol.
87:12949–12956. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen Z, Zhu M, Pan X, Zhu Y, Yan H, Jiang
T, Shen Y, Dong X, Zheng N, Lu J, et al: Inhibition of Hepatitis B
virus replication by SAMHD1. Biochem Biophys Res Commun.
450:1462–1468. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang S, Shan T, Zhou Y, Jiang Y, Tong W,
Liu F, Wen F, Zhang Q and Tong G: Molecular cloning and
characterizations of porcine SAMHD1 and its roles in replication of
highly pathogenic porcine reproductive and respiratory syndrome
virus. Dev Comp Immunol. 47:234–246. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Harris RS, Hultquist JF and Evans DT: The
restriction factors of human immunodeficiency virus. J Biol Chem.
287:40875–40883. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ivashkiv LB and Donlin LT: Regulation of
type I interferon responses. Nat Rev Immunol. 14:36–49. 2014.
View Article : Google Scholar :
|
|
19
|
Fink K and Grandvaux N: STAT2 and IRF9:
Beyond ISGF3. JAKSTAT. 2:e275212013.
|
|
20
|
Sarkis PT, Ying S, Xu R and Yu XF:
STAT1-independent cell type-specific regulation of antiviral
APOBEC3G by IFN-alpha. J Immunol. 177:4530–4540. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang F, Shang D, Zhang Y and Tian Y:
Interleukin-22 suppresses the growth of A498 renal cell carcinoma
cells via regulation of STAT1 pathway. PLoS One. 6:e203822011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fink K, Martin L, Mukawera E, Chartier S,
De Deken X, Brochiero E, Miot F and Grandvaux N: IFNβ/TNFα
synergism induces a non-canonical STAT2/IRF9-dependent pathway
triggering a novel DUOX2 NADPH oxidase-mediated airway antiviral
response. Cell Res. 23:673–690. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tsuno T, Mejido J, Zhao T, Schmeisser H,
Morrow A and Zoon KC: IRF9 is a key factor for eliciting the
antiproliferative activity of IFN-alpha. J Immunother. 32:803–816.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Schmittgen TD1 and Livak KJ: Analyzing
real-time PCR data by the comparative C (T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar
|
|
25
|
Chen H, Wang LW, Huang YQ and Gong ZJ:
Interferon-alpha induces high expression of APOBEC3G and STAT-1 in
vitro and in vivo. Int J Mol Sci. 11:3501–3512. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Berger A, Sommer AF, Zwarg J, Hamdorf M,
Welzel K, Esly N, Panitz S, Reuter A, Ramos I, Jatiani A, et al:
SAMHD1-deficient CD14+ cells from individuals with
Aicardi-Goutières syndrome are highly susceptible to HIV-1
infection. PLoS Pathog. 7:e10024252011. View Article : Google Scholar
|
|
27
|
Goujon C, Schaller T, Galão RP, Amie SM,
Kim B, Olivieri K, Neil SJ and Malim MH: Evidence for IFNα-induced,
SAMHD1-independent inhibitors of early HIV-1 infection.
Retrovirology. 10:232013. View Article : Google Scholar
|
|
28
|
St Gelais C, de Silva S, Amie SM, Coleman
CM, Hoy H, Hollenbaugh JA, Kim B and Wu L: SAMHD1 restricts HIV-1
infection in dendritic cells (DCs) by dNTP depletion, but its
expression in DCs and primary CD4+ T-lymphocytes cannot be
upregulated by interferons. Retrovirology. 9:1052012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang Q, Wang Y, Wei L, Jiang D, Wang JH,
Rao HY, Zhu L, Chen H, Fei R and Cong X: Role of ISGF3 in
modulating the anti-hepatitis B virus activity of interferon-alpha
in vitro. J Gastroenterol Hepatol. 23:1747–1761. 2008. View Article : Google Scholar
|
|
30
|
Lehtonen A, Matikainen S and Julkunen I:
Interferons up-regulate STAT1, STAT2, and IRF family transcription
factor gene expression in human peripheral blood mononuclear cells
and macrophages. J Immunol. 159:794–803. 1997.PubMed/NCBI
|
|
31
|
Tenoever BR, Ng SL, Chua MA, McWhirter SM,
García-Sastre A and Maniatis T: Multiple functions of the
IKK-related kinase IKKepsilon in interferon-mediated antiviral
immunity. Science. 315:1274–1278. 2007. View Article : Google Scholar : PubMed/NCBI
|