Introduction
Caveolin-1 (Cav-1) is an important structural and
functional component of caveolae, and is known to directly interact
via its scaffolding domain with multiple signaling molecules
(1). Cav-1 appears to act as a
tumor suppressor and an oncogene, depending on the context and type
of cancer. Cav-1 reportedly produces inhibitory effects on breast
cancer, as it is associated with breast cancer development and
progression (2,3). Under normal physiological conditions,
Cav-1 is abundantly expressed in breast stromal fibroblasts
(4,5). However, Cav-1 expression is reduced
in stromal fibroblasts of the breast cancer microenvironment, and
negatively correlated with the malignant potential of tumor cells.
Breast cancer patients with low or negative Cav-1 expression in
stromal fibroblasts often present a low survival rate, whereas the
survival rates of those with high stromal Cav-1 expression levels
are higher (4,6).
Although the prognostic values of the downregulation
of stromal Cav-1 in patients with breast cancer have been reported,
the exact mechanism is unclear (7). In order to fully assess the function
of Cav-1 as a tumor suppressor, further research into the
mechanisms of its expression is required. Additionally, the
correlations between Cav-1 expression, tumor stromal fibroblasts
and cancer cells must be verified.
Fibroblasts are major stromal cells for cancer and
are central to tumorigenesis, tumor growth and metastasis; they
secrete multiple factors that may prevent apoptosis, induce
proliferation and stimulate tumor angiogenesis (8,9).
Thus, a precise understanding of how stromal fibroblasts promote
tumor progression is important. Cav-1 downregulation may be a
mechanism implicated in the oncogenic transformation of
fibroblasts. Decreased expression levels or deleted Cav-1 in
fibroblasts can create a tumorigenic microenvironment, but the
relevant molecules are not fully clear (10).
Tumor protein 53-induced glycolysis and apoptosis
regulator (TIGAR) was discovered in 2005, following p53 activation
and detection with microarray analysis (11). The overexpression of TIGAR during
cancer development has been noted in various types of tumor.
Furthermore, cancer development is often delayed in the case of
TIGAR deletion. Recent research has highlighted that the expression
and activity of TIGAR can be disengaged from the p53 response,
narrowing the focus of its role in cancer development (12). Nevertheless, the activity of TIGAR
and the underlying mechanisms of regulation require further
investigation to allow for a more complete understanding of its
role in tumor pathology.
The present study aimed to clarify the potential
molecular mechanism of decreased Cav-1 in promoting tumor growth
through an investigation of Cav-1-targeted molecules in tumor
stromal fibroblasts and breast cancer cells. Using siRNA,
downregulation of the expression of Cav-1 was performed, and the
levels of certain growth factors were assessed, including stromal
cell-derived factor-1 (SDF-1), epidermal growth factor (EGF),
fibroblast-specific protein-1 (FSP-1) and TIGAR. The current study
provides evidence for the role of Cav-1 in tumor suppression.
Materials and methods
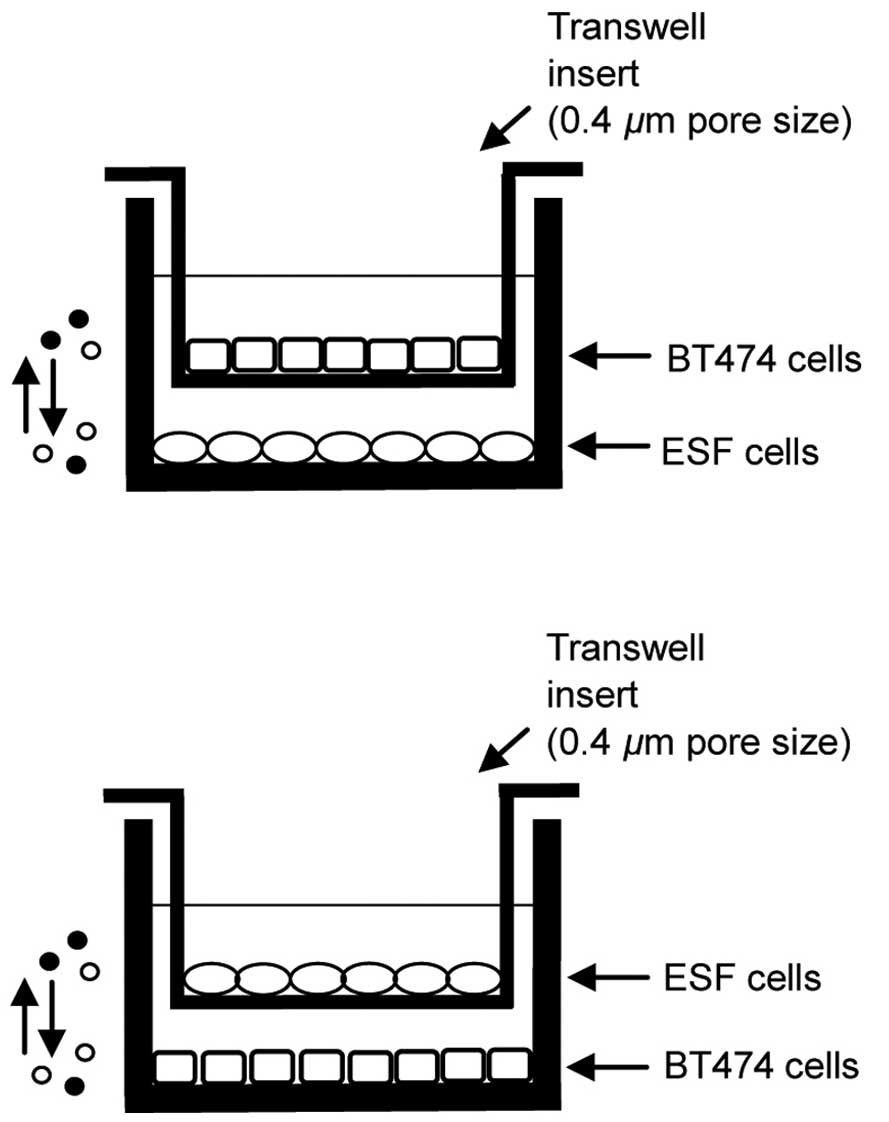
Cell culture and co-culture
The human skin fibroblast line CCC-ESF-1 (ESF) and
human breast cancer cell line BT474 were obtained from the Type
Culture Collection of the Chinese Academy of Sciences (Shanghai,
China). ESF or BT474 cells were cultured in Dulbecco's modified
Eagle's medium (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) with 10% fetal bovine serum (GE Healthcare Life
Sciences, Logan, UT, USA), 10 µg/ml streptomycin and 100
U/ml penicillin (Invitrogen; Thermo Fisher Scientific, Inc.) at
37°C in a humidified atmosphere with 5% CO2. ESF and
BT474 cells were co-cultured using polyester Transwell inserts (0.4
µm pore size; Thermo Fisher Scientific, Inc.). Cells
cultured on 6-well culture plates were used to detect the
expression of proteins. Cells cultured on 24-well culture plates
were used to assess levels of reactive oxygen species (ROS), cell
proliferation and apoptosis. ESF cells were plated at the bottom of
each well of the companion culture plates and allowed to adhere for
a minimum of 2 h without apical Transwell inserts. Subsequent to
plating, ESF cells were exposed to BT474 cell-conditioned media by
placing the BT474 Transwell inserts into the wells previously
plated with ESF cells. This method allowed the ESF and BT474 cells
to grow in the same medium without direct contact between them. The
co-culture models are presented in Fig. 1.
Cav-1 siRNA synthesis and
transfection
Cav-1 siRNAs were synthesized by GenePharma Co.,
Ltd. (Shanghai, China). The following sequences were used: Cav-1
siRNA-1, sense 5′-GCG ACC CUA AAC ACC UCA ATT-3′ and antisense
5′-UUG AGG UGU UUA GGG UCG CTT-3′; Cav-1 siRNA-2, sense 5′-CCU UCA
CUG UGA CGA AAUA TT-3′ and antisense 5′-UAU UUC GUC ACA GUG AAG
GTT-3′; Cav-1 siRNA-3, sense 5′-GCC GUG UCU AUU CCA UCU ATT-3′ and
antisense 5′-UAG AUG GAA UAG ACA CGG CTT-3′; negative control
siRNA, sense 5′-GCC GUG UCU AUU CCA UCU ATT-3′ and anti-sense
5′-ACG UGA CA C GUU CGG AGA ATT-3′. ESF-1 cells at 70–80%
confluence were transfected with the Cav-1 small interfering RNA or
the negative control siRNA by Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. Total RNA and total cellular protein were extracted at 24
and 48 h after transfection, respectively, to verify the effects of
the Cav-1 siRNAs.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). RNA concentrations
were measured using an SMA4000 spectrophotometer (Merinton
Instrument, Ltd., Beijing, China) at wavelengths of 260 and 280 nm.
The extracted RNA was reverse-transcribed into the cDNA using a
RevertAid First Strand cDNA Synthesis kit (Fermentas; Thermo Fisher
Scientific, Inc.). qPCR analysis of the cDNA was performed in
triplicate using an IQ SYBR Green Supermix (Bio-Rad Laboratories,
Inc., Hercules, CA, USA) and Super Real Premix Plus (Tiangen
Biotech Co., Ltd., Beijing, China). Relative changes were
calculated using the 2−ΔΔCq method. The primer sequences
used were as follows: Cav-1, sense 5′-ACGTAGA CTC GGA GGG ACATC-3′
and antisense 5′-GCA GAC AGC AAG CGG TAAA-3′; FSP-1, sense 5′-CCC
CAA GAA CAT CCA AAGT-3′ and antisense 5′-TTC AGG AAC AGC CAC
CAGT-3′; SDF-1, sense 5′-CAG GTG GTG GCTT AAC AGG-3′ and antisense
5′-AAG AGG AGG TGA AGG CAGTG-3′; EGF, sense 5′-CAA AAC GCCGAAG ACT
TACC-3′ and antisense 5′-GAC CAT CCA GAG CCA GACAC-3′; TIGAR, sense
5′-CAG CGG TAT TCC AGG ATTAG-3′ and anti-sense 5′-ACC TTA GCG AGT
TTC AGT CAG-3′. The PCR products were analyzed using CFX Manager
1.6 software (Bio-Rad Laboratories, Inc.).
Western blot analysis
Cells were lysed in 10 mM Tris-HCl (pH 7.4), 150 mM
NaCl, 1% NP-40, 0.1% sodium dodecyl sulfate (SDS), 1% sodium
deoxycholate and protease inhibitor cocktail tablet (Roche
Diagnostics, Basel, Switzerland). The protein content in each
sample was determined using a Bio-Rad Protein assay kit (Bio-Rad
Laboratories, Inc.). Equal amounts of protein were separated by
electrophoresis on 7.5~10% SDS-polyacrylamide gels (Beyotime
Institute of Biotechnology, Haimen, China). The electrophoresed
proteins were transferred to a nitrocellulose membrane (Bio-Rad
Laboratories, Inc.). Subsequent to overnight incubation at 4°C in a
blocking buffer [5% non-fat dry milk and 0.05% Tween-20 in
Tris-buffered saline (TBST)], the membranes were immunoblotted with
antibodies against rabbit anti-human polyclonal Cav-1 and mouse
anti-human monoclonal TIGAR (cat. nos. sc-894 and sc-377065; Santa
Cruz Biotechnology, Inc., Dallas, TX, USA) for 2 h at room
temperature. Following three washes with TBST for 5 min each, the
membranes were incubated with horseradish peroxidase-conjugated
secondary antibodies (goat anti-rabbit IgG, sc-2004 and goat
anti-mouse IgG, sc-2005; Santa Cruz Biotechnology, Inc.) at room
temperature for 1 h. Finally, the membranes were developed using
Beyotime ECL Plus (Beyotime Institute of Biotechnology).
Flow cytometry analysis
Cells were cultured for 72 h following transfection
with Cav-1 siRNA-2, harvested and then fixed with 4%
paraformaldehyde. Following a wash with 0.1% Triton X-100, the
cells were resuspended in 5% bovine serum albumin (Beyotime
Institute of Biotechnology) and incubated with the relevant primary
antibody for 1 h at 4°C. The antibodies against rabbit anti-human
polyclonal SDF-1 (cat. no. sc-28876) and rabbit anti-human
monoclonal FSP-1 (cat. no. ab124805) were purchased from Santa Cruz
Biotechnology, Inc., and Abcam (Cambridge, UK), respectively. The
antibody against rabbit anti-human polyclonal EGF (cat. no. BS3549)
was procured from Bioworld Technology, Inc. (St. Louis Park, MN,
USA). The cells were washed with phosphate-buffered saline (PBS)
and then incubated with the appropriate fluorescein
isothiocyanate-conjugated secondary antibody (goat anti-rabbit IgG,
sc-2012; Santa Cruz Biotechnology, Inc.) for 30 min at 4°C. The
cells were extensively washed with PBS and analyzed on an BDAccuri
C6 flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA).
Enzyme-linked immunosorbent assay
(ELISA)
The Cav-1 siRNA-2-transfected cells (ESFsiCav-1
cells) were co-cultured and mono-cultured for 72, 96 or 120 h
subsequent to transfection. The culture supernatants were collected
and stored at −70°C. The concentrations of growth factors in the
supernatants were determined using a Quantikine ELISA kit and a
human SDF-1α Quantikine ELISA kit (R&D Systems, Inc.,
Minneapolis, MN, USA) according to the manufacturer's protocol.
Standards and sample proteins were added and incubated at room
temperature for 2 h. Subsequent to washing, the conjugate was added
to the wells for 2 h at room temperature. The reaction was stopped
with a stop solution. The optical density (OD) of each well was
measured at 450 nm (SMA4000 spectrophotometer) and values were
correlated to the standard curve to determine protein
concentration.
Measurement of intracellular ROS
production
The intracellular generation of ROS in BT474 cells
was detected using the fluorescent probe
2′,7′-dichlorofluorescein-diacetate (DCFH-DA; Santa Cruz
Biotechnology, Inc.). BT474 cells were mono- and co-cultured with
ESF cells or ESFsiCav-1 cells for 72 h, then incubated with DCFH-DA
(20 mM final concentration) at 37°C for 35 min. The fluorescence
was analyzed using an FLx800TBID fluorescence reader (BioTek
Instruments, Inc., Winooski, VT, USA) using excitation and emission
wavelengths of 475 and 525 nm, respectively. The levels of ROS
production were expressed in relative fluorescence units (RFU).
Annexin V binding assay
When BT474 cells reached 80–90% confluence, they
were collected by trypsinization (Gibco; Thermo Fisher Scientific,
Inc.) and centrifugation at 1,200 × g, and washed once in PBS. An
annexin V binding assay was performed with 5×105 BT474
cells for each sample, according to the manufacturer's protocol
(Oncogene Science; Nuclea Biotechnologies, Inc., Cambridge, MA,
USA), followed by a FACScan cytometry analysis. In brief, cells
were incubated in 1.5 µl biotin-conjugated annexin V in 0.5
ml binding buffer for 15 min, and then incubated with
phycoerythrin-conjugated streptavidin for 5 min. To distinguish the
apoptosis from the other types of cell death, 7-amino-actinomycin
(7-AAD) was added prior to FACScan detection on a BDAccuri C6 flow
cytometer (BD Biosciences).
Cell counting kit-8 (CCK-8) assay
BT474 cells were seeded in 24-well culture plates
and ESF or ESFsiCav-1 cells in Transwell inserts in triplicate. The
Transwell inserts plated with ESF or ESFsiCav-1 cells were placed
into the wells containing BT474 cells. Fresh medium (100 µl)
and CCK-8 solution (40 µl; Wuhan Boster Biological
Technology, Ltd., Wuhan, China) were added to each well of the
culture plates following culture for 24, 48, 72, 96 and 120 h. The
plates were then incubated at 37°C for 1 h. A 100-µl aliquot
of the reactive solution was removed from each sample and then
placed in 96-well culture plates. The OD was measured at 450 nm
using an ELx800 microplate reader (BioTek Instruments, Inc.).
Statistical analysis
Data are presented as the mean ± standard deviation.
Statistical analysis was conducted using Student's t-test using
SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
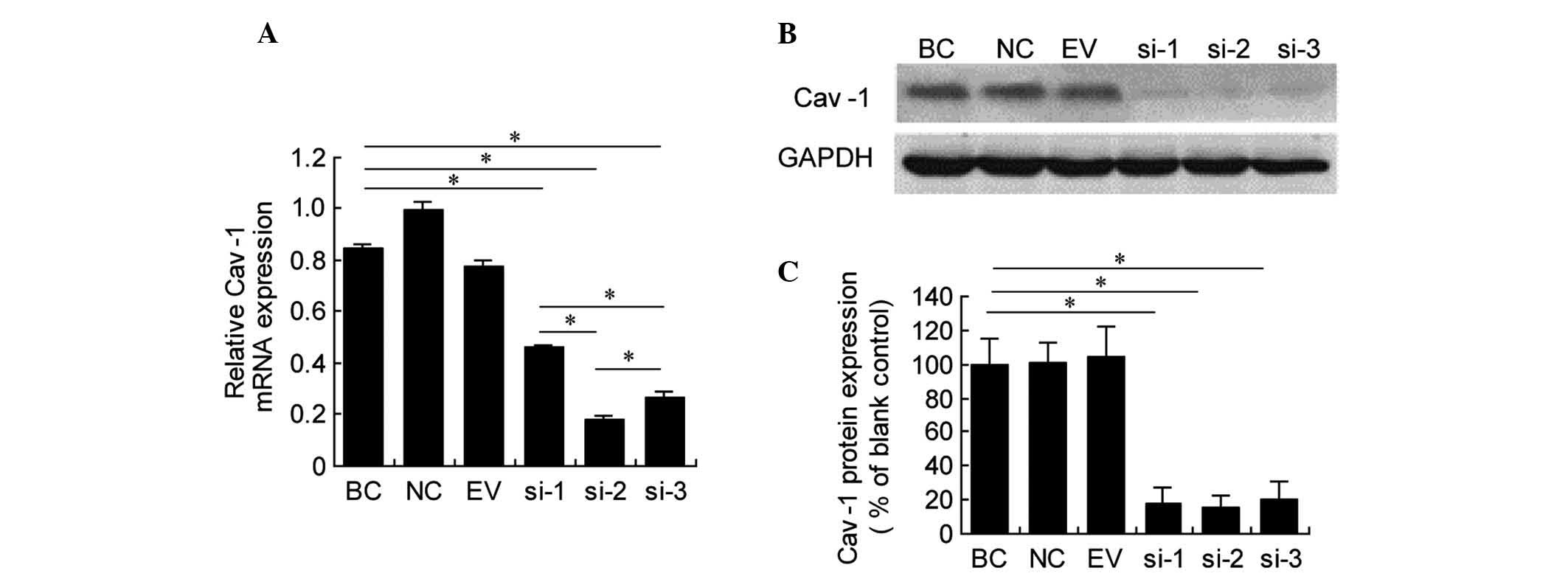
Interference effects of Cav-1 siRNA on
ESF cells
RT-qPCR and western blot analyses were performed to
determine the interference effects of Cav-1 siRNA on the ESF
fibroblast cell line. The results indicated that the Cav-1 mRNA
expression levels in ESF cells were significantly reduced in the
siRNA-1, -2 and -3 groups compared with the blank control group at
24 h following transfection with 100 nM Cav-1 siRNA (P<0.05;
Fig. 2A). Cav-1 mRNA expression
levels following siRNA interference were significantly lower in the
siRNA-2 group compared with those in siRNA-1 or -3 groups
(P<0.05; Fig. 2A).
Western blot analysis demonstrated that the Cav-1
protein expression levels were significantly reduced in the
siRNA-1, -2 and -3 groups compared with those in the blank control
group at 48 h subsequent to transfection with 100 nM Cav-1 siRNA
(P<0.05; Fig. 2B and C). Cav-1
protein expression in the siRNA-3 group was higher than in the
siRNA-1 and -2 groups, however no significant differences were
identified. These results indicate the specificity of the siRNA
used to target Cav-1. Since the RT-qPCR and western blot results
indicated that the siRNA-2 group was the most successful in
reducing Cav-1 expression, it was used as the Cav-1-specific
interference sequence for the sequential study.
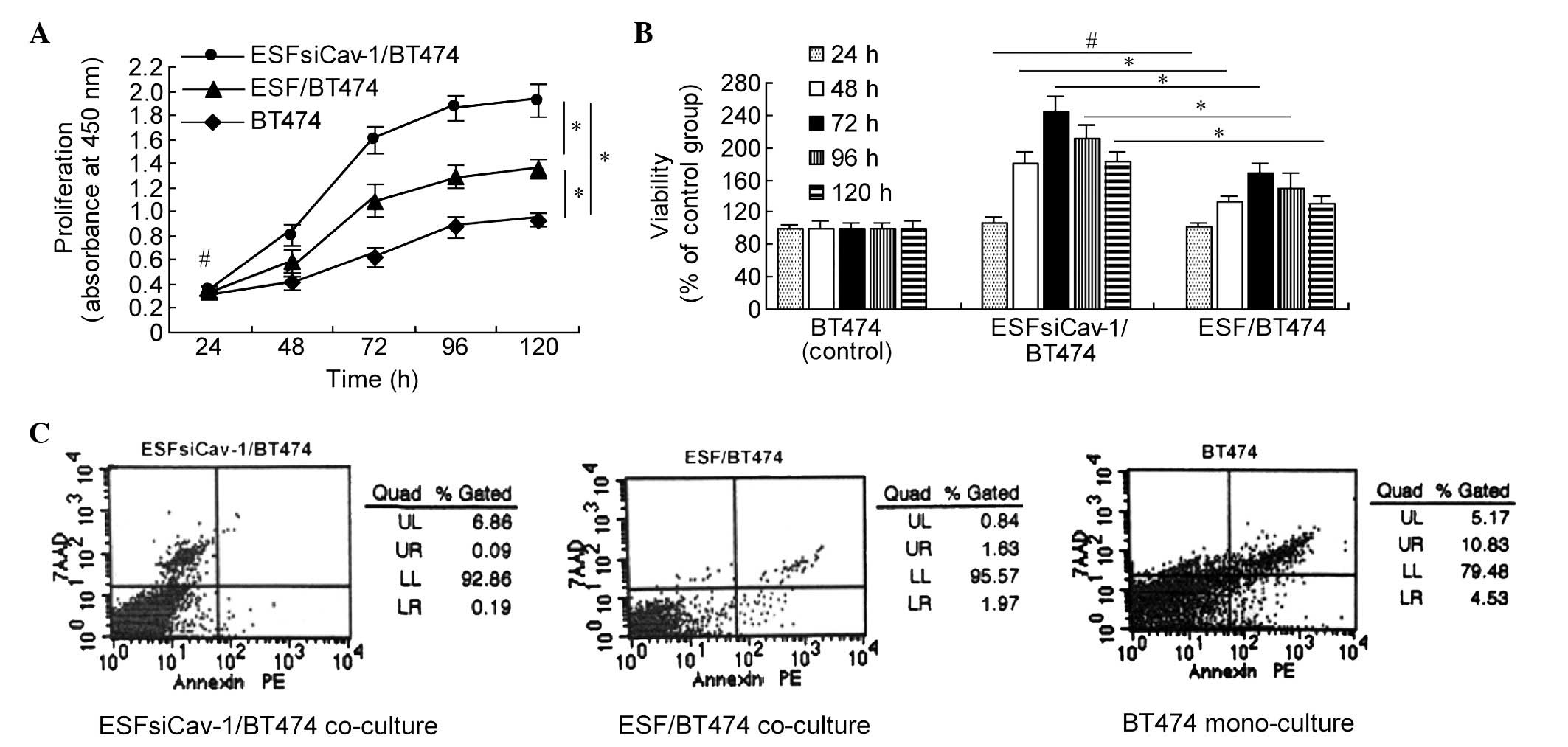
Downregulation of Cav-1 in ESF cells
promotes the growth of BT474 cells
CCK-8 assays from 24 to 120 h following mono- or
co-culture were performed in the BT474 breast cancer cell line to
determine the effects of Cav-1 downregulation on the proliferation
and viability of the BT474 cells. The groups did not significantly
differ in the observed levels of cell proliferation at 24 h.
However, BT474 cell proliferation was significantly greater in the
ESFsiCav-1/BT474 co-culture group than in the ESF/BT474 co-culture
or BT474 mono-culture groups at 48, 72, 96 and 120 h (P<0.05;
Fig. 3A).
Compared with the BT474 control group, the viability
of BT474 cells of the ESFsiCav-1/BT474 co-culture group increased
by 80% (48 h), 144% (72 h), 111% (96 h) and 82% (120 h) and those
of the ESF/BT474 co-culture group increased by 33% (48 h), 68% (72
h), 49% (96 h) and 31% (120 h). The percentage increases in the
ESFsiCav-1/BT747 cells were significantly greater than those in the
ESF/BT474 cells (Fig. 3B).
To investigate the effect of Cav-1 downregulation on
apoptosis in BT474 cells co-cultured with ESFsiCav-1 cells, an
annexin V binding assay was performed as the BT474 cells reached
80–90% confluence. A 10-fold reduction in the early apoptosis of
BT474 cells in the ESFsiCav-1/BT474 co-culture group was observed,
compared with the ESF/BT474 cell co-culture group. Furthermore, a
23-fold reduction in the early apoptotic cells in the
ESFsiCav-1/BT474 co-culture group was detected, compared with the
BT474 cell mono-culture group (Fig.
3C).
Proliferation of BT474 cells was
associated with the increase in levels of SDF-1, EGF and FSP-1 in
the ESFsiCav-1 cells
The downregulation of Cav-1 in ESF cells promoted
the proliferation and viability of BT474 cells. Therefore, the
expression of certain proliferation-associated molecules, including
SDF-1, EGF and FSP-1, was investigated. ESFsiCav-1 cells were mono-
and co-cultured with BT474 cells and the mRNA and protein
expression levels of the target molecules were examined by RT-qPCR
and flow cytometry. RT-qPCR assay demonstrated that Cav-1
downregulation significantly increased the mRNA expression levels
of SDF-1, EGF and FSP-1 in the ESF cells 48 h subsequent to
transfection with Cav-1 siRNA-2. Compared with the mono-culture of
ESFsiCav-1, the co-culture of ESFsiCav-1 with BT474 exhibited
enhanced SDF-1, EGF and FSP-1 mRNA expression, hence exhibiting a
synergistic effect (P<0.05; Fig.
4A).
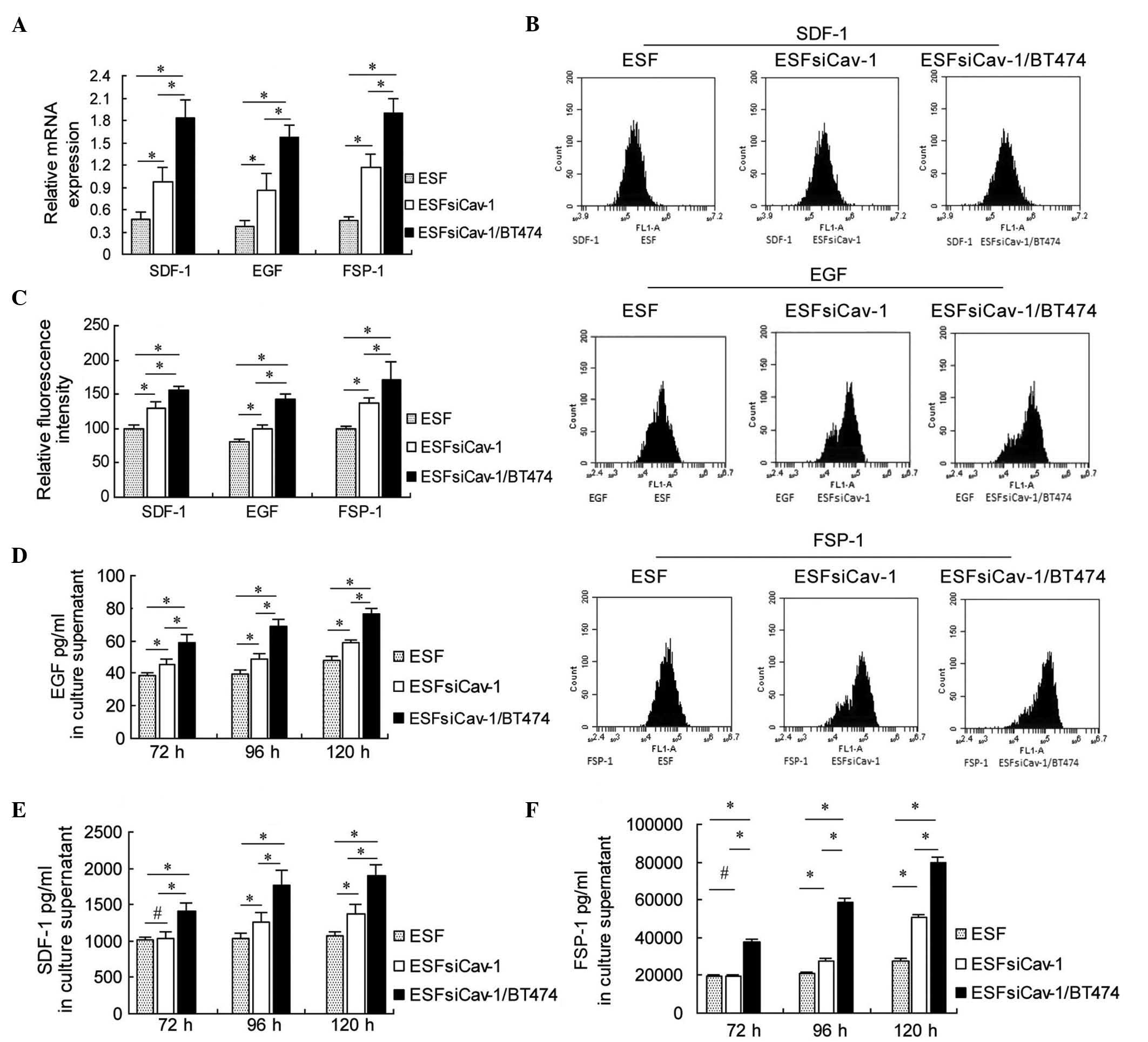 | Figure 4Upregulation of SDF-1, EGF and FSP-1
mRNA and protein levels in ESF cells by downregulation of Cav-1.
(A) Reverse transcription-quantitative polymerase chain reaction
analysis of the mRNA expression levels of SDF-1, EGF and FSP-1 at
48 h after transfection with Cav-1 siRNA-2. (B) Flow cytometry
analysis of the protein expression levels of SDF-1, EGF and FSP-1
at 72 h subsequent to transfection with Cav-1 siRNA-2. (C) Relative
fluorescence intensity of SDF-1, EGF and FSP-1 at 72 h after
transfection with Cav-1 siRNA-2. (D) EGF (E) SDF-1 and (F) FSP-1
were measured using ELISA at 72, 96 and 120 h after transfection
with Cav-1 siRNA-2. *P<0.05 and
#P>0.05, comparison shown by brackets. SDF-1, stromal
cell-derived factor-1; EGF, epidermal growth factor; FSP-1,
fibroblast-specific protein-1; Cav-1, caveolin-1. |
The flow cytometry results were consistent with the
RT-qPCR results. SDF-1, EGF and FSP-1 protein expression levels
were increased following Cav-1 downregulation, and were
significantly higher in the ESFsiCav-1/BT474 co-culture group,
compared with the ESF mono-culture group or ESFsiCav-1 mono-culture
group at 72 h after transfection with Cav-1 siRNA-2 (P<0.05;
Fig. 4B and C).
The concentrations of these molecules in the culture
supernatant were determined using ELISA. The results of ELISA
indicated that Cav-1 siRNA transfection increased SDF-1, EGF and
FSP-1 production in the supernatant of the ESFsiCav-1/BT474
co-culture at 72, 96 and 120 h compared with the control groups
(P<0.05; Fig. 4D–F). These data
suggest that SDF-1, EGF and FSP-1 are Cav-1-targeted molecules that
promote the proliferation of BT474 cells.
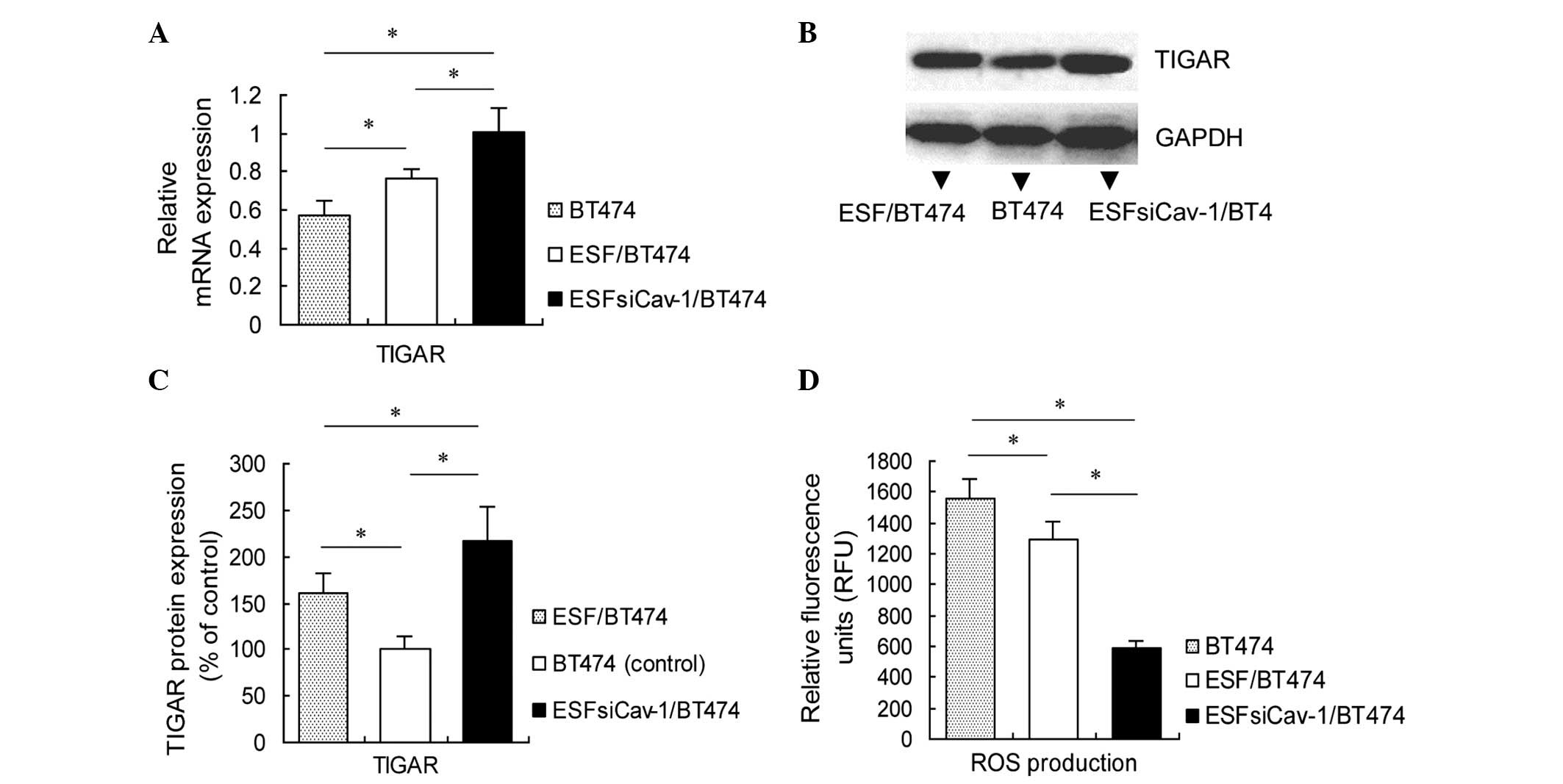
Downregulation of Cav-1 promotes TIGAR
expression in BT474 cells, alongside inhibition of apoptosis
Apoptosis of BT474 cells was reduced in the
co-culture with ESFsiCav-1 cells. Thus, the effects of the
downregulation of Cav-1 on the expression of apoptosis regulators
in breast cancer cells were investigated. RT-qPCR was used to
measure mRNA levels of TIGAR in BT474 cells 48 h after mono-culture
and co-culture. The results demonstrated that the BT474 cells from
the ESFsiCav-1/BT474 co-culture group expressed significantly
higher levels of TIGAR than the cells from the ESF/BT474 co-culture
group and those from the BT474 mono-culture group (P<0.05;
Fig. 5A). TIGAR protein expression
levels were then assessed using western blot analysis 72 h after
mono-culture and co-culture, and the results indicated that the
TIGAR protein levels were significantly increased in the
ESFsiCav-1/BT474 co-culture group compared with the ESF/BT474
co-culture group or BT474 mono-culture group (P<0.05; Fig. 5B and C). The effects of TIGAR
expression on ROS regulation can depend, at least in part, on the
cell type and context. To elucidate whether the upregulation of
TIGAR impacts on ROS production in BT474 cells, the intracellular
generation of ROS in BT474 cells was investigated using the
fluorescent probe DCFH-DA. As presented in Fig. 5D, co-culture of BT474 and
ESFsiCav-1 cells led to a reduction in the fluorescent signal in
these cells, compared with the ESF/BT474 co-culture group (589±50
vs. 1298±115; P<0.05) and the BT474 mono-culture group (589±50
vs. 1560±127; P<0.05). Collectively, these results indicate that
TIGAR expression is associated with Cav-1 downregulation, and that
the upregulation of TIGAR contributes to the inhibition of BT474
cell apoptosis mediated by Cav-1 downregulation.
Discussion
The results from the present study demonstrated that
the downregulation of Cav-1 in fibroblasts led to a significant
increase in the expression and secretion of the growth factors,
SDF-1, EGF and FSP-1. Furthermore, it upregulated the expression of
TIGAR, which may accelerate tumor cell proliferation and suppress
tumor cell apoptosis.
Fibroblasts from tumor stroma may be more likely to
trigger tumor growth compared with normal stroma. These fibroblasts
secrete high levels of growth factors, extracellular matrix
components and matrix metalloproteinases, but the relevant factors
and components are not fully understood (13). The downregulation or loss of Cav-1
expression in stromal fibroblasts is associated with tumor
prognosis (14). It has been
indicated that Cav-1 loss in stromal fibroblasts of patients with
breast cancer may be used as a predictor of the relapse of breast
cancer, lymph node metastasis and tamoxifen resistance (15,16).
This has not been associated with the expression of the estrogen
receptor (ER), progesterone receptor (PR) or human epidermal growth
factor receptor-2 (HER2) (15). In
patients with ER−/PR−/HER2− breast
cancer or ER−/PR−/HER2− ductal
carcinoma, the loss of Cav-1 in stromal fibroblasts has been used
as an indicator of unfavorable clinical outcome (4). Cav-1 expression in tumor cells is not
correlated with breast cancer prognosis (17). Thus, loss of stromal Cav-1 is a key
predictor of a 'lethal' cancer microenvironment.
Loss of stromal Cav-1 is also linked with the poor
prognosis of prostate cancer and metastasis of bone and lymph nodes
(18); and decreased Cav-1 levels
in fibroblasts results in increased levels of myofibroblast markers
and extracellular matrix proteins in co-cultured human breast
cancer cells with fibroblasts, suggesting that Cav-1 downregulation
initiates fibroblast activation in tumorigenesis (19). Myofibroblast markers and glycolytic
enzymes were observed to be upregulated in a model of
cancer-associated Cav-1-deficient fibroblasts under normoxic
conditions (20). However, the
mechanisms of phenotype transformation from benign to heterogeneous
fibroblasts are unclear. Further investigation is required into the
molecules associated with Cav-1 expression and tumor stromal
fibroblasts and cancer cells, in order to establish multiple
Cav-1-specific therapies and further clarify the mechanisms of
Cav-1 in tumor growth.
In the present study, fibroblasts were transfected
with synthetic siRNA Cav-1 sequences to determine the effect of the
downregulation of Cav-1 on tumor stromal and cancer cells. The
results indicated that Cav-1 expression was down-regulated in the
Cav-1 siRNA-transfected cells (Fig.
2), thus the Cav-1 siRNA sequences effectively interfered with
Cav-1 gene expression. The siRNA-2 exhibited a higher interference
efficacy than the siRNA-1 or the siRNA-3. Therefore, siRNA-2 was
selected as the Cav-1-specific interference sequence for the
current study.
Tumor occurrence and development are strongly
associated with stromal microenvironment. Additionally,
cancer-associated fibroblasts are the major stromal cells in this
microenvironment. These fibroblasts are derived from the
transdifferentiation of various cells, including quiescent
fibroblasts, epithelial cells, endothelial cells, mesenchymal stem
cells and pericytes (21,22). Cancer-associated fibroblasts in
direct contact with tumor cells secrete various paracrine factors,
synthesize oncogenic components and connect oncogenic signal
pathways to promote the development and progression of tumor cells
(23). In the present study, the
co-culture models of fibroblasts with breast cancer cells were
established to simulate the breast cancer microenvironment. The
reduced levels of Cav-1 in the co-culture were utilized to
investigate the association between Cav-1 and fibroblasts and
cancer cells through analyzing the expression of cancer-associated
molecules in fibroblasts and breast cancer cells.
SDF-1, also termed CXCL12, is a chemotactic cytokine
belonging to the large family of CXC chemokines (24–28).
SDF-1 induces cell migration, cell adhesion, neutrophil activation
and inflammation. Previous studies have reported that SDF-1 is
associated with tumor occurrence, metastasis and growth (24,25,29).
Stromal cells are key sources of SDF-1, and an increase in SDF-1
expression may be associated with tumor growth. The recruitment of
endothelial progenitor cells by SDF-1 and its direct effect on
cancer cells may promote tumor angiogenesis (26,30).
In the current study, the Cav-1 siRNA fibroblasts/breast cancer
cell co-culture group was the most effective in increasing SDF-1
expression amongst the groups investigated. This suggests that
downregulated Cav-1 and the co-culture with breast cancer cells
synergistically increased SDF-1 expression in fibroblasts, and the
tumor inhibition effect of Cav-1 may be associated with the
inhibition of the signaling pathways in which SDF-1
participates.
EGF is a peptide consisting 53 amino acids, with a
variety of biological functions. It stimulates epithelial cell
motility, and is thus required for re-epithelialization. It is also
a major stimulator of fibroblast migration and wound contraction,
and is hypothesized to affect cell proliferation, embryo
development and tumorigenesis (31–33).
The effect of Cav-1 downregulation on EGF expression in fibroblasts
was investigated in the present study. Downregulation of Cav-1
significantly upregulated EGF expression in the fibroblasts. This
indicates the antagonistic relationship between Cav-1 upregulation
and EGF expression. The microenvironment of the co-cultured Cav-1
siRNA fibroblasts with breast cancer cells was able to enhance the
expression of EGF.
FSP-1 (also termed S100A4) is implicated in numerous
stages of tumor progression, including motility, invasion and
apoptosis, however, its function remains uncertain (34,35).
A previous study demonstrated that the co-injection of
FSP-1+/+ fibroblasts with tumor cells restores tumor
development and metastasis in FSP-1−/− animals, whereas
co-injection with FSP-1−/− fibroblasts does not
(36). The stromal
microenvironment can be altered by FSP-1, in order to favor tumor
progression. In the current study, the expression of FSP-1 was
significantly higher in the Cav-1 siRNA-transfected fibroblasts
than in the control-transfected fibroblasts, which suggests that
the downregulation of Cav-1 is an upstream event of FSP-1. The
Cav-1 siRNA-induced upregulation of SDF-1, EGF and FSP-1 alters the
phenotypes of fibroblasts, causing them to become 'reactive'. The
microenvironment of reactive fibroblasts is beneficial to tumor
growth. The increased concentrations of SDF-1, EGF and FSP-1 in the
culture supernatant of Cav-1 siRNA fibroblasts can accelerate the
proliferation of tumor cells. The alterations in proliferation of
breast cancer cells were consistent with changes in SDF-1, EGF and
FSP-1 expression in the current study, which suggests that high
expression levels of SDF-1, EGF and FSP-1 can promote breast cancer
cell proliferation.
TIGAR may protect cells from ROS-associated
apoptosis, and thus, downregulation of the expression of TIGAR may
lead to p53-induced cell death (11,37).
It has been determined that p53 is not required for TIGAR
expression and activity (12).
Therefore, in order to identify the function of TIGAR in cancer
development, the factors regulating it require further study. The
present study identified that the breast cancer cells from the
Cav-1 siRNA fibroblasts/breast cancer cell co-culture group
presented the highest increase in the expression levels of TIGAR.
Downregulation of Cav-1 in fibroblasts influenced the surrounding
tumor cells via SDF-1, EGF, FSP-1 and TIGAR. Initially,
downregulation of Cav-1 increased the concentrations of the
tumor-associated molecules SDF-1, EGF and FSP-1 in tumor stroma.
This triggered the accelerated proliferation of tumor cells, which
may synergistically influence the expression of TIGAR in cancer
cells, suppressing cancer cell apoptosis. The downregulation of
Cav-1 in fibroblasts may not produce direct effects in tumor cells.
However, the resulting altered stromal microenvironment (with
increased expression levels of SDF-1, EGF and FSP-1) demonstrates
its importance in tumor suppression.
Cancer cells rapidly proliferate, and TIGAR
expression levels are upregulated in cancer cells (38). TIGAR functions to limit ROS, thus
protecting cells against ROS-induced death. As demonstrated in the
current study, the upregulation of TIGAR expression was accompanied
by low levels of ROS. The Cav-1-targeted cascade reactions observed
in the present study may be the hallmark of a malignant breast
tumor.
In summary, the current study highlighted
Cav-1-targeted molecules and their regulatory events, including the
regulation of SDF-1, EGF and FSP-1 expression and secretion in
stromal fibroblasts. Downregulation of Cav-1 promotes the
upregulation of TIGAR expression in breast cancer cells, resulting
in cancer cell proliferation and the suppression of cancer cell
apoptosis. These results provide novel insight into the
tumor-suppressor mechanism of Cav-1, indicating that
Cav-1-dependent signaling involves SDF-1, EGF, FSP-1 and TIGAR.
Acknowledgments
The current study was supported by the National
Natural Science Foundation of China (grant nos. 91229118 and
30860118).
References
|
1
|
Qian N, Ueno T, Kawaguchi-Sakita N,
Kawashima M, Yoshida N, Mikami Y, Wakasa T, Shintaku M, Tsuyuki S,
Inamoto T and Toi M: Prognostic significance of tumor/stromal
caveolin-1 expression in breast cancer patients. Cancer Sci.
102:1590–1596. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chiu WT, Lee HT, Huang FJ, Aldape KD, Tao
J, Steeg PS, Chou CY, Lu Z, Xie K and Huang S: Caveolin-1
upregulation mediates suppression of primary breast tumor growth
and brain metastases by stat3 inhibition. Cancer Res. 71:4932–4943.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sotgia F, Martinez-Outschoorn UE, Pavlides
S, Howell A, Pestell RG and Lisanti MP: Understanding the Warburg
effect and the prognostic value of stromal caveolin-1 as a marker
of a lethal tumor microenvironment. Breast Cancer Res. 13:213–225.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Witkiewicz AK, Dasgupta A, Nguyen KH, Liu
C, Kovatich AJ, Schwartz GF, Pestell RG, Sotiga F, Rui H and
Lisanti MP: Stromal caveolin-1 levels predict early DCIS
progression to invasive breast cancer. Cancer Biol Ther.
8:1071–1079. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Patani N, Martin LA, Reis-Filho JS and
Dowsett M: The role of caveolin-1 in human breast cancer. Brest
Cancer Res Treat. 131:1–15. 2013. View Article : Google Scholar
|
|
6
|
Sotgia F, Martinez-Outschoorn UE, Howell
A, Prestell RG, Pavlides S and Lisanti MP: Caveolin-1 and cancer
metabolism in the tumor microenvironment: Markers, models, and
mechanisms. Annu Rev Pathol. 7:423–467. 2012. View Article : Google Scholar
|
|
7
|
Du C, Chen L, Zhang HJ, Wang ZC, Liu WC,
Xie XD and Xie MJ: Caveolin-1 limits the contribution of BKCa
channel to MCF-7 breast cancer cell proliferation and invasion. Int
J Mol Sci. 15:20706–20722. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Paola C: Cancer associated fibroblasts:
The dark side of the coin. Am J Cancer Res. 1:482–497. 2011.
|
|
9
|
Buckley CD: Why does chronic inflammation
persist: An unexpected role for fibroblasts. Immunol Lett.
138:12–14. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang Z, Wang N, Li W, Liu P, Chen Q, Situ
H, Zhong S, Guo L, Lin Y, Shen J and Chen J: Caveolin-1 mediates
chemoresistance in breast cancer stem cells via β-catenin/ABCG2
signaling pathway. Carcinogenesis. 35:2346–2356. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jen KY and Cheung VG: Identification of
novel p53 target genes in ionizing radiation response. Cancer Res.
65:7666–7673. 2005.PubMed/NCBI
|
|
12
|
Lee P, Vousden KH and Cheung EC: TIGAR,
TIGAR, burning bright. Cancer Metab. 2:12014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yamaguchi H and Sakai R: Direct
interaction between carcinoma cells and cancer associated
fibroblasts for the regulation of cancer invasion. Cancers.
7:2054–2062. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Witkiewicz AK, Dasgupta A, Sammons S, Er
O, Potoczek MB, Guiles F, Sotgia F, Brody JR, Mitchell EP and
Lisanti MP: Loss of stromal caveolin-1 expression predicts poor
clinical outcome in triple negative and basal-like breast cancers.
Cancer Biol Ther. 10:135–43. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Witkiewicz AK, Dasgupta A, Sotgia F,
Mercier I, Pestell RG, Sabel M, Kleer CG, Brody JR and Lisanti MP:
An absence of stromal caveolin-1 expression predicts early tumor
recurrence and poor clinical outcome in human breast cancers. Am J
Pathol. 74:2023–2034. 2009. View Article : Google Scholar
|
|
16
|
EI-Gendi SM, Mostafa MF and EI-Gendi AM:
Stromal caveolin-1 expression in breast carcinoma. Correlation with
early tumor recurrence and clinical outcome. Pathol Oncol Res.
18:459–469. 2012. View Article : Google Scholar
|
|
17
|
Shan-Wei W, Kan-Lun X, Shu-Qin R, Li-Li Z
and Li-Rong C: Overexpression of caveolin-1 in cancer-associated
fibroblasts predicts good outcome in breast cancer. Breast Care
(Basel). 7:477–483. 2012. View Article : Google Scholar
|
|
18
|
Di Vizio D, Morello M, Sotgia F, Pestell
RG, Freeman MR and Lisanti MP: An absence of stromal caveolin-1 is
associated with advanced prostate cancer, metastatic disease and
epithelial Akt activation. Cell Cycle. 8:2420–2424. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Martinez-Outschoorn UE, Pavlides S,
Whitaker-Menezes D, Daumer KM, Milliman JN, Chiavarina B, Migneco
G, Wiktkiewicz AK, Martinez-Cantarin MP, Flomenberg N, et al: Tumor
cells induce the cancer associated fibroblast phenotype via
Caveolin-1 degradation: Implications for breast cancer and DCIS
therapy with autophagy inhibitors. Cell Cycle. 9:2423–2433. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pavlides S, Whitaker-Menezes D,
Castello-Cros R, Flomenberg N, Witkiewicz AK, Frank PG, Casimiro
MC, Wang C, Fortina P, Addya S, et al: The reverse Warburg effect:
Aerobic glycolysis in cancer associated fibroblasts and the tumor
stroma. Cell Cycle. 8:3984–4001. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kidd S, Spaeth E, Dembinski JL, Dietrich
M, Watson K, Klopp A, Battula VL, Weil M, Andreeff M and Marini FC:
Direct evidence of mesenchymal stem cell tropism for tumor and
wounding microenvironments using in vivo bioluminescent imaging.
Stem Cells. 27:2614–2623. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Crisan M, Yap S, Casteilla L, Chen CW,
Corselli M, Park TS, Andriolo G, Sun B, Zheng B, Zhang L, et al: A
perivascular origin for mesenchymal stem cells in multiple human
organs. Cell Stem Cell. 3:301–313. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jezierska-Drutel A, Rosenzweig SA and
Neumann CA: Role of oxidative stress and the microenvironment in
breast cancer development and progression. Adv Cancer Res.
119:107–125. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Engl T, Relja B, Marian D, Blumenberg C,
Müller I, Beecken WD, Jones J, Ringel EM, Bereiter-Hahn J, Jonas D
and Blaheta RA: CXCR4 chemokine receptor mediates prostate tumor
cell adhesion through alpha5 and beta3 integrins. Neoplasia.
8:290–301. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Peng SB, Peek V, Zhai Y, Paul DC, Lou Q,
Xia X, Eesalu T, Kohn W and Tang S: Akt activation, but not
extracellular bignal-reguhted kinase activation,is required for
SDF-lalpha/CXCR4-mediated migration of epitheloid carcinoma cells.
Mol Cancer Res. 3:227–236. 2005.PubMed/NCBI
|
|
26
|
Orimo A, Gupta PB, Sgroi DC,
Arenzana-Seisdedos F, Delaunay T, Naeem R, Carey VJ, Richardson AL
and Weinberg RA: Stromal fibroblasts present in invasive human
breast carcinomas promote tumor growth and angiogenesis through
elevated SDF-1/CXCLl2 secretion. Cell. 121:335–348. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nagasawa T, Hirota S, Tachibana K,
Takakura N, Nishikawa S, Kitamura Y, Yoshida N, Kikutani H and
Kishimoto T: Defects of B-cell lymphopoiesis and bone-marrow
myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature.
382:635–638. 1996. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lewellis SW and Knaut H: Attractive
guidance: how the chemokine SDF1/CXCL12 guides different cells to
different locations. Semin Cell Dev Bio. 23:333–340. 2012.
View Article : Google Scholar
|
|
29
|
Balkwill F: The significance of cancer
cell expression of the chemokine receptor CXCR4. Semin Cancer Biol.
14:171–179. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Allinen M, Beroukhim R, Cai L, Brennan C,
Lahti-Domenici J, Juang H, Porter D, Hu M, Chin L, Richardson A, et
al: Molecular characterization of the tumor microenvironment in
breast cancer. Cancer Cell. 6:17–32. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li CF, Ma Y, Wei YZ and Xue Y:
Relationship between VEGF, EGF and invasion, metastasis of gastric
cancer cells. Chin J Cancer Res. 21:122–129. 2009. View Article : Google Scholar
|
|
32
|
Baek MK, Kim MH, Jang HJ, Park JS, Chung
IJ, Shin BA, Ahn BW and Jung YD: EGF stimulates uPAR expression and
cell invasiveness through ERK, AP-l, and NF-kappaB signaling in
human gastric carcinoma cells. Oncol Rep. 20:1569–1575.
2008.PubMed/NCBI
|
|
33
|
Hardwicke J, Schmaljohann D, Boyce D and
Thomas D: Epidermal growth factor therapy and wound healing-past,
present and future perspectives. Surgeon. 6:172–177. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Helfman DM, Kim EJ, Lukanidin E and
Grigorian M: The metastasis associated protein S100A4: role in
tumour progression and metastasis. Br J Cancer. 92:1955–1958. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tarabykina S, Griffiths TR, Tulchinsky E,
Mellon JK, Bronstein IB and Kriajevska M: Metastasis-associated
protein S100A4: spotlight on its role in cell migration. Curr
Cancer Drug Targets. 7:217–228. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Grum-Schwensen B, Klingelhofer J, Berg CH,
El-Naaman C, Grigorian M, Lukanidin E and Ambartsumian N:
Suppression of tumor development and metastasis formation in mice
lacking the S100A4(mts1) gene. Cancer Res. 65:3772–3780. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Green DR and Chipuk JE: p53 and
metabolism: Inside the TIGAR. Cell. 126:30–32. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bensaad K, Tsuruta A, Selak MA, Vidal MN,
Nakano K, Bartrons R, Gottlieb E and Vousden KH: TIGAR, a
p53-inducible regulator of glycolysis and apoptosis. Cell.
126:107–120. 2006. View Article : Google Scholar : PubMed/NCBI
|