Introduction
Psoriasis is an inherent and inflammatory skin
disease with scaly erythematous lesions, affecting 2–3% of the
Caucasian population (1). The
scales are a result of a hyperproliferative epidermis, as well as
parakeratosis, and the highly mitotic rate of psoriatic
keratinocytes results in a thickened epidermis. Complex pathogenic
mechanisms associated with genetic expression variances and
different immune responses result in psoriasis aggravation
(2).
In addition to hyperproliferation, the psoriatic
keratinocytes exhibit an abnormal resistance to apoptosis.
Deceleration in keratinocyte apoptosis is the most significant
pathological change in psoriasis (3). The decrease in apoptosis, which
causes imbalances in skin homeostasis, may be associated with the
induction of psoriatic hyperplasia (3). Multiple factors have been identified
to contribute to anti-apoptosis in psoriatic keratinocytes,
including the activation of mitogenic signaling pathways and the
inactivation of certain apoptotic molecules (4,5). In
the present study, various PageRank hub genes were identified to be
associated with anti-apoptosis, as well as with the regulation of
apoptosis in psoriasis. Furthermore, the current study identified
that the target genes in anti-apoptosis and apoptosis regulation
are tightly associated with the ESR1 gene. The ESR1
gene encodes an estrogen receptor α (ERα). The estrogen stimulates
a rapid relocation of ERα to induce high mitochondrial gene
expression (6). It has been
clearly demonstrated that activation of the mitochondrial signaling
pathway, as well as the extrinsic apoptosis signaling pathway, is
key in the process of many forms of programmed cell death (5). Thus, it was hypothesized in the
present study that ERα may exert a protective role against
psoriatic proliferation.
The cytokine related Toll-like receptor (TLR)
signaling pathway, a classical signaling pathway, is considered to
be important in the progression of psoriasis (7). However, based on the analyses of the
present study, the pathways in cancer was the most significant
pathway that correlated with psoriasis, demonstrating a more
significant association than the TLR signaling pathway. Pathways in
cancer affect tumor proliferation by protecting the tumor from
cytokine-induced apoptosis. The current results suggest that the
activation of pathways in cancer may inhibit psoriatic keratinocyte
apoptosis.
To analyze the disease-associated genes and gene
functions, the differentially expressed genes (DEGs) associated
with psoriasis were screened and compared with healthy controls
from the DNA microarray database. A gene interaction network was
generated for topological analysis of relevant genes associated
with psoriasis using PageRank and the gene enrichment functions
were subsequently evaluated. The top-ranked PageRank hub gene,
ESR1, which is down-regulated in psoriasis, exhibited
binding sites enriched with anti-apoptotic genes that provides
support for the negative association between estrogen and
apoptosis. The current results demonstrate that reduced levels of
ERα expression result in the anti-apoptotic behavior of psoriatic
keratinocytes by activating anti-apoptotic gene expression.
Pathways in cancer may protect the psoriatic cell from apoptosis by
inhibiting ESR1 expression. The present study provides
evidence regarding ESR1 gene function and demonstrates that
the interaction with anti-apoptotic genes is involved in the
underlying biological mechanisms of apoptosis resistance in
psoriasis.
Materials and methods
Affymetrix microarray data and
preprocessing
The microarray gene expression profiles were
extracted from the Gene Expression Omnibus (GEO) database (GEO
accession no. GSE13355; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE13355)
(8,9), which contained gene expression
profiles for 58 psoriatic patients and 64 healthy control subjects.
All samples were run on GeneChip® Human Genome U133 Plus
2.0 Array (Affymetrix, Inc., Santa Clara, CA, USA), microarrays
containing >54,000 gene probes. The raw data were processed
using the Robust Multichip Average (RMA) method (5). The expression values were adjusted
using RMA expression values (on the log scale) to account for batch
and gender effects.
DEG identification
DEGs between psoriasis and normal skin were
identified using the limma R package (10). The raw P-value was corrected by the
Benjamin and Hochberg method to circumvent the multi-test bias
(11). Finally, the false
discovery rate (FDR) <0.01 and |log fold change (FC)| >1 were
used as the cut-off for DEG identification.
Interaction network construction of
DEGs
A gene interaction network was generated by
mapping all the DEGs in the Search Tool for the Retrieval of
Interacting Genes (STRING) database, which is a database for
predicting functional associations between proteins (12). The interactions include direct
(physical) and indirect (functional) associations, and all of the
associations are derived from four sources: Genomic context,
high-throughput experiments, coexpression (conserved) and previous
knowledge (13). All of the
associating pairs were selected based on a combined probabilistic
confidence score.
Identification of top-ranked hub genes
using Google PageRank score
In order to identify hub genes (key genes), the
PageRank score was calculated in all DEGs. The PageRank analysis
was initially applied to Google (the web search engine) to
identifying significant web pages (14–16)
and has been used during the robust analysis of protein networks to
identify important nodes (17).
Unlike simply calculating the degree of each gene, the PageRank
score measures the importance or popularity of a gene based solely
on the interaction (link) structure of the interaction network. It
selects the genes that exhibit a high degree, whilst also
maintaining the important low-degree genes, which link to other
important genes in the protein-protein interaction network. It is
expected that the genes with more interaction links, especially
those from important genes, are given higher PageRank scores.
Gene-annotation enrichment analysis using
the Database for Annotation, Visualization and Integrated Discovery
(David)
DAVID software (Laboratory of Human Retrovirology
and Immunoinformatics, Clinical Services Program; Leidos Biomedical
Research, Inc., Frederick, MD, USA) (18,19),
a gene-annotation enrichment analysis tool, adopts a common core
strategy to systematically map a hubs gene list to the associated
biological annotation [for example, gene ontology (GO) terms or
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways], and then
statistically highlight the most overrepresented (enriched)
biological annotation out of thousands of linked terms and
contents. In the present analysis, GO and KEGG pathway annotations
were integrated to evaluate the function enrichment of the hub
genes.
Statistical analysis
DAVID software (18,19)
was used for all statistical analyses, and GO terms and KEGG
pathways were selected using a standard parameter setting
(FDR<0.05 and P<0.1). All the known human genes were used as
the background for Fisher's exact test, and a P-value was
calculated and the Bonferroni correction was applied.
Results
DEG analysis
After extracting the gene expression data from the
public dataset, GSE13355, the DEGs between psoriasis lesions and
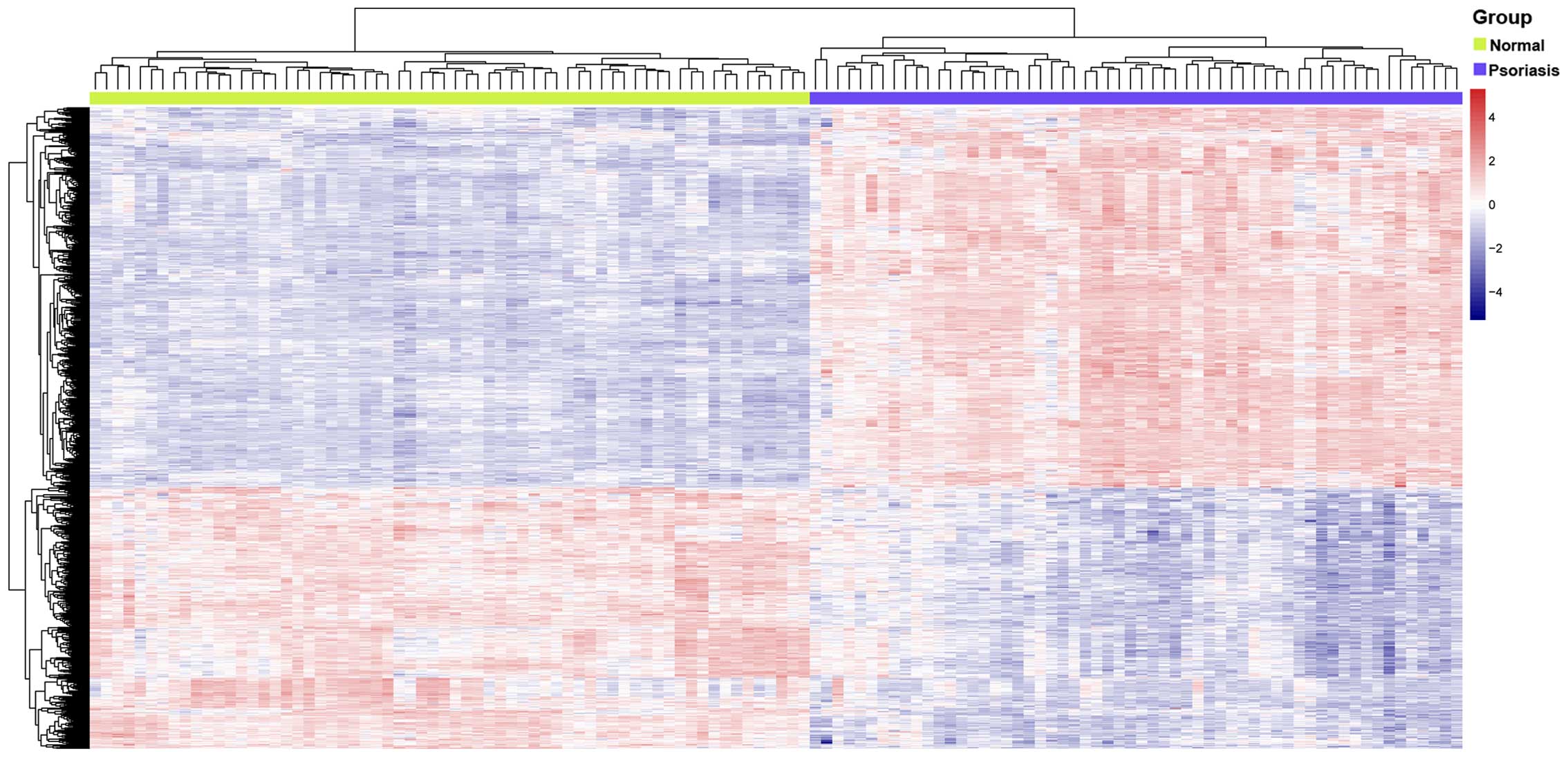
normal skin were identified. DEGs (n=927; 449 upregulated and 478
downregulated) were selected (FDR <0.01 and |logFC| >1) and a
heat map of the DEG expression level between the two groups is
presented in Fig. 1.
Construction and topological analysis of
an interaction network
The interaction network was constructed using STRING
and the DEGs that had been identified. In total, 640 genes were
identified and the interaction network of the DEGs is presented in
Fig. 2. Based on the network
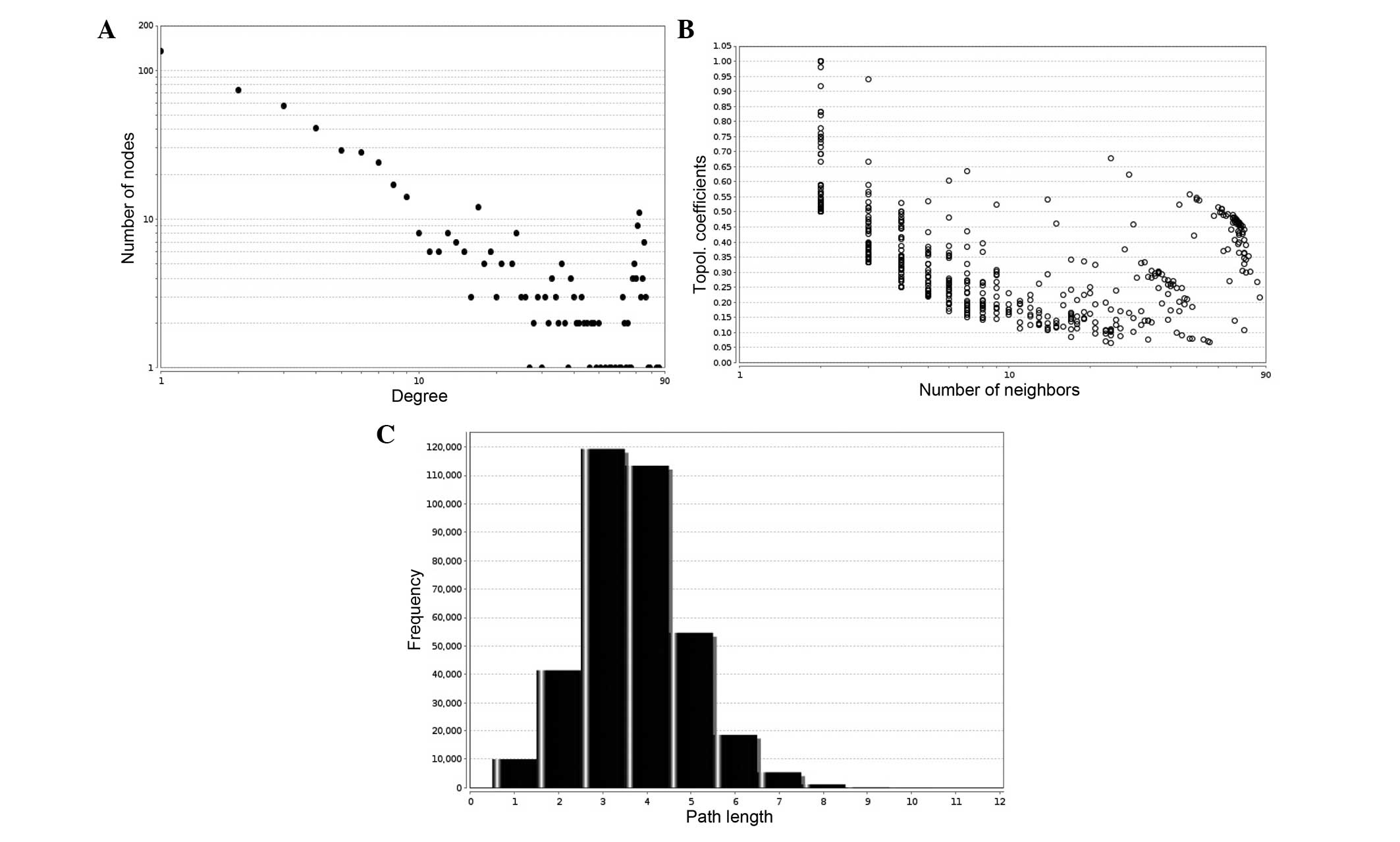
topological analysis, it was found that the protein interaction
network exhibited scale-free attributes. Three network topologies
were processed using statistical analyses and are presented in
Fig. 3. Fig. 3A shows the node degree distribution
in the network, the x-axis represents the node degrees (the number
of direct connections the node has with other nodes) and the y-axis
represents the number of nodes with the different degrees. The node
degree distribution indicates that both loosely and closely
connected nodes exist in the network. Fig. 3B demonstrates the clustering
coefficient distribution in the network, and the clustering
coefficient displays the aggregation degree of nodes; the
distribution range of the clustering coefficient is between 0 and
1. Fig. 3C shows the path length
of the network, indicating that the predominant path length was
~3–4. Subsequent to the analysis of the three topological property
parameters of the interaction network, the present study was able
to demonstrate the small-world effect of the network, which is
subject to scale-free property (20).
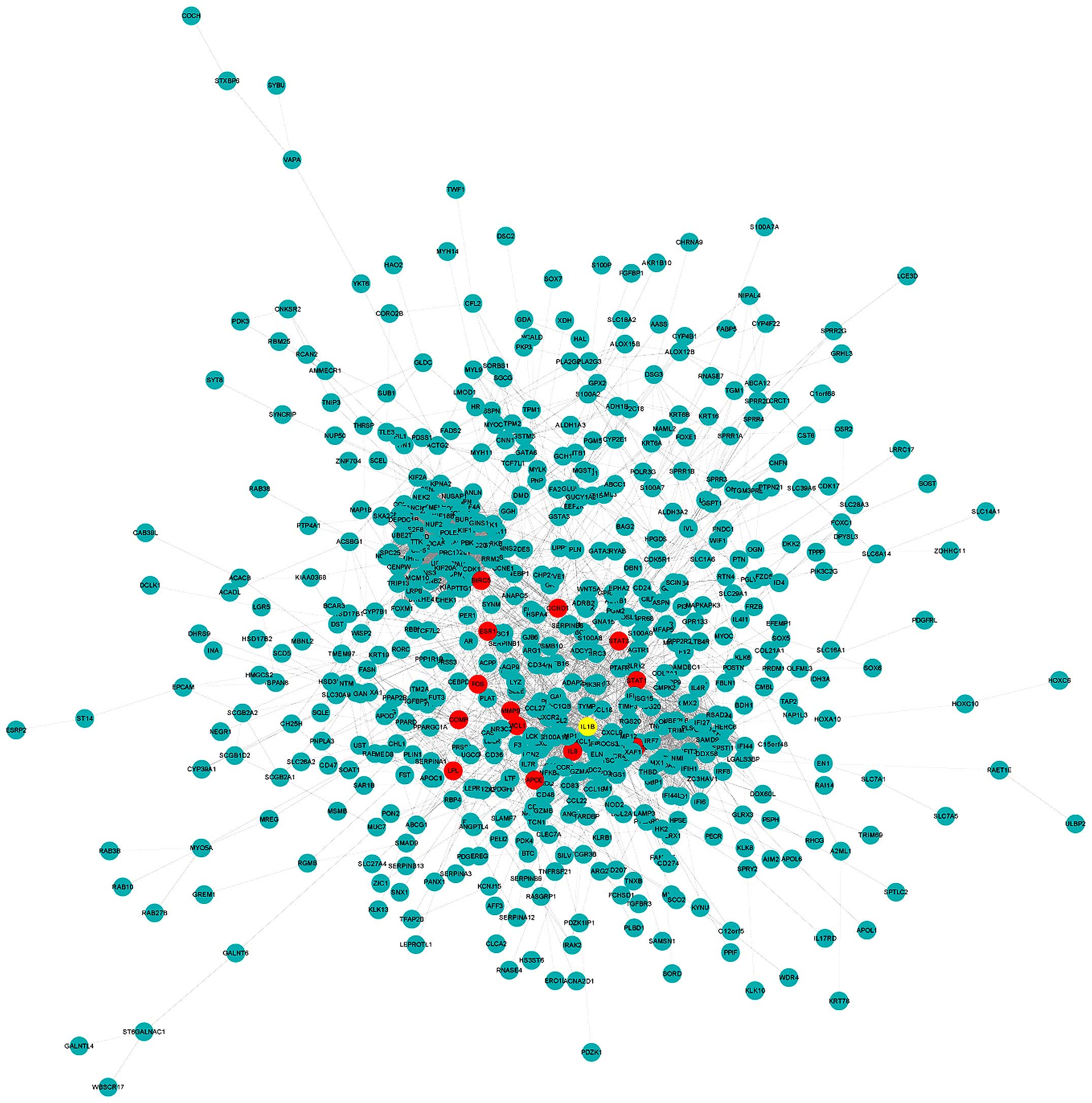 | Figure 2Interaction network of the
differentially expressed genes. The yellow-colored node is the top
hub gene, ESR1, with highest PageRank score. The 13 nodes
highlighted red are the hub genes with the next highest PageRank
scores, including IL-1B, STAT1, IL-8,
CXCL10, FOS, MMP9, STAT3, APOE,
CCND1, COMP, MCL1, LPL and
BIRC5. IL-1B, interleukin-1β; STAT1, signal transducer and
activator of transcription-1; IL-8, interleukin-8; CXCL10,
chemokine (C-X-C motif) ligand 10; FOS, FBJ murine osteosarcoma
viral oncogene homolog; MMP9, matrix metalloproteinase-9; STAT3,
signal transducer and activator of transcription-3; APOE,
apolipoprotein E; CCDN1, cyclin D1; COMP, cartilage oligomeric
matrix protein; MCL1, myeloid cell leukemia-1; LPL, lipoprotein
lipase; BIRC5, baculoviral IAP repeat containing-5. |
PageRank hub genes and enrichment
analysis
PageRank analysis was adopted to select the most
popular and important genes within the interaction network. A high
PageRank score is associated with a high degree. According to
analysis of the PageRank score, 14 genes with the highest PageRank
scores were selected as the top hub genes (Table I). Thirteen of the top 14 hub genes
are highlighted in red and ESR1, identified as the top hub
gene, was highlighted in yellow in the interaction network
(Fig. 2). ESR1,
representing high relativity with psoriasis, demonstrates a
downregulated level of expression in Table I. The other 13 of the top hub
genes, demonstrating various levels of expression, are presented in
Table I.
 | Table IPageRank, degree distribution and the
FC of the top 14 nodes in the interaction network. |
Table I
PageRank, degree distribution and the
FC of the top 14 nodes in the interaction network.
| Gene name | Page-rank
score | Degree
distribution | logFC |
|---|
| ESR1 | 0.007521607 | 56 | −1.149321612 |
| IL-1B | 0.007016809 | 55 | 1.097904425 |
| STAT1 | 0.006989954 | 75 | 2.906284146 |
| IL-8 | 0.006069015 | 53 | 4.192048124 |
| CXCL10 | 0.00604473 | 69 | 2.828564993 |
| FOS | 0.006031463 | 48 | −1.125965959 |
| MMP9 | 0.005700515 | 47 | 1.565431608 |
| STAT3 | 0.00500142 | 44 | 1.102377065 |
| APOE | 0.004996554 | 33 | −1.088425518 |
| CCND1 | 0.0048065 | 42 | −1.00144553 |
| COMP | 0.00457533 | 23 | 1.397789198 |
| MCL1 | 0.004510315 | 24 | 1.207643197 |
| LPL | 0.004289353 | 26 | −1.162938926 |
| BIRC5 | 0.004181229 | 86 | 1.746154734 |
To gain further insight into the functions of the
top 14 hub genes, the enrichment of GO functions and KEGG pathway
analysis were performed using the DAVID tool. The significant GO
enrichment terms are presented in Table II. The function of anti-apoptosis
was confirmed to be highly associated with psoriasis. Six of the
top hub genes [ESR1, apolipoprotein E (APOE), myeloid
cell leukemia-1 (MCL1), cartilage oligomeric matrix protein
(COMP), baculoviral IAP repeat containing-5 (BIRC5)
and interleukin-1β (IL-1B)] were identified to be involved
in the anti-apoptosis effects of psoriasis (Table II). The other functions, such as
regulation of apoptosis, programmed cell death and cell death, also
represent the association with psoriasis. The same eight top hub
genes [ESR1, APOE, MCL1, COMP,
BIRC5, IL-1B, matrix metalloproteinase-9
(MMP9) and signal transducer and activator of
transcription-1 (STAT1)] were observed to be correlated with
regulation of apoptosis, programmed cell death and cell death
(Table II).
 | Table IIGene ontology enrichment terms in the
top-ranked 14 hub genes. |
Table II
Gene ontology enrichment terms in the
top-ranked 14 hub genes.
| Term | Count | FDR | Genes |
|---|
| Anti-apoptosis | 6 | 0.001376778 | MCL1, APOE,
COMP, ESR1, IL-1B, BIRC5 |
| Response to organic
substance | 8 | 0.002326258 | FOS, CCND1,
MCL1, APOE, ESR1, IL-1B, STAT1, STAT3 |
| Regulation of
apoptosis | 8 | 0.004836793 | MCL1, APOE,
MMP9, COMP, ESR1, IL-1B, BIRC5, STAT1 |
| Regulation of
programmed cell death | 8 | 0.005168359 | MCL1, APOE,
MMP9, COMP, ESR1, IL-1B, BIRC5, STAT1 |
| Regulation of cell
death | 8 | 0.005297534 | MCL1, APOE,
MMP9, COMP, ESR1, IL-1B, BIRC5, STAT1 |
| Response to organic
cyclic substance | 5 | 0.006196132 | FOS, CCND1,
IL-1B, STAT1, STAT3 |
| Negative regulation
of apoptosis | 6 | 0.019556803 | MCL1, APOE,
COMP, ESR1, IL-1B, BIRC5 |
| Negative regulation
of programmed cell death | 6 | 0.020933116 | MCL1, APOE,
COMP, ESR1, IL-1B, BIRC5 |
| Negative regulation
of cell death | 6 | 0.021217271 | MCL1, APOE,
COMP, ESR1, IL-1B, BIRC5 |
| Pathways in
cancer | 7 | 0.022194793 | FOS, CCND1,
IL-8, MMP9, BIRC5, STAT1, STAT3 |
| Response to hormone
stimulus | 6 | 0.023292142 | FOS, CCND1,
ESR1, IL-1B, STAT1, STAT3 |
| Toll-like receptor
signaling pathway | 6 | 0.030280341 | FOS, CCND1,
MCL1, ESR1, BIRC5, STAT1 |
| Response to
endogenous stimulus | 6 | 0.03749322 | FOS, CCND1,
ESR1, IL-1B, STAT1, STAT3 |
| Response to steroid
hormone stimulus | 5 | 0.038520418 | FOS, CCND1,
ESR1, IL-1B, STAT3 |
In addition, the ESR1 topological neighbors
were selected from the interaction network; 56 pairs that directly
interacted with the ESR1 gene are shown in Fig. 4, including nine top-ranked hub
genes [APOE, BIRC5, cyclin D1 (CCDN1), FBJ
murine osteosarcoma viral oncogene homolog (FOS),
interleukin-8 (IL-8), lipoprotein lipase (LPL),
MMP9, STAT1, signal transducer and activator of
transcription-3 (STAT3)] highlighted in red. The five genes
(APOE, MCL1, COMP, BIRC5 and
IL-1B) with anti-apoptotic function were directly or
indirectly associated with the ESR1 gene (Fig. 4). Three genes were indirectly
linked with the ESR1 gene by binding to the ESR1 gene
topological neigbors, particularly by binding to the important
apoptosis-regulating genes, such as B-cell CLL/lymphoma 2
(BCL2), MMP9 and CCND1.
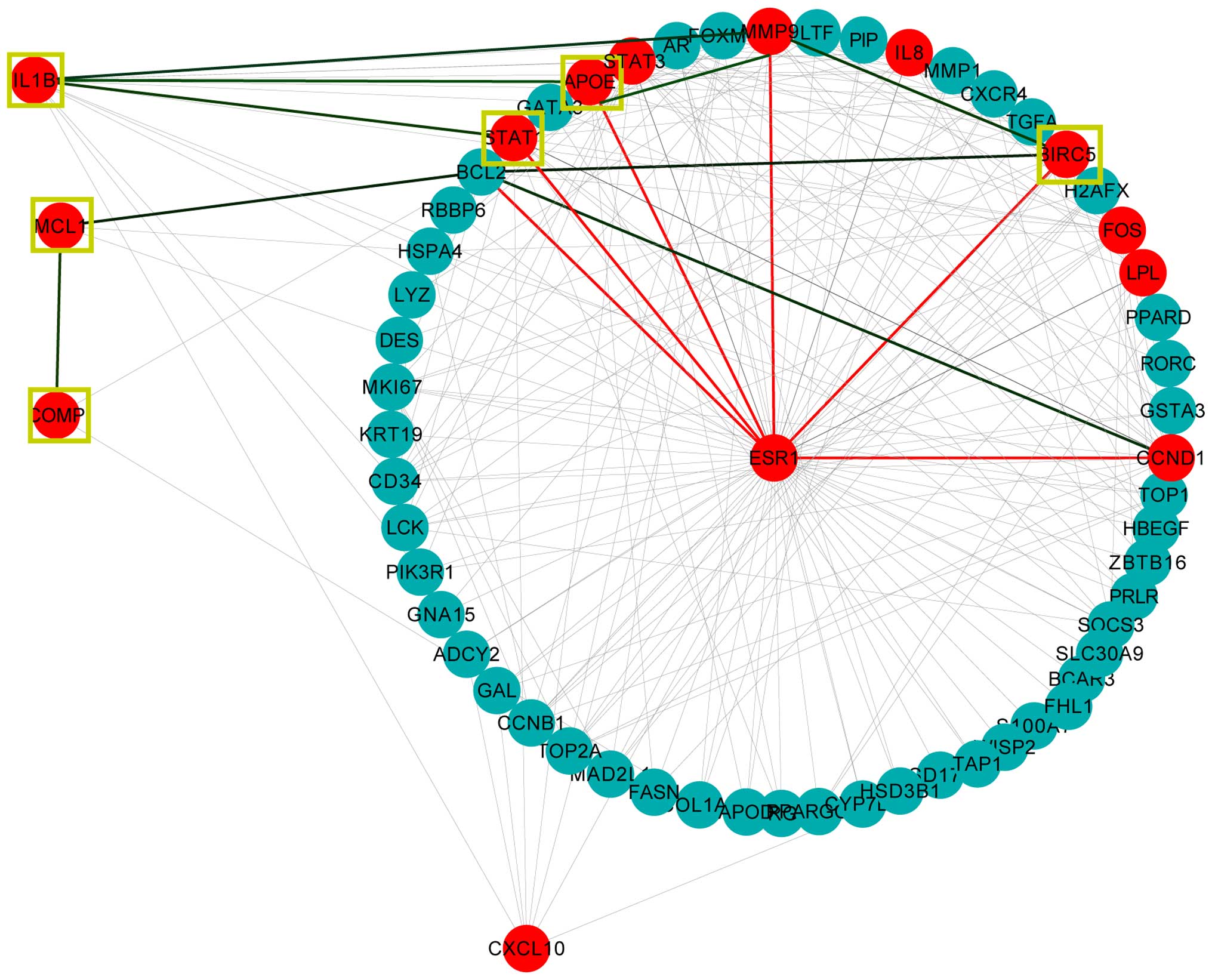 | Figure 4ESR1 topological neighbors.
The 56 nodes form a circle as they form ESR1 topological
neighbors. The 14 nodes that are highlighted in red are the top hub
genes that are associated with psoriasis, and include the
ESR1 gene. The yellow square represents the anti-apoptosis
gene ontology annotation node. The red line links genes to the
ESR1 gene and the green line links genes to the neighbors of
the ESR1 gene. ESR1, estrogen receptor-1; IL-1B,
interleukin-1β; STAT1, signal transducer and activator of
transcription-1; IL-8, interleukin-8; CXCL10, chemokine (C-X-C
motif) ligand 10; FOS, FBJ murine osteosarcoma viral oncogene
homolog; MMP9, matrix metalloproteinase-9; STAT3, signal transducer
and activator of transcription-3; APOE, apolipoprotein E; CCDN1,
cyclin D1; COMP, cartilage oligomeric matrix protein; MCL1, myeloid
cell leukemia-1; LPL, lipoprotein lipase; BIRC5, baculoviral IAP
repeat containing-5. |
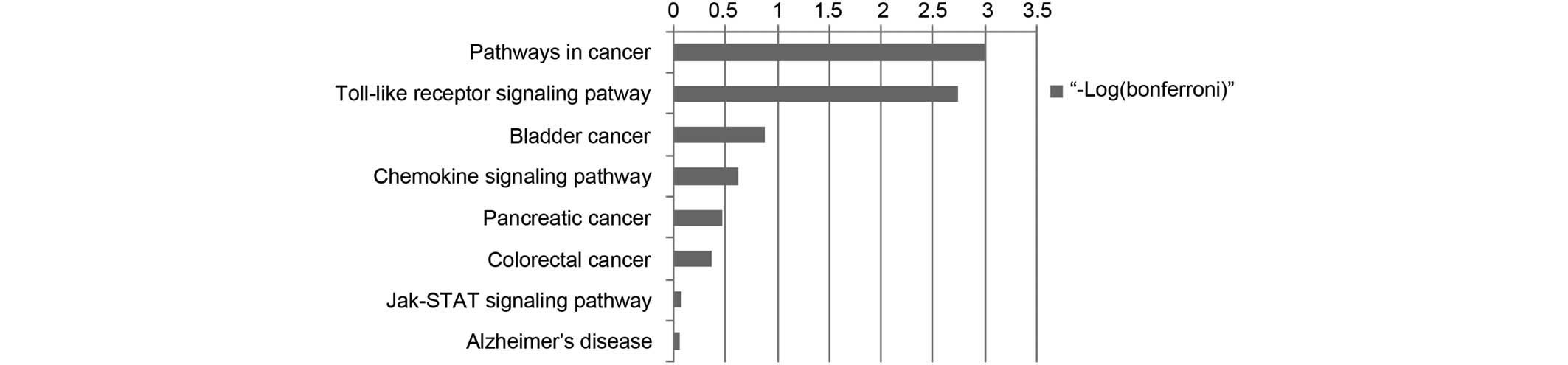
Using KEGG pathway analysis (FDR <0.05), the
pathways in cancer was detected to be most significantly associated
with psoriasis, more so than the TLR signaling pathway, as shown in
Fig. 5. Other pathways, including
the chemokine signaling pathway and Janus kinase/signal transducers
and activators of transcription signaling pathway were also
detected in the present study; however, the significance was
identified to be relatively lower. Seven of the top hub genes
(FOS, CCND1, IL-8, MMP9, BIRC5,
STAT1 and STAT3) were detected in response to
pathways in cancer and five of the top hub genes [FOS,
IL-8, IL-1B, STAT1 and chemokine (C-X-C motif)
ligand 10 (CXCL10)] exerted an effect on the TLR signaling
pathway (data not shown). All of the above-mentioned top hub genes,
apart from the CXCL10 gene, were involved in the first two
pathways, demonstrating a direct association with the ESR1
gene (Fig. 4). The present study
demonstrated that four of the genes (MMP9, BIRC5,
IL-1B and STAT1) were highly associated with
anti-apoptosis, regulation of apoptosis and programmed cell
death.
Discussion
Psoriasis is a chronic inflammatory skin disorder.
Significant decreases in keratinocyte apoptosis is a specific
pathogenic phenomenon in psoriasis lesions compared with normal
skin (3). In the present study,
927 DEGs were identified from the GEO database, GSE13355, which
contained gene expression profiles for 58 psoriatic lesion samples
and 64 normal skin samples. An interaction network with the 927
DEGs was then constructed (Fig.
2). Following PageRank analysis (Table I), ESR1 and another 13 genes
were selected as the top hub genes from 640 DEGs according to their
PageRank score. In addition, the functions of the 14 top hub genes
were analyzed by using GO term function analysis (Table II), indicating that the most
significant enrichment function was associated with anti-apoptosis.
Other functions, including response to organic substances, and
regulation of apoptosis, programmed cell death and cell death, were
also highly associated with psoriasis.
Apoptosis is a type of programmed cell death.
Multiple factors control the extrinsic and intrinsic apoptotic
signaling pathways. To maintain epidermal homeostasis, the complex
apoptotic process is regulated by numerous anti-apoptotic molecules
(5). The ESR1 gene, as well
as five other top hub genes, has been demonstrated to exert an
effect on anti-apoptosis in psoriasis according to the present
results. Four of the genes (MCL1, COMP, BIRC5
and IL-1B) demonstrate an upregulated expression level in
psoriasis. The other two genes (ESR1 and APOE)
demonstrate a downregulated level of expression. In addition to the
six top hub genes, a further two highly expressed genes,
MMP9 and STAT1, were identified to be involved in the
regulation of apoptosis, programmed cell death and cell death. The
MCL1 gene encodes an anti-apoptotic protein, myeloid cell
leukemia-1. Ultraviolet B radiation therapy inhibits growth and
induces apoptosis of human skin cells by downregulating MCL1
expression (21). IL-1B
induced IL-8 and p53 mRNA expression, as well as protein production
of IL-8 (22). p53 overexpression
has been described as a physiological reaction to
hyperproliferation in order to resist keratinocyte apoptosis.
Furthermore, knockdown of p53 expression levels in human
keratinocytes accelerates MCL1 reduction, thereby enhancing
apoptosis (23).
The ESR1 gene encodes an ERα (24), which is a ligand-activated
transcription factor that binds to estrogen (25). ERα is the receptor that transmits
estrogen signaling; however, the function of ERα in psoriasis
development remains largely unknown. In the present study, the
anti-apoptotic genes (BIRC5 and APOE) as topological
neighbors, demonstrated a direct association with the ESR1
gene (Fig. 4). Other
anti-apoptotic genes (MCL1, IL-1B and COMP)
demonstrated indirect connections with the ESR1 gene by
binding to the ESR1 topological neighbors, particularly by
binding to the important apoptosis regulation genes, such as
BCL-2, MMP9 and CCND1 (Fig. 4). The MMP9 gene, an
important gene in apoptosis regulation, has been demonstrated to be
involved in the cell death pathways (26). Psoriasin (termed, S100
calcium-binding protein A7), which is highly expressed in tumors
and hyperproliferative skin, enhances anti-apoptotic nuclear
factor-κB (NF-κB)-mediated MMP-9 secretion in ERα−
breast cancer cells, but has the opposite effect in ERα+
cells (27,28). CCDN1 and BCL2, at
lower expression levels, were also associated with the ESR1
gene (Fig. 4; Table I). A previous study regarding
colorectal cancer demonstrated that 17β-estradiol activates p53 to
inhibit cell proliferation by inhibiting CCDN1 (29). In addition, various studies have
reported that a lower expression level of B-cell lymphoma 2 (Bcl-2)
was strongly associated with psoriasis (4,30–32).
The strong bcl-2-like protein 4 (BAX) expression, as well as the
concomitant decrease in the level of Bcl-2 expression indicated a
resistance to BAX-mediated apoptosis in psoriasis (30). Thus, the reduced expression level
of ESR1 may lead to the upregulated expression of
anti-apoptotic genes via mediation of the genes that are associated
with apoptosis regulation, such as MMP9, BCL2 and
CCND1. This provides evidence that the ESR1 gene may
be implicated in psoriatic apoptotic resistance and that ERα may
exert a protective role against psoriatic proliferation. To the
best of our knowledge, the underlying mechanism of the association
between ESR1 and the anti-apoptosis signaling pathway in
psoriasis has not been fully elucidated; however, the present
findings indicate that the reduced level of ESR1 gene
expression associated with psoriasis, is highly correlated with
apoptosis resistance.
Using KEGG pathway analysis for the top 14 hub
genes, the signaling pathway most significantly linked with
psoriasis is the pathways in cancer, as well as the TLR signaling
pathway (Fig. 5). As a classical
pathway in psoriasis, the TRL signaling pathway has been shown to
be important in psoriasis and is involved in detecting invading
micro-organisms and producing pro-inflammatory stimuli to the skin
(7). TLRs drive aberrant
activation and unrestricted inflammatory responses (33).
In the current study, it is the first time, to the
best of our knowledge, that the pathways in cancer has been
detected to have the most significant association with psoriasis,
more so than the TLR signaling pathway. Pathways in cancer exerts
an effect on tumor proliferation by protecting tumor cells from
cytokine-induced apoptosis. A previous study demonstrated that the
thickened psoriatic skin displays a particular resistance of
keratinocytes to interferon-γ (INF-γ)- and tumor necrosis factor-α
(TNF-α)-induced apoptosis by sustaining the activation of the
phosphatidylinositol 3-kinase/Akt signaling pathway and
consequently activating the anti-apoptotic NF-κB cascade (4). Thus, the mechanisms of cell
proliferation result in activation of anti-apoptotic molecules,
which contribute to suppressing cytokine-induced apoptosis and
maintaining the response to pro-inflammatory stimuli in
keratinocytes (34). Hsu et
al (35) reported that ERα
inhibited bladder cancer cell growth by inhibiting Akt activity
(35). Therefore, the present
study hypothesized that ERα expression may activate INF-γ and
TNF-α-induced apoptosis in cells by inhibiting Akt activity in
psoriasis and various types of cancer, including bladder
cancer.
The current study demonstrated that seven of the top
hub genes that were involved in the pathways in cancer were
directly associated with the ESR1 gene. The above-mentioned
evidence suggests that activating the pathways in cancer may
protect the psoriatic cell from cytokine-induced apoptosis by
inhibiting ERα expression. Further investigations are required to
establish why the pathways in cancer is more significant than the
cytokine-related TLR signaling pathway in apoptosis resistance in
psoriasis.
In conclusion, activating the pathways in cancer, as
well as reducing ERα expression promotes proliferation and
resistance to apoptosis. The present results indicate that a lower
ERα expression level may induce psoriatic keratinocyte
proliferation and anti-apoptosis effects via anti-apoptotic genes
modulated by activation of the pathways in cancer. However, the
contribution of individual factors requires elucidation. Future
studies are required to investigate the functions of ERα, the
association between anti-apoptosis genes in psoriasis and whether
these genes are involved in the biological mechanisms underlying
the coordination of psoriatic keratinocyte apoptosis
resistance.
Acknowledgments
The authors would like to thank Wilson Liao
(Assistant Professor of Dermatology; Department of Dermatology,
University of California, San Francisco, CA, USA) for providing the
information processing methods. The present study was supported by
grants from the National Natural Science Foundation of China (grant
no. 30973758).
Abbreviations:
|
ERα
|
estrogen receptor α
|
|
DEGs
|
differentially expressed genes
|
|
STRING
|
search tool for the retrieval of
interacting genes
|
|
GEO
|
gene expression omnibus database
|
|
GO
|
gene ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
ESR1
|
estrogen receptor-1
|
|
MCL1
|
myeloid cell leukemia-1
|
|
BIRC5
|
baculoviral IAP repeat
containing-5
|
|
MMP9
|
matrix metalloproteinase-9
|
|
IL-1B
|
interleukin-1β
|
|
IL-8
|
interleukin-8
|
|
APOE
|
apolipoprotein E
|
|
CCDN1
|
cyclin D1
|
|
BCL-2
|
B-cell lymphoma-2
|
|
BAX
|
BCL2-associated X protein
|
|
STAT1
|
signal transducer and activator of
transcription-1
|
|
STAT3
|
signal transducer and activator of
transcription-3
|
|
FOS
|
FBJ murine osteosarcoma viral oncogene
homolog
|
|
CXCL10
|
chemokine (C-X-C motif) ligand 10
|
|
E2
|
17β-estradiol
|
|
TLRs
|
toll-like receptors
|
|
NF-κB
|
nuclear factor-κB
|
References
|
1
|
Christophers E: Psoriasis-epidemiology and
clinical spectrum. Clin Exp Dermatol. 26:314–320. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nestle FO, Kaplan DH and Barker J:
Psoriasis. N Engl J Med. 361:496–509. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Laporte M, Galand P, Fokan D, de Graef C
and Heenen M: Apoptosis in established and healing psoriasis.
Dermatology. 200:314–316. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Madonna S, Scarponi C, Pallotta S, Cavani
A and Albanesi C: Anti-apoptotic effects of suppressor of cytokine
signaling 3 and 1 in psoriasis. Cell Death Dis. 3:e3342012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Portt L, Norman G, Clapp C, Greenwood M
and Greenwood MT: Anti-apoptosis and cell survival: A review.
Biochim Biophys Acta. 1813:238–259. 2011. View Article : Google Scholar
|
|
6
|
Sanchez MI, Shearwood AM, Chia T, Davies
SM, Rackham O and Filipovska A: Estrogen-mediated regulation of
mitochondrial gene expression. Mol Endocrinol. 29:14–27. 2015.
View Article : Google Scholar
|
|
7
|
Miller LS: Toll-like receptors in skin.
Adv Dermatol. 24:71–87. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nair RP, Duffin KC, Helms C, Ding J,
Stuart PE, Goldgar D, Gudjonsson JE, Li Y, Tejasvi T, Feng BJ, et
al: Genome-wide scan reveals association of psoriasis with IL-23
and NF-kappaB pathways. Nat Genet. 41:199–204. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Swindell WR, Johnston A, Carbajal S, Han
G, Wohn C, Lu J, Xing X, Nair RP, Voorhees JJ, Elder JT, et al:
Genome-wide expression profiling of five mouse models identifies
similarities and differences with human psoriasis. PloS One.
6:e182662011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kerr MK: Linear models for microarray data
analysis: Hidden similarities and differences. J Computat Biol.
10:891–901. 2003. View Article : Google Scholar
|
|
11
|
Bhat-Nakshatri P, Song EK, Collins NR,
Uversky VN, Dunker AK, O'Malley BW, Geistlinger TR, Carroll JS,
Brown M and Nakshatri H: Interplay between estrogen receptor and
AKT in estradiol-induced alternative splicing. BMC Med Genomics.
6:212013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res.
41(Database Issue): D808–D815. 2013. View Article : Google Scholar :
|
|
13
|
Snel B, Lehmann G, Bork P and Huynen MA:
STRING: A web-server to retrieve and display the repeatedly
occurring neighbourhood of a gene. Nucleic Acids Res. 28:3442–3444.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Page L, Brin S, Motwani R and Winograd T:
The Page Rank citation ranking: Bringing order to the web. Stanford
Info Lab. 1999.
|
|
15
|
Dellavalle RP, Schilling LM, Rodriguez MA,
Van de Sompel H and Bollen J: Refining dermatology journal impact
factors using Page Rank. J Am Acad Dermatol. 57:116–119. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Griffiths TL, Steyvers M and Firl A:
Google and the mind: Predicting fluency with Page Rank. Psychol
Sci. 18:1069–1076. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bánky D, Iván G and Grolmusz V: Equal
opportunity for low-degree network nodes: A Page Rank-based method
for protein target identification in metabolic graphs. PloS One.
8:e542042013. View Article : Google Scholar
|
|
18
|
Huang DW, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar
|
|
19
|
Huang DW, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar :
|
|
20
|
Albert R and Barabási AL: Statistical
mechanics of complex networks. Rev Mod Phys. 74:47–97. 2002.
View Article : Google Scholar
|
|
21
|
Park YK and Jang BC: UVB-induced
anti-survival and pro-apoptotic effects on HaCaT human
keratinocytes via caspase- and PKC-dependent downregulation of PKB,
HIAP-1, Mcl-1, XIAP and ER stress. Int J Mol Med. 33:695–702.
2014.
|
|
22
|
Rasmussen MK, Iversen L, Johansen C,
Finnemann J, Olsen LS, Kragballe K and Gesser B: IL-8 and p53 are
inversely regulated through JNK, p38 and NF-kappaB p65 in HepG2
cells during an inflammatory response. Inflamm Res. 57:329–339.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chaturvedi V, Sitailo LA, Qin JZ, Bodner
B, Denning MF, Curry J, Zhang W, Brash D and Nickoloff BJ:
Knockdown of p53 levels in human keratinocytes accelerates Mcl-1
and Bcl-x(L) reduction thereby enhancing UV-light induced
apoptosis. Oncogene. 24:5299–5312. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zang YC, Halder JB, Hong J, Rivera VM and
Zhang JZ: Regulatory effects of estriol on T cell migration and
cytokine profile: Inhibition of transcription factor NF-kappa B. J
Neuroimmunol. 124:106–114. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chang KC, Wang Y, Oh IG, Jenkins S,
Freedman LP, Thompson CC, Chung JH and Nagpal S: Estrogen receptor
beta is a novel therapeutic target for photoaging. Mol Pharmacol.
77:744–750. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Paolillo N, Piccirilli S, Giardina E,
Rispoli V, Colica C and Nisticò S: Effects of paraquat and
capsaicin on the expression of genes related to inflammatory,
immune responses and cell death in immortalized human HaCat
keratinocytes. Int J Immunopathol Pharmacol. 24:861–868. 2011.
|
|
27
|
Petersson S, Bylander A, Yhr M and
Enerback C: S100A7 (Psoriasin), highly expressed in ductal
carcinoma in situ (DCIS), is regulated by IFN-gamma in mammary
epithelial cells. BMC Cancer. 7:2052007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sneh A, Deol YS, Ganju A, Shilo K, Rosol
TJ, Nasser MW and Ganju RK: Differential role of psoriasin (S100A7)
in estrogen receptor alpha positive and negative breast cancer
cells occur through actin remodeling. Breast Cancer Res Treat.
138:727–739. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hsu HH, Kuo WW, Ju DT, Yeh YL, Tu CC, Tsai
YL, Shen CY, Chang SH, Chung LC and Huang CY: Estradiol agonists
inhibit human LoVo colorectal-cancer cell proliferation and
migration through p53. World J Gastroenterol. 20:16665–16673. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Koçak M, Bozdogan O, Erkek E, Atasoy P and
Birol A: Examination of Bcl-2, Bcl-X and bax protein expression in
psoriasis. Int J Dermatol. 42:789–793. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gabr SA, Berika MY and Alghadir AH:
Apoptosis and clinical severity in patients with psoriasis and HCV
infection. Indian J Dermatol. 59:230–236. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gündüz K, Demireli P, Vatansever S and
Inanir I: Examination of bcl-2 and p53 expressions and apoptotic
index by TUNEL method in psoriasis. J Cutan Pathol. 33:788–792.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu Y, Yin H, Zhao M and Lu Q: TLR2 and
TLR4 in autoimmune diseases: A comprehensive review. Clin Rev
Allergy Immunol. 47:136–147. 2014. View Article : Google Scholar
|
|
34
|
Dallaglio K, Marconi A and Pincelli C:
Survivin: A dual player in healthy and diseased skin. J Invest
Dermatol. 132:18–27. 2012. View Article : Google Scholar
|
|
35
|
Hsu I, Yeh CR, Slavin S, Miyamoto H, Netto
GJ, Tsai YC, Muyan M, Wu XR, Messing EM, Guancial EA and Yeh S:
Estrogen receptor alpha prevents bladder cancer via INPP4B
inhibited akt pathway in vitro and in vivo. Oncotarget.
5:7917–7935. 2014. View Article : Google Scholar : PubMed/NCBI
|