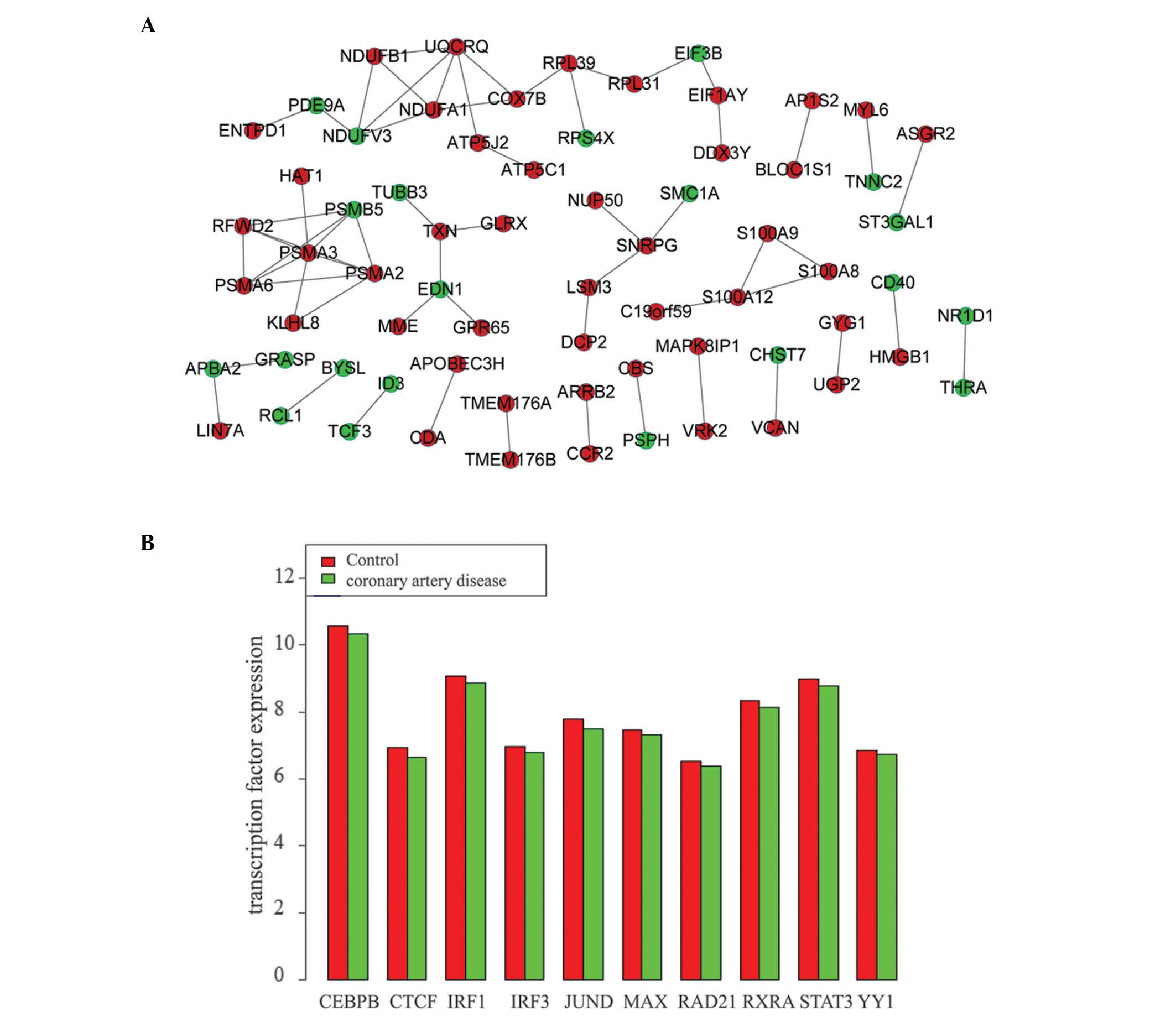
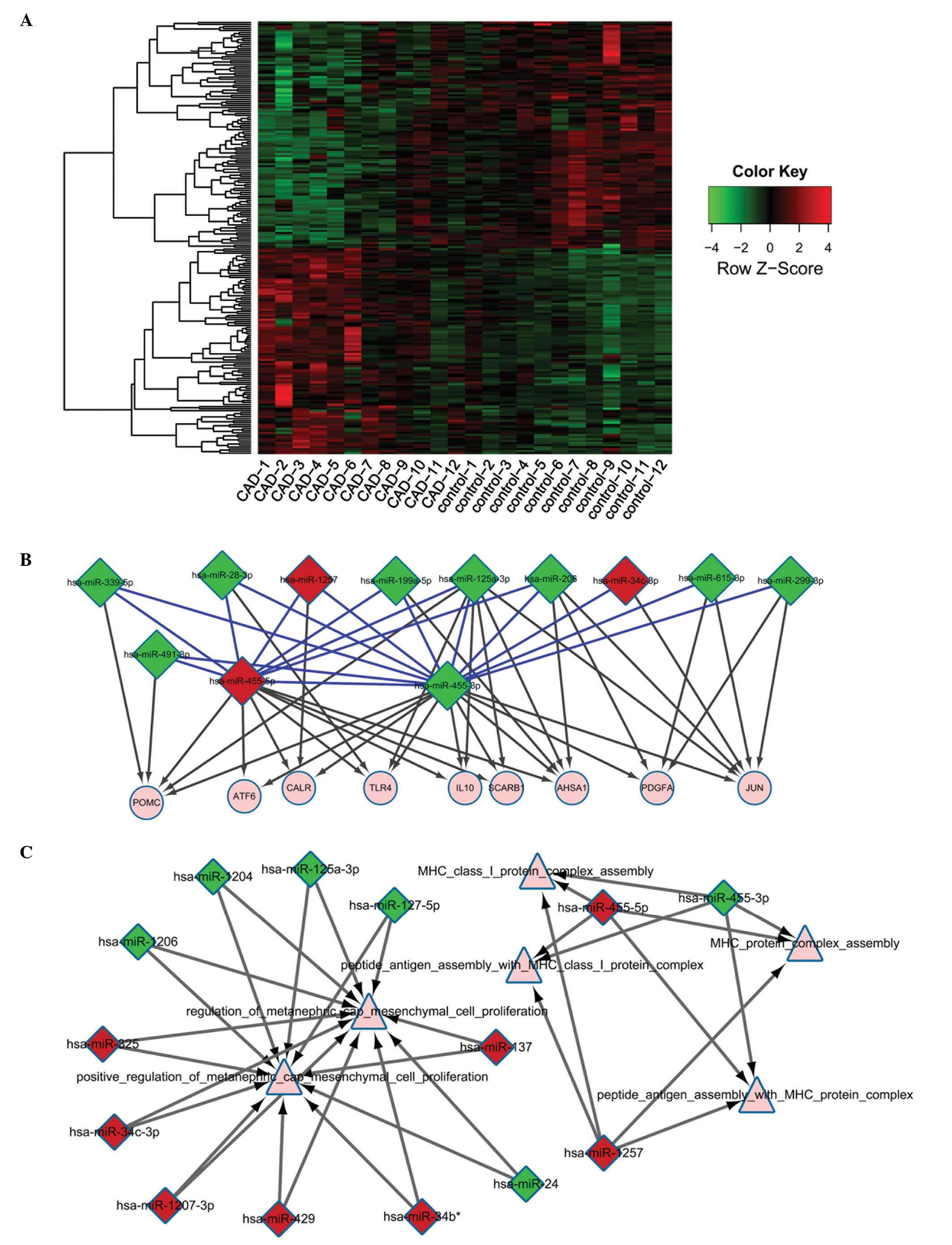
|
1
|
Sun JL, Huang WM, Guo JH, Li XY, Ma XL and
Wang CY: Relationship between myocardial bridging and coronary
arteriosclerosis. Cell Biochem Biophys. 65:485–489. 2013.
View Article : Google Scholar
|
|
2
|
Ahmadi N, Hajsadeghi F, Mirshkarlo HB,
Budoff M, Yehuda R and Ebrahimi R: Post-traumatic stress disorder,
coronary atherosclerosis, and mortality. Am J Cardiol. 108:29–33.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dean JC and Ilvento CC: Improved cancer
detection using computer-aided detection with diagnostic and
screening mammography: Prospective study of 104 cancers. AJR Am J
Roentgenol. 187:20–28. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Velazquez EJ, Lee KL, Deja MA, Jain A,
Sopko G, Marchenko A, Ali IS, Pohost G, Gradinac S, Abraham WT, et
al: STICH Investigators: Coronary-artery bypass surgery in patients
with left ventricular dysfunction. N Engl J Med. 364:1607–1616.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cooper-DeHoff RM, Gong Y, Handberg EM,
Bavry AA, Denardo SJ, Bakris GL and Pepine CJ: Tight blood pressure
control and cardiovascular outcomes among hypertensive patients
with diabetes and coronary artery disease. JAMA. 304:61–68. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Walldius G and Jungner I: The IL/apoA-I
ratio: A strong, new risk factor for cardiovascular disease and a
target for lipid-lowering therapy - a review of the evidence. J
Intern Med. 259:493–519. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hartford M, Wiklund O, Hultén LM, Persson
A, Karlsson T, Herlitz J, Hulthe J and Caidahl K: Interleukin-18 as
a predictor of future events in patients with acute coronary
syndromes. Arterioscler Thromb Vasc Biol. 30:2039–2046. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Contu R, Latronico MV and Condorelli G:
Circulating microRNAs as potential biomarkers of coronary artery
disease A promise to be fulfilled? Circ Res. 107:573–574. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tang Y, Zheng J, Sun Y, Wu Z, Liu Z and
Huang G: MicroRNA-1 regulates cardiomyocyte apoptosis by targeting
Bcl-2. Int Heart J. 50:377–387. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kim HW, Haider HK, Jiang S and Ashraf M:
Ischemic preconditioning augments survival of stem cells via
miR-210 expression by targeting caspase-8-associated protein 2. J
Biol Chem. 284:33161–33168. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Taurino C, Miller WH, McBride MW, McClure
JD, Khanin R, Moreno MU, Dymott JA, Delles C and Dominiczak AF:
Gene expression profiling in whole blood of patients with coronary
artery disease. Clin Sci (Lond). 119:335–343. 2010. View Article : Google Scholar
|
|
12
|
Liew CC, Ma J, Tang HC, Zheng R and
Dempsey AA: The peripheral blood transcriptome dynamically reflects
system wide biology: A potential diagnostic tool. J Lab Clin Med.
147:126–132. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Blankenberg S, Rupprecht HJ, Bickel C,
Peetz D, Hafner G, Tiret L and Meyer J: Circulating cell adhesion
molecules and death in patients with coronary artery disease.
Circulation. 104:1336–1342. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Diodati JG, Cannon RO III, Hussain N and
Quyyumi AA: Inhibitory effect of nitroglycerin and sodium
nitroprusside on platelet activation across the coronary
circulation in stable angina pectoris. Am J Cardiol. 75:443–448.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang X, Cheng X, Liu H, Zheng C, Rao K,
Fang Y, Zhou H and Xiong S: Identification of key genes and crucial
modules associated with coronary artery disease by bioinformatics
analysis. Int J Mol Med. 34:863–869. 2014.PubMed/NCBI
|
|
16
|
Chen F, Zhao X, Peng J, Bo L, Fan B and Ma
D: Integrated microRNA-mRNA analysis of coronary artery disease.
Mol Biol Rep. 41:5505–5511. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hua L, Yang Z and Liu H: Explore coronary
artery disease related microRNA clusters by combing single
nucleotide polymor phisms with microRNA microar ray. Fifth
International Conference on Biomedical Engineering and Infromatics
(BMEI). 1193–1197. 2012.
|
|
18
|
Elashoff MR, Wingrove JA, Beineke P,
Daniels SE, Tingley WG, Rosenberg S, Voros S, Kraus WE, Ginsburg
GS, Schwartz RS, et al: Development of a blood-based gene
expression algorithm for assessment of obstructive coronary artery
disease in non-diabetic patients. BMC Medical Genomics. 4:262011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sinnaeve PR, Donahue MP, Grass P, Seo D,
Vonderscher J, Chibout SD, Kraus WE, Sketch M Jr, Nelson C,
Ginsburg GS, et al: Gene expression patterns in peripheral blood
correlate with the extent of coronary artery disease. PLoS One.
4:e70372009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sondermeijer BM, Bakker A, Halliani A, de
Ronde MW, Marquart AA, Tijsen AJ, Mulders TA, Kok MG, Battjes S,
Maiwald S, et al: Platelets in patients with premature coronary
artery disease exhibit upregulation of miRNA340* and miRNA624*.
PLoS One. 6:e259462011. View Article : Google Scholar
|
|
21
|
Patterson TA, Lobenhofer EK,
Fulmer-Smentek SB, Collins PJ, Chu T-M, Bao W, Fang H, Kawasaki ES,
Hager J, Tikhonova IR, et al: Performance comparison of one-color
and two-color platforms within the MicroArray Quality Control
(MAQC) project. Nat Biotechnol. 24:1140–1150. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
McCall MN and Irizarry RA: Thawing frozen
robust multi-array analysis (fRMA). BMC Bioinformatics. 12:3692011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Efron B and Tibshirani R: On testing the
significance of sets of genes. Ann Appl Stat. 1:107–129. 2007.
View Article : Google Scholar
|
|
24
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization, and integrated discovery. Genome Biol. 4:32003.
View Article : Google Scholar
|
|
25
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Altermann E and Klaenhammer TR:
PathwayVoyager: Pathway mapping using the Kyoto Encyclopedia of
Genes and Genomes (KEGG) database. BMC Genomics. 6:602005.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res.
41(Database Issue): D808–D815. 2013. View Article : Google Scholar :
|
|
28
|
Spinelli L, Gambette P, Chapple CE,
Robisson B, Baudot A, Garreta H, Tichit L, Guénoche A and Brun C:
Clust&See: A Cytoscape plugin for the identification,
visualization and manipulation of network clusters. Biosystems.
113:91–95. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Weinmann AS and Farnham PJ: Identification
of unknown target genes of human transcription factors using
chromatin immunoprecipitation. Methods. 26:37–47. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Strauer BE, Brehm M, Zeus T, Bartsch T,
Schannwell C, Antke C, Sorg RV, Kögler G, Wernet P, Müller HW and
Köstering M: Regeneration of human infarcted heart muscle by
intracoronary autologous bone marrow cell transplantation in
chronic coronary artery disease: The IACT Study. J Am Coll Cardiol.
46:1651–1658. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar :
|
|
32
|
Xiao F, Zuo Z, Cai G, Kang S, Gao X and Li
T: miRecords: An integrated resource for microRNA-target
interactions. Nucleic Acids Res. 37(Database Issue): D105–D110.
2009. View Article : Google Scholar :
|
|
33
|
Dweep H, Sticht C, Pandey P and Gretz N:
miRWalk - database: Prediction of possible miRNA binding sites by
'walking' the genes of three genomes. J Biomed Inform. 44:839–847.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Davis AP, King BL, Mockus S, Murphy CG,
Saraceni-Richards C, Rosenstein M, Wiegers T and Mattingly CJ: The
comparative toxicogenomics database: Update 2011. Nucleic Acids
Res. 9(Database Issue): D1067–D1072. 2011. View Article : Google Scholar
|
|
35
|
Balthasar N, Coppari R, McMinn J, Liu SM,
Lee CE, Tang V, Kenny CD, McGovern RA, Chua SC Jr, Elmquist JK and
Lowell BB: Leptin receptor signaling in POMC neurons is required
for normal body weight homeostasis. Neuron. 42:983–991. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Schwartz MW, Seeley RJ, Woods SC, Weigle
DS, Campfield LA, Burn P and Baskin DG: Leptin increases
hypothalamic pro-opiomelanocortin mRNA expression in the rostral
arcuate nucleus. Diabetes. 46:2119–2123. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
McDaniel FK, Molden BM, Mohammad S and
Baldini G, McPike L, Narducci P, Granell S and Baldini G:
Constitutive cholesterol-dependent endocytosis of melanocortin-4
receptor (MC4R) is essential to maintain receptor responsiveness to
α-melanocyte-stimulating hormone (α-MSH). J Biol Chem.
287:21873–21890. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Logue J, Murray HM, Welsh P, Shepherd J,
Packard C, Macfarlane P, Cobbe S, Ford I and Sattar N: Obesity is
associated with fatal coronary heart disease independently of
traditional risk factors and deprivation. Heart. 97:564–568. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Vickers KC, Rye K-A and Tabet F: MicroRNAs
in the onset and development of cardiovascular disease. Clin Sci
(Lond). 126:183–194. 2014. View Article : Google Scholar
|
|
40
|
Cannon CP, Shah S, Dansky HM, Davidson M,
Brinton EA, Gotto AM Jr, Stepanavage M, Liu SX, Gibbons P, Ashraf
TB, et al: Determining the Efficacy and Tolerability Investigators:
Safety of anacetrapib in patients with or at high risk for coronary
heart disease. N Engl J Med. 363:2406–2415. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
O'Neill LA and Bowie AG: The family of
five: TIR-domain-containing adaptors in Toll-like receptor
signalling. Nat Rev Immunol. 7:353–364. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Otsui K, Inoue N, Kobayashi S, Shiraki R,
Honjo T, Takahashi M, Hirata K, Kawashima S and Yokoyama M:
Enhanced expression of TLR4 in smooth muscle cells in human
atherosclerotic coronary arteries. Heart Vessels. 22:416–422. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zeuke S, Ulmer AJ, Kusumoto S, Katus HA
and Heine H: TLR4-mediated inflammatory activation of human
coronary artery endothelial cells by LPS. Cardiovasc Res.
56:126–134. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Meder B, Keller A, Vogel B, Haas J,
Sedaghat-Hamedani F, Kayvanpour E, Just S, Borries A, Rudloff J,
Leidinger P, et al: MicroRNA signatures in total peripheral blood
as novel biomarkers for acute myocardial infarction. Basic Res
Cardiol. 106:13–23. 2011. View Article : Google Scholar
|
|
45
|
Nabel EG and Braunwald E: A tale of
coronary artery disease and myocardial infarction. N Engl J Med.
366:54–63. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Király R, Demény M and Fésüs L: Protein
transamidation by transglutaminase 2 in cells: A disputed
Ca2+-dependent action of a multifunctional protein. FEBS
J. 278:4717–4739. 2011. View Article : Google Scholar
|
|
47
|
Lee D, Oka T, Hunter B, Robinson A, Papp
S, Nakamura K, Srisakuldee W, Nickel BE, Light PE, Dyck JR, et al:
Calreticulin induces dilated cardiomyopathy. PLoS One.
8:e563872013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Basso C, Thiene G, Corrado D, Buja G,
Melacini P and Nava A: Hypertrophic cardiomyopathy and sudden death
in the young: Pathologic evidence of myocardial ischemia. Hum
Pathol. 31:988–998. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kamiński MJ, Kamińska M, Skorupa I,
Kazimierczyk R, Musiał WJ and Kamiński KA: In-silico identification
of cardiovascular disease-related SNPs affecting predicted microRNA
target sites. Pol Arch Med Wewn. 123:355–363. 2013.
|
|
50
|
Yamagata T, Nishida J, Tanaka S, Sakai R,
Mitani K, Yoshida M, Taniguchi T, Yazaki Y and Hirai H: A novel
interferon regulatory factor family transcription factor,
ICSAT/Pip/LSIRF, that negatively regulates the activity of
interferon-regulated genes. Mol Cell Biol. 16:1283–1294. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ridker PM, Rifai N, Stampfer MJ and
Hennekens CH: Plasma concentration of interleukin-6 and the risk of
future myocardial infarction among apparently healthy men.
Circulation. 101:1767–1772. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Guo M, Mao X, Ji Q, Lang M, Li S, Peng Y,
Zhou W, Xiong B and Zeng Q: Inhibition of IFN regulatory factor-1
downregulate Th1 cell function in patients with acute coronary
syndrome. J Clin Immunol. 30:241–252. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Fischer P and Hilfiker-Kleiner D: Survival
pathways in hypertrophy and heart failure: The gp130-STAT3 axis.
Basic Res Cardiol. 102:279–297. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhang W, Young AC, Imarai M, Nathenson SG
and Sacchettini JC: Crystal structure of the major
histocompatibility complex class I H-2Kb molecule containing a
single viral peptide: Implications for peptide binding and T-cell
receptor recognition. Proc Natl Acad Sci USA. 89:8403–8407. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Mor A, Planer D, Luboshits G, Afek A,
Metzger S, Chajek-Shaul T, Keren G and George J: Role of naturally
occurring CD4+ CD25+ regulatory T cells in
experimental atherosclerosis. Arterioscler Thromb Vasc Biol.
27:893–900. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Yilmaz A, Reiss C, Tantawi O, Weng A,
Stumpf C, Raaz D, Ludwig J, Berger T, Steinkasserer A, Daniel WG
and Garlichs CD: HMG-CoA reductase inhibitors suppress maturation
of human dendritic cells: New implications for atherosclerosis.
Atherosclerosis. 172:85–93. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Chen X, Du Y and Huang Z:
CD4+CD25+ Treg derived from hepatocellular
carcinoma mice inhibits tumor immunity. Immunol Lett. 148:83–89.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Döring Y, Manthey HD, Drechsler M, Lievens
D, Megens RT, Soehnlein O, Busch M, Manca M, Koenen RR, Pelisek J,
et al: Auto-antigenic protein-DNA complexes stimulate plasmacytoid
dendritic cells to promote atherosclerosis. Circulation.
125:1673–1683. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Rosano GM, Vitale C and Fragasso G:
Metabolic therapy for patients with diabetes mellitus and coronary
artery disease. Am J Cardiol. 98:14J–18J. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Fichtlscherer S, De Rosa S, Fox H,
Schwietz T, Fischer A, Liebetrau C, Weber M, Hamm CW, Roxe T, Röxe
T, Müller-Ardogan M, et al: Circulating microRNAs in patients with
coronary artery disease. Circ Res. 107:677–684. 2010. View Article : Google Scholar : PubMed/NCBI
|