Introduction
Breast cancer is one of the most common types of
malignancies among females worldwide and has become a serious
health burden for women with 1,380,000 new cases and 458,000 breast
cancer-related fatalities annually (1,2).
However, mechanisms underlying the onset and development of breast
cancer remain to be fully elucidated. Like other malignancies,
breast cancer development is the result of a combination of genetic
and environmental factors (3-6). The
most frequent type of genetic alteration, single nucleotide
polymorphisms (SNPs), are known to be important carcinogenesis and
cancer development (7,8). It is well known that breast cancer is
associated with SNPs in a number of genes, such as BCL-2 and XRCC3
(9,10).
Deleted in liver cancer 1 (DLC1), mapped to
chromosome 8p22, is an important tumor suppressor gene originally
identified to be deleted in hepatocellular carcinoma (HCC).
Recently, increased attention has been paid to its actions in other
types of malignancies, including breast, prostate, cervical and
lung (11) cancer. DLC1 can
regulate the structure of the actin cytoskeleton and inhibit cell
proliferation, migration, invasion and angiogenesis, which
predominantly depends on its homology to rat RhoGAP, which acts as
a catalyzing enzyme in the conversion of the active GTP-bound rho
complex to the inactive GDP-bound form (12-14).
Mutation screening of DLC1 was performed in 159 patients with
prostate cancer indicated 8 exonic and 23 intronic SNPs of DLC1;
however, no association with cancer was identified (15). Among these mutations, rs621554
(IVS19+108C>T) polymorphisms, a synonymous SNP located 108 bp
downstream of exon 19, was found to be correlated with HCC
susceptibility in patients with hepatitis B virus (16). However, little is known regarding
the influence of the DLC1 rs621554 polymorphism in breast cancer.
Thus, the present study aimed to determine the association between
the rs621554 polymorphism and breast cancer susceptibility,
clinicopathological variables and prognosis.
Materials and methods
Patients and samples
This study involved 453 patients who were diagnosed
with breast cancer and 330 healthy females who were recruited from
annual physical examinations between 2008 and 2012 at the Qilu
hospital of Shandong University (Jinan, China). The average age of
the case and control group was 50.00±11.40 and 48.23±7.93 years,
respectively, and there was significant difference in the age of
the two groups as determined by Student′s t-test (P=0.367). In
order to investigate the correlation between the DLC1 rs621554
polymorphism and changes in DLC1 mRNA levels, 20 tumor tissue
samples were obtained from the patient group during tumor
resection. To detect whether the rs621554 polymorphism is
associated with breast cancer prognosis, 245 patients with
long-term clinical follow-up were enrolled. The patients were
followed up every three months, and disease free survival and
overall survival were analyzed. Whole blood samples (200 µl)
were obtained from each individual in the case and control groups
and were stored at −80°C until use. The following
clinicopathological characteristics were determined from patient
notes: Age at diagnosis, pathological type, tumor size,
histological grade, lymph node status, as well as biological
markers, such as the presence of estrogen receptor (ER),
progesterone receptor (PR) and human epidermal growth factor
receptor 2 (HER2). This study was approved by ethical committee of
Shandong University. Written informed consent was signed by each
subject involved in the study.
DNA extraction
DNA was extracted from whole blood cells using the
TIANamp Genomic DNA kit (Tiangen, Beijing, China) according to the
manufacturer′s instructions. DNA concentration and purity were
measured using an ultraviolet spectrophotometer (GE Healthcare,
Pittsburgh, PA, USA). The extracted DNA samples were routinely
stored at −20°C.
SNP genotyping analysis
Cycling probes were synthesized by Takara
Biotechnologies (Dalian), Co. Ltd. (Dalian, China), and an allelic
discrimination assay [Cycleave PCR Core kit, Takara Biotechnologies
(Dalian), Co. Ltd] was performed using an ABI 7900HT thermal
cycler. The PCR amplification conditions were conducted according
to the manufacturer′s instructions: 95°C for 30 sec, 45 cycles of
95°C for 5 sec, 55°C for 10 sec, and 72°C for 25 sec. Several DNA
samples were randomly selected for DNA sequencing to confirm the
genotyping.
Reverse transcription-quantitative
polymerase chain reaction analysis (RT-qPCR)
Total RNA was extracted using TRIzol reagent [Takara
Biotechnologies (Dalian), Co. Ltd.]. The cDNA was reverse
transcribed from 1 µg total RNA using the PrimerScript RT
Reagent kit [Takara Biotechnologies (Dalian), Co. Ltd.]. mRNA
quantification was performed using SYBR PCR mix [Takara
Biotechnologies (Dalian), Co. Ltd.] on an ABI 7900HT platform
(Applied Biosystems, Foster City, CA, USA). Primer sequences were
as follows: Forward: 5′-GGGCTGCTTTTAACTCTGGTAAAG-3′ and reverse:
5′-CCATGGGTGGAATCATATTGG-3′ for glyceraldehyde 3-phosphate
dehydrogenase; and forward: 5′-CCAAACCCAAGACTACGGCTA-3′ and
reverse: 5′-GCTGTGCATACTGGGGGAA-3′ for DLC1. Primers were obtained
from Sangon Biological Engineering, Co., Ltd. (Shanghai, China). A
20-µl reaction system was used and contained 1X Cycleave PCR
reaction mixture, 0.4 µM cycling probe, 0.2 µM PCR
forward primer, 0.2 µM PCR reverse primer and 50 ng DNA
template. The cycling probe method was used, the CC genotype
presented strong fluorescence, TT genotype presented no
fluorescence and CT genotype present weak fluorescence (17). Sequence detection system software
1.2.1 (SDS1.2.1, Applied Biosystems) was used for analysis. The
experiment was repeated in triplicate to confirm the results.
Statistical analysis
SPSS 18.0 software (SPSS Inc., Chicago, IL, USA) was
used to analyze data in this study. The genotype and allele
frequencies were tested using the public statistical web-tool
http://www.oege.org/software/hwe-mr-calc.shtml for
Hardy-Weinburg equilibrium (HWE). P>0.05 was considered to
indicate non-deviation from HWE. Logistic regression models were
used to analyze the association between the rs621554 polymorphism
and breast cancer susceptibility, as well as the
clinicopathological characteristics. Differences in patient
survival rates were analyzed using the Kaplan-Meier method. The Cox
hazard regression model was used to analyze the independent
prognostic value of the rs621554 polymorphism. P<0.05 was
considered to indicate a statistically significantly
difference.
Results
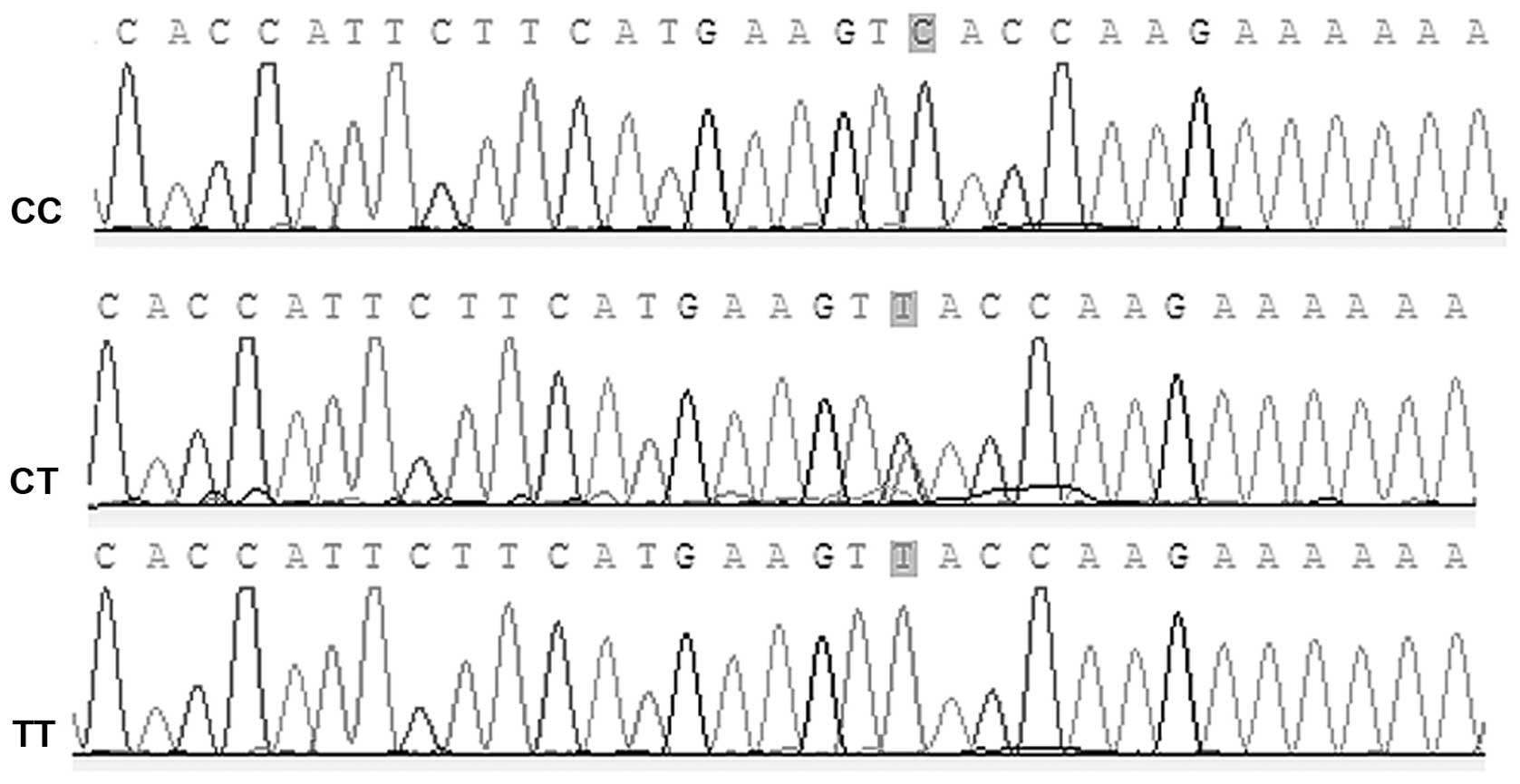
DNA sequencing shows complete reliability
of genotyping using cycling probe method
Ten DNA samples randomly selected from the patient
group were sent to Sangon Biological Engineering Co., Ltd. for DNA
sequencing. As shown in Fig. 1,
DNA sequencing showed consistent results, which suggest reliability
of the genotyping method.
rs621554 polymorphisms are associated
with breast cancer susceptibility
In total, 453 patients with breast cancer and 330
healthy females were involved in this study. Subjects from the two
groups were confirmed to meet HWE using the χ2 test. The
genotype and allele frequencies were tested using Hardy-Weinburg
equilibrium (HWE). χ2 test was performed to confirm the
representativeness of each group. P>0.05 was considered as
non-deviation from HWE. The χ2 result was 3.06 for the
patient group and 0.43 for control group, respectively
(P>0.05).
Table I shows a
significantly different distribution of rs621554 polymorphism in
the patient and control groups. A higher percentage of CT or CC
genotypes were observed in the patient group compared with the
control group (CT: 83.33 vs. 75.45%; CC: 81.12 vs. 67.07%,
respectively). Logistic regression analysis showed significant
differences between the two groups (P=0.032 and P=0.001,
respectively). Moreover, the CT and CC genotypes were associated
with a higher risk of breast cancer [Odds ratio (OR), 1.627; 95%
confidence interval (CI), 1.041–2.541; and OR, 2.109; 95% CI,
1.328–3.348, respectively]. Based on the above, our study indicated
that the C allele may be a risk factor for breast cancer
susceptibility. Accordingly, the CT and CC genotypes were taken
together and compared with the TT group.
 | Table IGenotype distribution in cancer cases
and controls. |
Table I
Genotype distribution in cancer cases
and controls.
| Genotype | No. of patients
| P-value | OR (95% CI) |
|---|
| Control (n=330) | Cancer (n=453) |
|---|
| TT | 54 | 44 | | |
| CT | 166 | 220 | 0.032 | 1.627 (1.041,
2.541) |
| CC | 110 | 189 | 0.001 | 2.109 (1.328,
3.348) |
Association between rs621554
polymorphisms and breast cancer clinicopathological variables
Table II shows the
association between the TT and CT+CC genotype and
clinicopathological characteristics of breast cancer patients,
including age at diagnosis, pathological type, tumor size,
histological grade, lymph node status, and biological markers, such
as ER, PR and HER2. It was demonstrated that the CT and CC genotype
were strongly associated with tumor size (P=0.035), lymph node
metastasis (P=0.047) and PR status (P=0.031). No significant
association was found between genotype and age at diagnosis,
pathological type, histological grade, and status of ER and
HER2.
 | Table IIAssociation between rs621554 and
clinicopathological variables. |
Table II
Association between rs621554 and
clinicopathological variables.
| Variable | Genotype
| P-value |
|---|
| TT | CC+CT |
|---|
| Age (years) | | | 0.561 |
| ≤45 | 20 | 148 | |
| >45 | 24 | 214 | |
| Pathological
type | | | 0.759 |
| Ductal | 37 | 296 | |
| Other | 7 | 64 | |
| Tumor size (cm) | | | 0.035 |
| ≤2 | 24 | 136 | |
| >2 | 15 | 175 | |
| Histological
grade | | | 0.242 |
| Low | 23 | 165 | |
| High | 6 | 75 | |
| Lymph node
metastasis | | | 0.047 |
| No | 28 | 170 | |
| Yes | 14 | 167 | |
| ER | | | 0.359 |
| Negative | 26 | 178 | |
| Positive | 16 | 149 | |
| PR | | | 0.031 |
| Negative | 12 | 151 | |
| Positive | 30 | 176 | |
| HER2 | | | 0.476 |
| Negative | 38 | 283 | |
| Positive | 4 | 44 | |
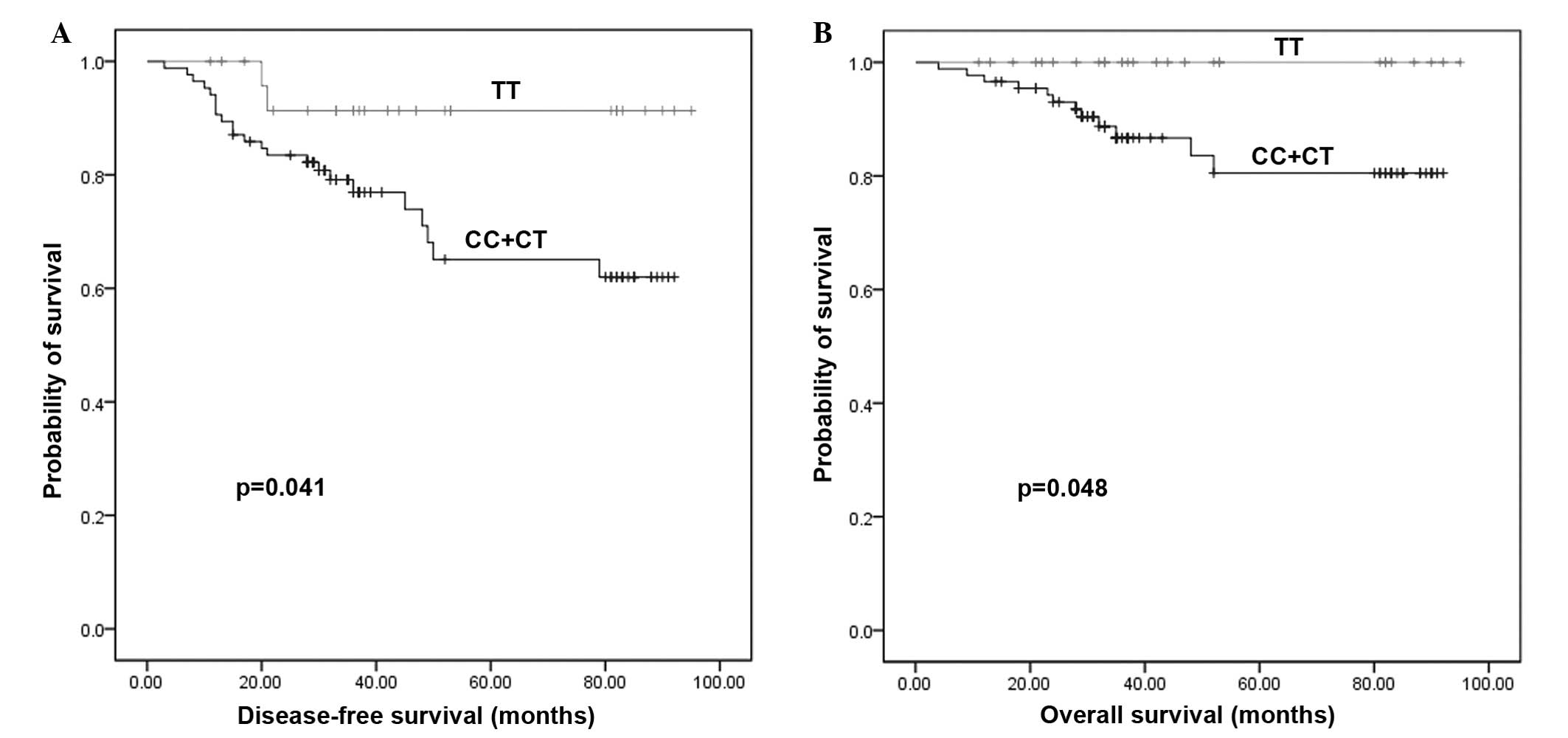
rs621554 polymorphisms are correlated
with breast cancer prognosis
The association between rs621554 polymorphisms and
prognosis was also analyzed using the Kaplan-Meier method. As shown
in Fig. 2, compared with patients
with the TT genotype, patients with CT and CC genotypes had shorter
disease-free survival (P=0.041) and overall survival (P=0.048)
rates. Additionally, COX hazard logistic regression analysis was
performed to demonstrate that rs621554 polymorphisms are not an
independent prognostic factor in breast cancer, with P=0.192 for
disease-free survival and P=0.131 for overall survival (data not
shown).
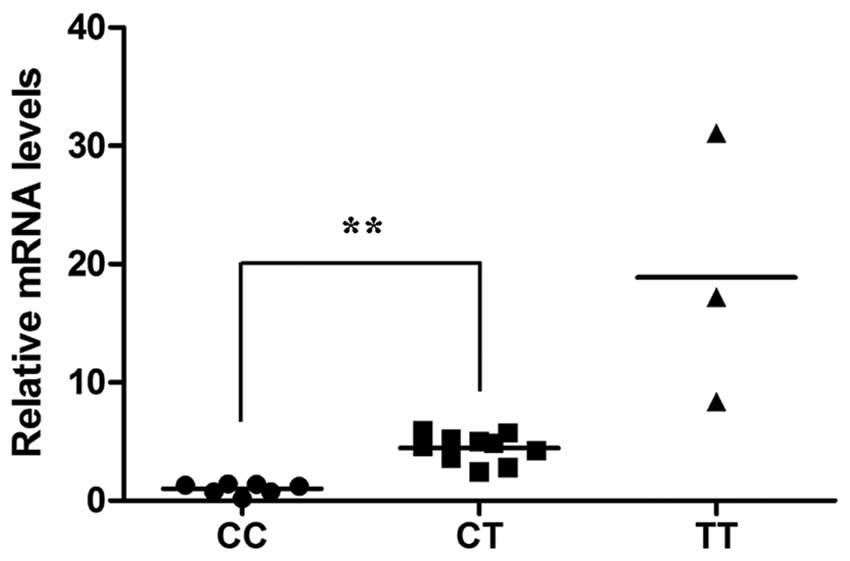
rs621554 polymorphisms are associated
with the mRNA level of DLC1 in vivo
The possible correlation between the rs621554
polymorphism and the mRNA level of DLC1 was also investigated. In
total, 20 samples of breast cancer tissue were taken for analysis
and the result is shown in Fig. 3.
The relative mRNA level of DLC1 was 1.000±0.451 for the CC
genotype, 4.452±1.18 for the CT genotype, and 18.886±11.433 for the
TT genotype. Significant differences were identified in the mRNA
level between patients with the CC and CT genotype (P<0.001),
and no significance was found between the CC and TT genotypes
(P=0.113) or the CT and TT genotypes (P=0.160).
Discussion
As with other types of malignancies, genetic and
environmental factors are involved in the development of breast
cancer. SNPs are the most frequent gene mutation, and certain SNPs
have been shown to result in a higher risk of cancer (18). Since the 6q25.1 rs2046210 was first
reported by Zheng et al (19) as a novel SNP associated with breast
cancer risk, increased attention has been paid to cancer-associated
SNPs with the aim of identifying sensitive genetic markers to
assess cancer risk and to monitor therapeutic response.
The human DLC1 gene is a tumor suppressor that acts
in a Rho-dependent manner, since the protein it encodes is highly
homologous to the rat p122-RhoGAP (20,21).
It is found to be crucial in regulating the structure of the actin
cytoskeleton, and inhibit cell proliferation, migration, invasion
and angio-genesis (12-14). It has been shown to be
downregulated or absent in a number of different types of cancer
(22). According to previous
studies, genetic variants widely exist in DLC1 and may influence
the antitumor effect of DLC1 (16,23,24).
rs621554, located in intron 19, is a synonymous SNP in DLC1, which
is associated with HCC (16).
Nevertheless, whether the rs621554 polymorphism is associated with
breast cancer remains to be determined.
In the present study, the CT and CC genotypes (CT+CC
genotype) of rs621554 DLC1 were shown to be associated with a
significantly higher risk of breast cancer compared with that of
the TT genotype. It appears that the T allele of rs621554 DLC1 is
protective against breast cancer. Since clinocopatho-logical
characteristics are vital in evaluating cancer prognosis their
correlation with the rs621554 polymorphism was investigated, and it
was demonstrated that the CT+CC genotype was associated with larger
tumor size, lymph node metastasis and negative PR status, but was
not associated with ER or HER2 status. This may due to the limited
number of samples, which suggests that a larger number of cases is
required in a further study. In addition, it was demonstrated that
the CT+CC genotype was a prognostic indicator for disease-free
survival and overall survival in breast cancer, but a COX hazard
regression analysis indicated that, with the limited sample size,
CT+CC genotype was not shown to be an independent indicator.
Despite this, however, a conclusion can be made that the CT+CC
genotype is associated with breast cancer susceptibility,
development and prognosis.
Next, in order to determine the possible correlation
between the rs621554 polymorphism and DLC1 expression, DLC1 mRNA
expression was detected. It was demonstrated that it is higher in
patients with the CT genotype compared with the CC genotype,
suggesting the protective effect of the T allele. However, the DLC1
expression level in patients with the TT genotype is not
significantly different compared with that in CT and CC group. This
phenomenon may due to small number patients with the TT genotype
(only 3 samples) and large standard error. It was hypothesized that
the change in the mRNA level of DLC1 due to the SNP may be
responsible for the development and progression of breast cancer.
The specific mechanism underlying this effect may involve
alterations in the binding site of transcriptional factors
(25), since rs621554 polymorphism
is located in the intron of DLC1, where a variety of cis elements
are involved. However, further investigation is required to confirm
this.
In conclusion, the results demonstrated that
individuals with the CT and CC genotype of the DLC1 rs621554
polymorphism are more susceptible to breast cancer compared with
those with the TT genotype and the two genotypes together are
associated with larger tumor size, lymph node metastasis and
negative PR. In addition, the CT or CC genotype may be associated
with a poor prognosis. Further research is required to determine
the underlying molecular mechanisms with larger patient cohorts to
further confirm rs621554 polymorphism as a clinical biomarker for
early diagnosis and prognosis of breast cancer.
Abbreviations:
|
DLC1
|
deleted in liver cancer 1
|
|
SNPs
|
single nucleotide polymorphisms
|
|
HWE
|
Hardy-Weinburg equilibrium
|
Acknowledgments
This study was supported by the National Natural
Science Foundation of China (grant nos. 81272903 and 81172529); the
Shandong Science and Technology Development Plan (grant no.
2013GRC31801) and the Special Support Plan for National High Level
Talents (Ten Thousand Talents Program) to Dr Qifeng Yang.
References
|
1
|
Basu S, Nachat-Kappes R, Caldefie-Chézet F
and Vasson MP: Eicosanoids and adipokines in breast cancer: from
molecular mechanisms to clinical considerations. Antioxid Redox
Signal. 18:323–360. 2013. View Article : Google Scholar
|
|
2
|
Bray F, Jemal A, Grey N, Ferlay J and
Forman D: Global cancer transitions according to the Human
Development Index (2008–2030): a population-based study. Lancet
Oncol. 13:790–801. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Børresen AL: Role of genetic factors in
breast cancer susceptibility. Acta Oncol. 31:151–155. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bishop DT: The involvement of genetic
factors in susceptibility to breast cancer. Eur J Cancer Prev.
2(Suppl 3): 125–130. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mavaddat N, Antoniou AC, Easton DF and
Garcia-Closas M: Genetic susceptibility to breast cancer. Molr
Oncol. 4:174–191. 2010. View Article : Google Scholar
|
|
6
|
Nickels S, Truong T, Hein R, et al:
Evidence of gene-environment interactions between common breast
cancer susceptibility loci and established environmental risk
factors. PLoS Genet. 9:e10032842013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Czerska K, Nawara M and Bal J: I. Single
nucleotide polymorphism in human genetic analyses. Med Wieku
Rozwoj. 7:531–546. 2003.In Polish.
|
|
8
|
Erichsen HC and Chanock SJ: SNPs in cancer
research and treatment. Br J Cancer. 90:747–751. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang N, Li X, Tao K, et al: BCL-2 (-938C
> A) polymorphism is associated with breast cancer
susceptibility. BMC Med Genet. 12:482011. View Article : Google Scholar :
|
|
10
|
Romanowicz-Makowska H, Bryś M, Forma E, et
al: Single nucleotide polymorphism (SNP) Thr241Met in the XRCC3
gene and breast cancer risk in Polish women. Pol J Pathol.
63:121–125. 2012.PubMed/NCBI
|
|
11
|
Popescu NC and Goodison S: Deleted in
liver cancer-1 (DLC1): an emerging metastasis suppressor gene. Mol
Diagn Ther. 18:293–302. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yamaga M, Kawai K, Kiyota M, Homma Y and
Yagisawa H: Recruitment and activation of phospholipase C
(PLC)-delta1 in lipid rafts by muscarinic stimulation of PC12
cells: contribution of p122RhoGAP/DLC1, a tumor-suppressing
PLCdelta1 binding protein. Adv Enzyme Regul. 48:41–54. 2008.
View Article : Google Scholar
|
|
13
|
Shih YP, Liao YC, Lin Y and Lo SH: DLC1
negatively regulates angiogenesis in a paracrine fashion. Cancer
Res. 70:8270–8275. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhou X, Thorgeirsson SS and Popescu NC:
Restoration of DLC-1 gene expression induces apoptosis and inhibits
both cell growth and tumorigenicity in human hepatocellular
carcinoma cells. Oncogene. 23:1308–1313. 2004. View Article : Google Scholar
|
|
15
|
Zheng SL, Mychaleckyj JC, Hawkins GA, et
al: Evaluation of DLC1 as a prostate cancer susceptibility gene:
mutation screen and association study. Mutat Res. 528:45–53. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dong X, Zhou G, Zhai Y, et al: Association
of DLC1 gene polymorphism with susceptibility to hepatocellular
carcinoma in Chinese hepatitis B virus carriers. Cancer
Epidemiology. 33:265–270. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Suzuki Y, Saito R, Zaraket H, Dapat C,
Caperig-Dapat I and Suzuki H: Rapid and specific detection of
amantadine-resistant influenza A viruses with a Ser31Asn mutation
by the cycling probe method. J Clin Microbiol. 48:57–63. 2010.
View Article : Google Scholar :
|
|
18
|
Temin HM: Evolution of cancer genes as a
mutation-driven process. Cancer Res. 48:1697–1701. 1988.PubMed/NCBI
|
|
19
|
Zheng W, Long J, Gao YT, et al:
Genome-wide association study identifies a new breast cancer
susceptibility locus at 6q25.1. Nat Genet. 41:324–328. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yuan BZ, Miller MJ, Keck CL, Zimonjic DB,
Thorgeirsson SS and Popescu NC: Cloning, characterization, and
chromosomal localization of a gene frequently deleted in human
liver cancer (DLC-1) homologous to rat RhoGAP. Cancer Res.
58:2196–2199. 1998.PubMed/NCBI
|
|
21
|
Zhou X, Zimonjic DB, Park SW, Yang XY,
Durkin ME and Popescu NC: DLC1 suppresses distant dissemination of
human hepatocellular carcinoma cells in nude mice through reduction
of RhoA GTPase activity, actin cytoskeletal disruption and
down-regulation of genes involved in metastasis. Int J Oncol.
32:1285–1291. 2008.PubMed/NCBI
|
|
22
|
Ullmannova V and Popescu NC: Expression
profile of the tumor suppressor genes DLC-1 and DLC-2 in solid
tumors. Int J Oncol. 29:1127–1132. 2006.PubMed/NCBI
|
|
23
|
Xie CR, Sun HG, Sun Y, et al: Significance
of genetic variants in DLC1 and their association with
hepatocellular carcinoma. Mol Med Rep. 12:4203–4209.
2015.PubMed/NCBI
|
|
24
|
Dai X, Li L, Liu X, Hu W, Yang Y and Bai
Z: Cooperation of DLC1 and CDK6 affects breast cancer clinical
outcome. G3 (Bethesda). 5:81–91. 2014.
|
|
25
|
Lee PH and Shatkay H: F-SNP:
computationally predicted functional SNPs for disease association
studies. Nucleic Acids Res. 36:D820–D824. 2008. View Article : Google Scholar :
|