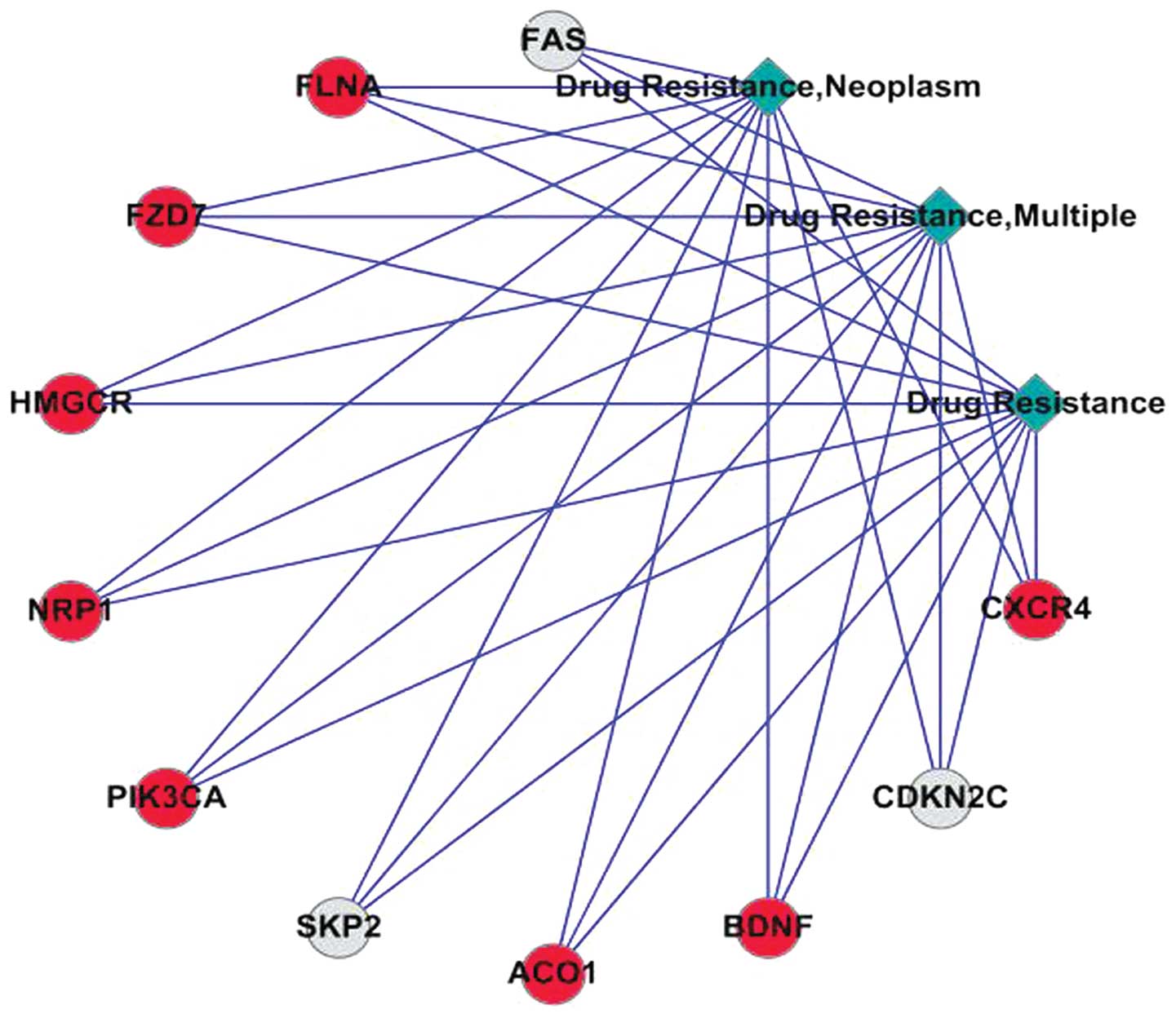
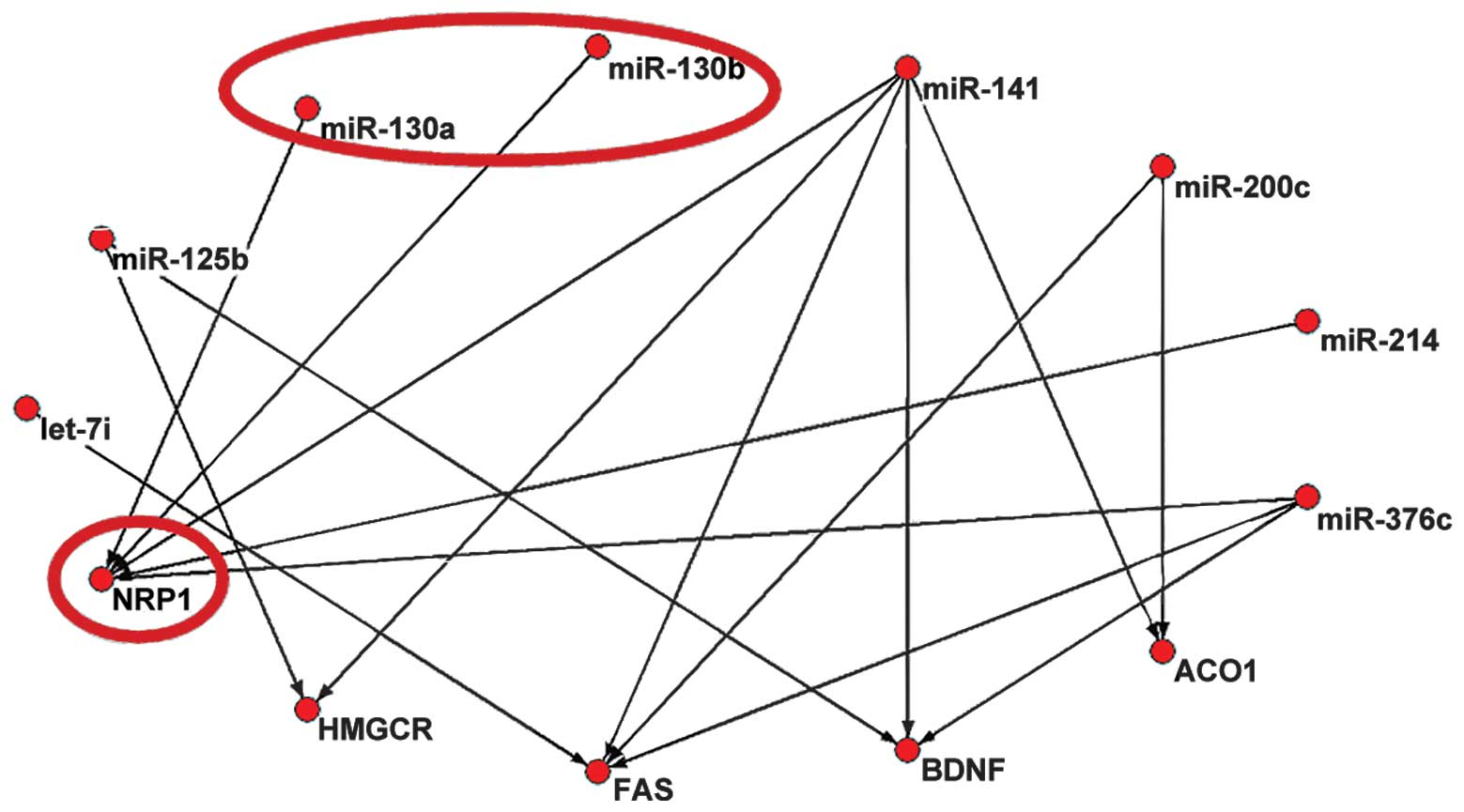
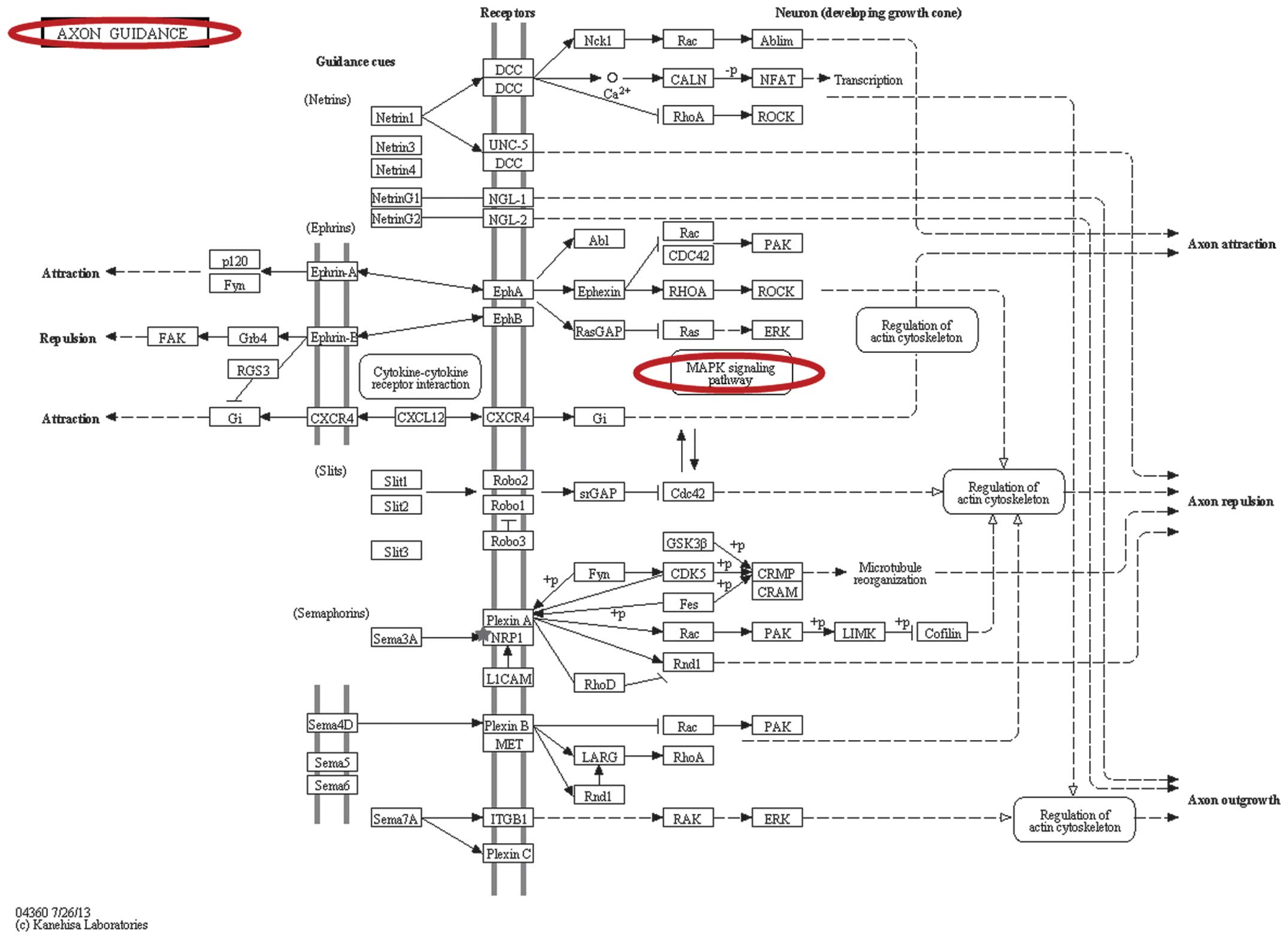
|
1
|
Oberaigner W, Minicozzi P, Bielska-Lasota
M, Allemani C, de Angelis R, Mangone L and Sant M; Eurocare Working
Group: Survival for ovarian cancer in Europe: The across-country
variation did not shrink in the past decade. Acta Oncol.
51:441–453. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Corney DC and Nikitin AY: MicroRNA and
ovarian cancer. Histol Histopathol. 23:1161–1169. 2008.PubMed/NCBI
|
|
3
|
Matsuda A and Katanoda K: Five-year
relative survival rate of ovarian cancer in the USA, Europe and
Japan. Jpn J Clin Oncol. 44:1962014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Samrao D, Wang D, Ough F, Lin YG, Liu S,
Menesses T, Yessaian A, Turner N, Pejovic T and Mhawech-Fauceglia
P: Histologic parameters predictive of disease outcome in women
with advanced stage ovarian carcinoma treated with neoadjuvant
chemotherapy. Transl Oncol. 5:469–474. 2012. View Article : Google Scholar
|
|
6
|
Rubin SC, Randall TC, Armstrong KA, Chi DS
and Hoskins WJ: Ten-year follow-up of ovarian cancer patients after
second-look laparotomy with negative findings. Obstet Gynecol.
93:21–24. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lin H and Changchien CC: Management of
relapsed/refractory epithelial ovarian cancer: Current standards
and novel approaches. Taiwan J Obstet Gynecol. 46:379–388. 2007.
View Article : Google Scholar
|
|
8
|
Faggad A, Darb-Esfahani S, Wirtz R, Sinn
B, Sehouli J, Könsgen D, Lage H, Noske A, Weichert W, Buckendahl
AC, Budczies J, Müller BM, Elwali NE, Dietel M and Denkert C:
Expression of multidrug resistance-associated protein 1 in invasive
ovarian carcinoma: implication for prognosis. Histopathology.
54:657–666. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zong C, Wang J and Shi TM: MicroRNA 130b
enhances drug resistance in human ovarian cancer cells. Tumour
Biol. 35:12151–12156. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jung HJ and Suh Y: MicroRNA in aging: From
discovery to biology. Curr Genomics. 13:548–557. 2012. View Article : Google Scholar :
|
|
11
|
Okamura K, Phillips MD, Tyler DM, Duan H,
Chou YT and Lai EC: The regulatory activity of microRNA* species
has substantial influence on microRNA and 3′UTR evolution. Nat
Struct Mol Biol. 15:354–363. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ro S, Park C, Young D, Sanders KM and Yan
W: Tissue-dependent paired expression of miRNAs. Nucleic Acids Res.
35:5944–5953. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Carrington JC and Ambros V: Role of
microRNAs in plant and animal development. Science. 301:336–338.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pitto L, Ripoli A, Cremisi F, Simili M and
Rainaldi G: microRNA(interference) networks are embedded in the
gene regulatory networks. Cell Cycle. 7:2458–2461. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ying H, Lv J, Ying T, Jin S, Shao J, Wang
L, Xu H, Yuan B and Yang Q: Gene-gene interaction network analysis
of ovarian cancer using TCGA data. J Ovarian Res. 6:882013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen L, Xuan J, Gu J, Wang Y, Zhang Z,
Wang TL and Shih IeM: Integrative network analysis to identify
aberrant pathway networks in ovarian cancer. Pac Symp Biocomput.
31:422012.
|
|
21
|
Liu H, Xiao F, Serebriiskii IG, O'Brien
SW, Maglaty MA, Astsaturov I, Litwin S, Martin LP, Proia DA,
Golemis EA and Connolly DC: Network analysis identifies an
HSP90-central hub susceptible in ovarian cancer. Clin Cancer Res.
19:5053–5067. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pan XH: Pathway crosstalk analysis based
on protein-protein network analysis in ovarian cancer. Asian Pac J
Cancer Prev. 13:3905–3909. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Schmeier S, Schaefer U, Essack M and Bajic
VB: Network analysis of microRNAs and their regulation in human
ovarian cancer. BMC Syst Biol. 5:1832011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Perumal M, Stronach EA, Gabra H and
Aboagye EO: Evaluation of 2-deoxy-2-[18F]fluoro-D-glucose- and
3′-deoxy-3′-[18F] fluorothymidine- positron emission tomography as
biomarkers of therapy response in platinum-resistant ovarian
cancer. Mol Imaging Biol. 14:753–761. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Haslehurst AM, Koti M, Dharsee M, Nuin P,
Evans K, Geraci J, Childs T, Chen J, Li J, Weberpals J, Davey S,
Squire J, Park PC and Feilotter H: EMT transcription factors snail
and slug directly contribute to cisplatin resistance in ovarian
cancer. BMC Cancer. 19:912012. View Article : Google Scholar
|
|
26
|
Li M, Balch C, Montgomery JS, Jeong M,
Chung JH, Yan P, Huang TH, Kim S and Nephew KP: Integrated analysis
of DNA methylation and gene expression reveals specific signaling
pathways associated with platinum resistance in ovarian cancer. BMC
Med Genomics. 8:342009. View Article : Google Scholar
|
|
27
|
Trinh XB, Tjalma WA, Dirix LY, Vermeulen
PB, Peeters DJ, Bachvarov D, Plante M, Berns EM, Helleman J, Van
Laere SJ and van Dam PA: Microarray-based oncogenic pathway
profiling in advanced serous papillary ovarian carcinoma. PLoS One.
6:e224692011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Edgar R, Domrachev M and Lash AE: Gene
expression omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar :
|
|
29
|
Sorrentino A, Liu CG, Addario A, Peschle
C, Scambia G and Ferlini C: Role of microRNAs in drug-resistant
ovarian cancer cells. Gynecol Oncol. 111:478–486. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang H, Kong W, He L, Zhao JJ, O'Donnell
JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV and Cheng JQ:
MicroRNA expression profiling in human ovarian cancer: miR-214
induces cell survival and cisplatin resistance by targeting PTEN.
Cancer Res. 68:425–433. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang N, Kaur S, Volinia S, Greshock J,
Lassus H, Hasegawa K, Liang S, Leminen A, Deng S, Smith L, et al:
MicroRNA microarray identifies Let-7i as a novel biomarker and
therapeutic target in human epithelial ovarian cancer. Cancer Res.
68:10307–10314. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kong F, Sun C, Wang Z, Han L, Weng D, Lu Y
and Chen G: miR-125b confers resistance of ovarian cancer cells to
cisplatin by targeting pro-apoptotic Bcl-2 antagonist killer 1. J
Huazhong Univ Sci Technolog Med Sci. 31:543–549. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ye G, Fu G, Cui S, Zhao S, Bernaudo S, Bai
Y, Ding Y, Zhang Y, Yang BB and Peng C: MicroRNA 376c enhances
ovarian cancer cell survival by targeting activin receptor-like
kinase 7: Implications for chemoresistance. J Cell Sci.
124:359–368. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cheng W, Liu T, Wan X, Gao Y and Wang H:
MicroRNA-199a targets CD44 to suppress the tumorigenicity and
multidrug resistance of ovarian cancer-initiating cells. FEBS J.
279:2047–2059. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fu X, Tian J, Zhang L, Chen Y and Hao Q:
Involvement of microRNA-93, a new regulator of PTEN/Akt signaling
pathway, in regulation of chemotherapeutic drug cisplatin
chemosensitivity in ovarian cancer cells. FEBS Lett. 586:1279–1286.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
van Jaarsveld MT, Helleman J, Boersma AW,
van Kuijk PF, van Ijcken WF, Despierre E, Vergote I, Mathijssen RH,
Berns EM, Verweij J, et al: miR-141 regulates KEAP1 and modulates
cisplatin sensitivity in ovarian cancer cells. Oncogene.
32:4284–4293. 2013. View Article : Google Scholar
|
|
37
|
Yang C, Cai J, Wang Q, Tang H, Cao J, Wu L
and Wang Z: Epigenetic silencing of miR-130b in ovarian cancer
promotes the development of multidrug resistance by targeting
colony-stimulating factor 1. Gynecol Oncol. 124:325–334. 2012.
View Article : Google Scholar
|
|
38
|
Ziliak D, Gamazon ER, Lacroix B, Kyung Im
H, Wen Y and Huang RS: Genetic variation that predicts platinum
sensitivity reveals the role of miR-193b* in chemotherapeutic
susceptibility. Mol Cancer Ther. 11:2054–2061. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Prislei S, Martinelli E, Mariani M,
Raspaglio G, Sieber S, Ferrandina G, Shahabi S, Scambia G and
Ferlini C: MiR-200c and HuR in ovarian cancer. BMC Cancer.
13:722013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Krek A, Grün D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M
and Rajewsky N: Combinatorial microRNA target predictions. Nat
Genet. 37:495–500. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
42
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J R Statist Soc B. 57:289–300. 1995.
|
|
43
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41(Database issue):
D991–D995. 2013. View Article : Google Scholar :
|
|
44
|
Melaiu O, Cristaudo A, Melissari E, Di
Russo M, Bonotti A, Bruno R, Foddis R, Gemignani F and Pellegrini
S: Review of transcriptome studies combined with data mining
reveals novel potential markers of malignant pleural mesothelioma.
Mutat Res. 750:132–140. 2012. View Article : Google Scholar
|
|
45
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar
|
|
46
|
Huang GW, Sherman BT, Tan Q, Kir J, Liu D,
Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC and Lempicki RA:
DAVID bioinformatics resources: Expanded annotation database and
novel algorithms to better extract biology from large gene lists.
Nucleic Acids Res. 35:W169–W175. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Vlachos IS, Kostoulas N, Vergoulis T,
Georgakilas G, Reczko M, Maragkakis M, Paraskevopoulou MD,
Prionidis K, Dalamagas T and Hatzigeorgiou AG: DIANA miRPath v.2.0:
Investigating the combinatorial effect of microRNAs in pathways.
Nucleic Acids Res. 40:W498–W504. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang F and Drabier R: IPAD: The
integrated pathway analysis database for systematic enrichment
analysis. BMC Bioinformatics. 13(Suppl 15): S72012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Batagelj V and Mrvar A: Pajek-Analysis and
Visualization of Large Networks. Mutzel P, Jünger M and Leipert S:
Graph Drawing. Lecture Notes in Computer Science. Springer Berlin
Heidelberg; 2265. pp. 477–478. 2002, View Article : Google Scholar
|
|
51
|
Ozgur A, Vu T, Erkan G and Radev DR:
Identifying gene-disease associations using centrality on a
literature mined gene-interaction network. Bioinformatics.
24:i277–i285. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42(Database issue): D92–D97. 2014. View Article : Google Scholar :
|
|
53
|
Biedler JL and Riehm H: Cellular
resistance to actinomycin D in Chinese hamster cells in vitro:
cross-resistance, radioautographic, and cytogenetic studies. Cancer
Res. 30:1174–1184. 1970.PubMed/NCBI
|
|
54
|
Ying H, Lv J, Ying T, Li J, Yang Q and Ma
Y: MicroRNA and transcription factor mediated regulatory network
for ovarian cancer: Regulatory network of ovarian cancer. Tumour
Biol. 34:3219–3225. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Chen C, Li M, Chai H, Yang H, Fisher WE
and Yao Q: Roles of neuropilins in neuronal development,
angiogenesis, and cancers. World J Surg. 29:271–275. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Geretti E and Klagsbrun M: Neuropilins:
novel targets for anti-angiogenesis therapies. Cell Adh Migr.
1:56–61. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Grandclement C and Borg C: Neuropilins: A
new target for cancer therapy. Cancers (Basel). 3:1899–1928. 2011.
View Article : Google Scholar
|
|
58
|
Lampropoulou A and Ruhrberg C: Neuropilin
regulation of angiogenesis. Biochem Soc Trans. 42:1623–1628. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Beck B, Driessens G, Goossens S, Youssef
KK, Kuchnio A, Caauwe A, Sotiropoulou PA, Loges S, Lapouge G, Candi
A, et al: A vascular niche and a VEGF-Nrp1 loop regulate the
initiation and stemness of skin tumours. Nature. 478:399–403. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Jubb AM, Strickland LA, Liu SD, Mak J,
Schmidt M and Koeppen H: Neuropilin-1 expression in cancer and
development. J Pathol. 226:50–60. 2012. View Article : Google Scholar
|
|
61
|
Pan Q, Chanthery Y, Liang WC, Stawicki S,
Mak J, Rathore N, Tong RK, Kowalski J, Yee SF, Pacheco G, et al:
Blocking neuropilin-1 function has an additive effect with
anti-VEGF to inhibit tumor growth. Cancer Cell. 11:53–67. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Delgoffe GM, Woo SR, Turnis ME, Gravano
DM, Guy C, Overacre AE, Bettini ML, Vogel P, Finkelstein D,
Bonnevier J, et al: Stability and function of regulatory T cells is
maintained by a neuropilin-1-semaphorin-4a axis. Nature.
501:252–256. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Ghosh S, Sullivan CA, Zerkowski MP,
Molinaro AM, Rimm DL, Camp RL and Chung GG: High levels of vascular
endothelial growth factor and its receptors (VEGFR-1, VEGFR-2,
neuropilin-1) are associated with worse outcome in breast cancer.
Hum Pathol. 39:1835–1843. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Staton CA, Koay I, Wu JM, Hoh L, Reed MW
and Brown NJ: Neuropilin-1 and neuropilin-2 expression in the
adenoma-carcinoma sequence of colorectal cancer. Histopathology.
62:908–915. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Piechnik A, Dmoszynska A, Omiotek M, Mlak
R, Kowal M, Stilgenbauer S, Bullinger L and Giannopoulos K: The
VEGF receptor, neuropilin-1, represents a promising novel target
for chronic lymphocytic leukemia patients. Int J Cancer.
133:1489–1496. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Chaudhary B, Khaled YS, Ammori BJ and
Elkord E: Neuropilin 1: Function and therapeutic potential in
cancer. Cancer Immunol Immunother. 63:81–99. 2014. View Article : Google Scholar
|
|
67
|
Baba T, Kariya M, Higuchi T, Mandai M,
Matsumura N, Kondoh E, Miyanishi M, Fukuhara K, Takakura K and
Fujii S: Neuropilin-1 promotes unlimited growth of ovarian cancer
by evading contact inhibition. Gynecol Oncol. 105:703–711. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Duman-Scheel M: Deleted in colorectal
cancer (DCC) path-finding: Axon guidance gene finally turned tumor
suppressor. Curr Drug Targets. 13:1445–1453. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Ge C, Li Q, Wang L and Xu X: The role of
axon guidance factor semaphorin 6B in the invasion and metastasis
of gastric cancer. J Int Med Res. 41:284–292. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Göhrig A, Detjen KM, Hilfenhaus G, Körner
JL, Welzel M, Arsenic R, Schmuck R, Bahra M, Wu JY, Wiedenmann B
and Fischer C: Axon guidance factor SLIT2 inhibits neural invasion
and metastasis in pancreatic cancer. Cancer Res. 74:1529–1540.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Sundaram MV: RTK/Ras/MAPK signaling.
WormBook. 1–19. 2006.
|
|
72
|
Wei SQ, Sui LH, Zheng JH, Zhang GM and Kao
YL: Role of ERK1/2 kinase in cisplatin-induced apoptosis in human
ovarian carcinoma cells. Chin Med Sci J. 19:125–129.
2004.PubMed/NCBI
|
|
73
|
Li F, Meng L, Zhou J, Xing H, Wang S, Xu
G, Zhu H, Wang B, Chen G, Lu YP and Ma D: Reversing chemoresistance
in cisplatin-resistant human ovarian cancer cells: A role of c-Jun
NH2-terminal kinase 1. Biochem Biophys Res Commun. 335:1070–1077.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Yang D, Sun Y, Hu L, Zheng H, Ji P, Pecot
CV, Zhao Y, Reynolds S, Cheng H, Rupaimoole R, Cogdell D, Nykter M,
Broaddus R, Rodriguez-Aguayo C, Lopez-Berestein G, Liu J,
Shmulevich I, Sood AK, Chen K and Zhang W: Integrated analyses
identify a master microRNA regulatory network for the mesenchymal
subtype in serous ovarian cancer. Cancer Cell. 23:186–199. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Han Y, Huang H, Xiao Z, Zhang W, Cao Y, Qu
L and Shou C: Integrated analysis of gene expression profiles
associated with response of platinum/paclitaxel-based treatment in
epithelial ovarian cancer. PLoS One. 7:e527452012. View Article : Google Scholar
|
|
76
|
Delfino KR and Rodriguez-Zas SL:
Transcription factor-microRNA-target gene networks associated with
ovarian cancer survival and recurrence. PLoS One. 8:e586082013.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Liu M1, Zhang X, Hu CF, Xu Q, Zhu HX and
Xu NZ: MicroRNA-mRNA functional pairs for cisplatin resistance in
ovarian cancer cells. Chin J Cancer. 33:285–294. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Yang L, Moss T, Mangala LS, Marini J, Zhao
H, Wahlig S, Armaiz-Pena G, Jiang D, Achreja A, Win J, Roopaimoole
R, Rodriguez-Aguayo C, Mercado-Uribe I, Lopez-Berestein G, Liu J,
Tsukamoto T, Sood AK, Ram PT and Nagrath D: Metabolic shifts toward
glutamine regulate tumor growth, invasion and bioenergetics in
ovarian cancer. Mol Syst Biol. 10:7282014. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Ibrahim FF, Jamal R, Syafruddin SE, Ab
Mutalib NS, Saidin S, MdZin RR, Hossain Mollah MM and Mokhtar NM:
MicroRNA-200c and microRNA-31 regulate proliferation, colony
formation, migration and invasion in serous ovarian. J Ovarian Res.
8:562015. View Article : Google Scholar
|