Introduction
Breast cancer is one of the most common types of
malignancies among women worldwide (1). Hereditary genetic factors are
responsible for ~25% of breast cancer according to twin studies
(2). Despite a study that involved
variants associated with a risk of breast cancer (3), less is known regarding the
significance of these prognostic genetic variants.
Receptor tyrosine-protein kinase ErbB4 is a
member of the epidermal growth factor receptor (EGFR)
subfamily as well as EGFR (ErbB1), ErbB2
(HER2, neu), ErbB3 (HER3) and
ErbB4 (HER4) (4).
Experimental studies have demonstrated oncogenic and tumor
suppressive functions of ErbB4 in breast cancer (5). In breast cancer, ErbB4
expression is associated with a favorable outcome; however,
ErbB4 expression combined with ErbB1 and ErbB2
upregulation may lead to an unfavorable outcome (6). Additionally, overexpression of
ErbB4 in conjunction with ESR1 expression may also
promote favorable outcome in breast cancer patients (7). Conversely, the pro-apoptotic BH3
domain of ErbB4 has been suggested to enhance the apoptosis
of breast cancer cells when overexpressed (8). MicroRNAs (miRNAs) are key in the
regulation of the expression of numerous genes, including
ErbB4. A number of miRNAs, such as miR-193a-3p, target
specific sites in the 3′-untranslated region (3′-UTR) of
ErbB4 mRNA in order to downregulate its expression (9).
Single nucleotide polymorphisms (SNPs) in growth
regulatory genes, such as ErbB4, may affect tumor growth in
breast cancer. Association between SNPs at the promoter and within
the intronic region of ErbB4 and the development of breast
cancer has been shown in previous studies (10,11).
In addition, functional SNPs (12), located in the 3′-UTR could directly
weaken or strengthen the interaction of miRNA with 3′-UTR consensus
sites (13), that functions in RNA
silencing. Numerous databases and online tools, including miRNASNP
(13), dbSMR (14) and PolymiRTS (15), have been developed to predict the
effect of SNPs in miRNA target sites, which may aid in clarifying
the role of SNPs in the development of certain types of cancer.
In the present study, in silico investigation
was used to predict the effect of SNPs in the 3′-UTR on
ErbB4 expression. According to predictions, SNP rs1836724 is
a putative functional polymorphism that alters the interaction of
miRNAs targeting ErbB4 mRNA. To the best of our knowledge,
this study is the first to investigate whether rs1836724 influences
susceptibility to breast cancer in the Iranian population.
Moreover, computational analysis was enhanced in order to interpret
experimental observations and achieve a molecular insight into
breast cancer.
Materials and methods
In silico analysis
miRNASNP (version 2.0) bioinformatics online tools
(bioguo.org/miRNASNP/) (13) were used in order to predict
putative SNPs in the 3′-UTR of the ErbB4 gene that could
alter miRNA interactions. All possible miRNA/SNP-variant mRNA
interactions and ΔΔG (the difference between the mRNA-miRNA hybrid
free energy connected to each allele), were provided by miRNASNP.
Results were categorized by the gain or loss of
miRNA/target-interaction ability, which was calculated based on
binding energy change between the two SNP variants. Additional
bioinformatics analysis was performed using the miRWalk (version
2.0) database (16) to determine
potential common miRNAs which target ESR1 and ErbB4
mRNAs.
Sampling, DNA extraction and
genotyping
Peripheral blood samples were retrieved from 70
patients who were recently diagnosed and histologically confirmed
to have breast cancer between August 2013 and August 2014 at the
Sayed-ol-Shohada Hospital (Isfahan, Iran). In total, 76 control
blood samples were obtained from female individuals undergoing a
regular health check at the hospital. In this study, control
samples with any history of cancer were excluded. The clinical and
pathological characteristics of the patients were collected from
the hospital and are summarized in Table I. Age ranges of case and control
subjects were 31–72 and 30–85 years old, respectively. Written
informed consent was collected from all involved participants. The
current study was approved by the ethics committee of
Sayed-ol-Shohada Hospital (Isfahan, Iran).
 | Table IClinicopathological characteristics of
the patients with breast carcinoma. |
Table I
Clinicopathological characteristics of
the patients with breast carcinoma.
| Characteristic | No. of patients |
|---|
| Early metastasis |
| Positive | 26 |
| Negative | 44 |
| Histological
grade |
| I | 10 |
| II | 26 |
| III | 18 |
| Unknown | 16 |
| Stage |
| I | 10 |
| II | 12 |
| III | 2 |
| IV | 40 |
| Unknown | 6 |
| Estrogen receptor
status |
| Positive | 32 |
| Negative | 12 |
| Unknown | 26 |
| Progesterone receptor
status |
| Positive | 30 |
| Negative | 14 |
| Unknown | 26 |
| HER2 status | |
| Positive | 16 |
| Negative | 28 |
| Unknown | 26 |
Genomic DNA was extracted using the PrimePrep
Genomic DNA Isolation kit (GeNetBio, Chungnam, South Korea),
according to the manufacturer's instructions. DNA purity and
concentration was determined using a spectrophotometer (NanoDrop
1000; Thermo Fisher Scientific Inc., Wilmington, DE, USA).
DNA fragments were amplified using the following
primers for rs1836724: Forward: 5′-TTAATAGAAATTTGAGTTTTGCGTT-3′ and
reverse: 5′-TATCAGATTCCAGAGGCCAAT-3′. Standard cycling was
performed in a thermocycler (ASTEC PC-818; ASTEC, Fukuoka, Japan)
under the following conditions: Initial denaturation at 96°C for 2
min followed by 35 cycles of 94°C for 30 sec, 56.5°C for 30 sec,
65°C for 30 sec, and finally 65°C for 7 min. It should be noted
that flanking region of rs1836724 is AT-rich and despite
alternative genotyping tools (17), the PCR program with reduced
extension temperature (65°C) and following restriction fragment
length polymorphism was the best strategy for AT-rich DNA
genotyping (18). The PCR products
were electrophoresed by 2.5% agarose gel electrophoresis in 1X
Tris-Borate-EDTA buffer at 100 V and stained with RedSafe Nucleic
Acid Staining solution (Boca Scientific, Inc., Boca Raton, FL, USA)
for visualization. Detection of allelic variations was enhanced by
digesting polymerase chain reaction products with the restriction
enzyme AlwNI (Thermo Fisher Scientific Inc.). The
AlwNI restriction enzyme does not cut PCR product containing
a T allele (band is 383 bp); furthermore, it yielded two fragments
of 272 bp and 111 bp, as there is a C allele in the original PCR
product. The accuracy of the genotypes was confirmed by randomly
performed Sanger sequencing using the Bioneer Sequencing Service
(Bioneer Corporation, Daejeon, South Korea).
Statistical analysis
Statistical analysis was assessed by comparing case
and control samples, and ER positive and ER negative samples.
SNPStats online tools (bioinfo.iconcologia.net/snpstats/) (17), was used to calculate allele
frequency, and genotype frequency.
Deviation from Hardy-Weinberg equilibrium (HWE),
odds ratios (ORs) with 95% confidence intervals (CIs), and the
Cochran-Armitage (CA) test for trend were executed using the
DeFinetti program (ihg.gsf.de/cgi-bin/hw/hwa1.pl) to analyze the
association between rs1836724 and breast cancer. The CA test
considers individuals' genotypes, as opposed to solely the alleles
for association assessment, using the guidelines provided by the
DeFinetti program. Consistency with Hardy-Weinberg equilibrium was
investigated using Pearson's χ2, Log likelihood ratio
(Llr) χ2, and exact tests. In addition, the association
test was evaluated using χ2 test. Logistic regression
models were used to determine if odds ratios (OR) are associated
with 95% confidence intervals (95% CI). P<0.05 was considered to
indicate a statistically significant difference. Additional
bioinformatics investigation was conducted to acquire estrogen
receptor (ESR1) targeted miRNAs using miRWalk V.2.0 database
(19).
Results
In silico analysis
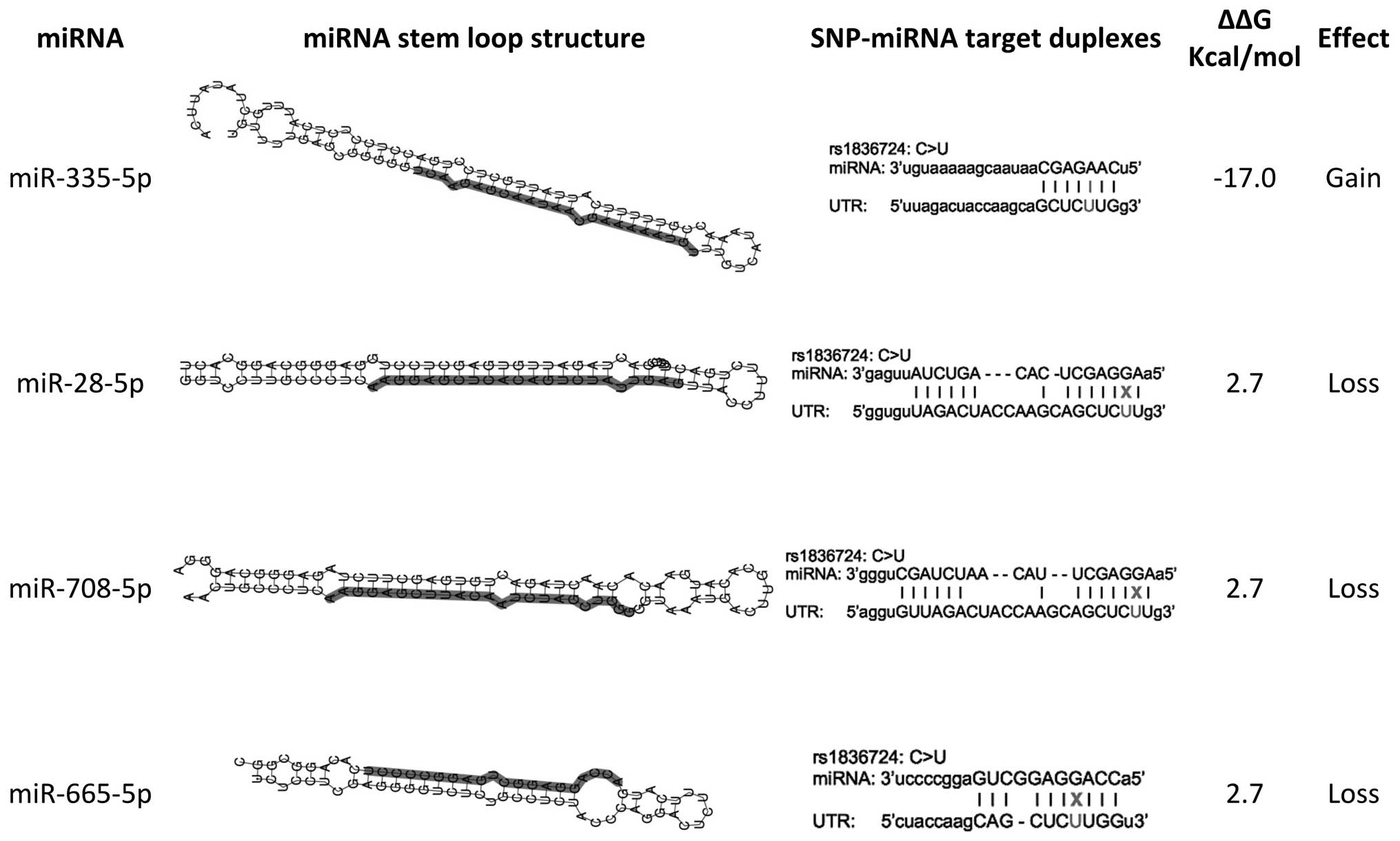
Computational predictions suggested that rs1836724
is located in ErbB4 3′-UTR within the potential target
sequence of has-miR-335-5p, hsa-miR-28-5p, has-miR-708-5p and
has-miR-665 (Fig. 1).
Statistical analysis
Allele frequencies, observed genotypes, expected
genotypes and HWE P-values are shown in Table II. No deviation from HWE was
observed in the groups. According to the allele frequency
comparison [OR (95% CI)=1.722 (1.056–2.808), P=0.02869) and the CA
test for trend (OR=1.697, P=0.2913), T allele of rs1836724 was
found to be associated with a risk of breast cancer (Table III). Moreover, the C/T genotype
of rs1836724 was significantly associated with an ER-positive
phenotype among patients [OR (95% CI)=6.000 (1.082–33.274),
P=0.02846] compared with the T/T genotype. Finally, in
silico algorithms demonstrated that ESR1, the gene
responsible for the ER phenotype in the case group, may be targeted
by similar miRNAs as ErbB4, including has-miR-28-5p,
has-miR-708-5p and has-miR-665 (Table
IV).
 | Table IIEstimation of allele frequency,
observed genotypes, expected genotypes and HWE P-values of
rs1836724 in control, case groups, and ER-positive, and ER-negative
subgroups. |
Table II
Estimation of allele frequency,
observed genotypes, expected genotypes and HWE P-values of
rs1836724 in control, case groups, and ER-positive, and ER-negative
subgroups.
| Group | Allele frequency
| Observed genotypes
| Expected genotypes
| HWE P-value
|
|---|
| T | C | T/T | T/C | C/C | T/T | T/C | C/C | Pearson's | Llr | Exact |
|---|
| Control | 0.59 | 0.41 | 26 | 38 | 12 | 26.64 | 36.71 | 12.64 | 0.759440 | 0.759217 | 0.816842 |
| Case | 0.71 | 0.29 | 36 | 28 | 6 | 35.71 | 28.57 | 5.71 | 0.867109 | 0.867489 | 1.000000 |
| ER negative | 0.75 | 0.25 | 8 | 2 | 2 | 6.75 | 4.50 | 0.75 | 0.054292 | 0.065276 | 0.089899 |
| ER positive | 0.66 | 0.34 | 12 | 18 | 2 | 13.78 | 14.44 | 3.78 | 0.162761 | 0.150196 | 0.252820 |
 | Table IIIAssociation analysis of rs1836724 and
risk of breast cancer. |
Table III
Association analysis of rs1836724 and
risk of breast cancer.
| Risk allele | Allele frequency
comparison
| Armitage's trend
test
|
|---|
| Odds ratio (95%
CI) | P-value | Common odds
ratio | P-value |
|---|
| C | 0.581
(0.356–0.947) | 0.02869 | 0.586 | 0.02913 |
| T | 1.722
(1.056–2.808) | | 1.697 | |
 | Table IVSuggested miRNAs with increased
binding possibility to the 3′-UTR ErbB4 by C allele may
target ESR1 mRNA according to the aforementioned
algorithms. |
Table IV
Suggested miRNAs with increased
binding possibility to the 3′-UTR ErbB4 by C allele may
target ESR1 mRNA according to the aforementioned
algorithms.
| Putative miRNA
binding site | Database used |
|---|
| hsa-miR-28-5p | miRWalk |
| Microt4 |
| miRMap |
| RNA22 |
| RNAhybrid |
| hsa-miR-665 | miRMap |
| RNA22 |
| RNAhybrid |
| hsa-miR-708-5p | miRWalk |
| Microt4 |
| miRMap |
| RNA22 |
| RNAhybrid |
Discussion
Thus far, altered expression of ErbB4 has
been reported in various studies of breast cancer (20,21).
miRNAs are important in regulating ErbB4 expression
(22). It was demonstrated that
polymorphisms in the 3′-UTR of genes could affect miRNA binding
sites, resulting in post-translational dysregulation of mRNA and a
predisposition to cancer (23,24).
Computational analysis scrutinized the 3′-UTR region of
ErbB4 for its cancer risk variants. Noticeably, the
rs1836724 SNP was identified within the 3′-UTR of ErbB4 and
target binding site of four miRNAs. The presence of SNP
rs1836724T/T would weaken the target sites of has-miR-28-5p,
has-miR-708-5p and has-miR-665, and strengthen the target site of
has-miR-335-5p (Fig. 1). A
significant calculated ΔΔG suggested rs1836724T/T as a possible
causative genetic factor in the development of breast tumor cells.
To the best of our knowledge, this is the first case-control study
conducted in an Iranian population attempting to examine the
correlation between rs1836724T/T and the risk of breast cancer.
In this case-control study, all female participants
selected belonged to the same ethnicity in order to eliminate
variation of alleles and genotype frequencies, as certain ones may
only occur in specific ethnic groups, thus skewing the results.
Analysis of allele frequencies suggested that the C allele is the
minor allele (allele frequency, 0.35) when observing all subjects
of the study, the same as that demonstrated in the NCBI SNP
databank reports (ncbi.nlm.nih.gov/snp/). However, comparing allele
frequencies of the T allele in the control group (0.65) and case
group (0.71), indicated that it is a risk factor for breast cancer.
The genotype distribution data suggested that C/C is the minor
genotype in the control and case groups (genotype frequency, 0.16
and 0.09, respectively). Notably, the case subgroup analysis
highlighted allele frequency differences between ER-negative (T,
0.75; C, 0.25) and ER-positive (T, 0.66; C, 0.34) that may lead to
the different effect of alleles on this phenotype. HWE P-value data
(Pearson, Llr, and exact test) were all >0.05 and no deviation
from HWE was identified.
This study determined an association between
rs1836724 and the susceptibility to breast cancer using allele
frequency comparison and the CA test for trend. Together, the
association between the T allele and the risk of breast cancer was
confirmed (1.722 OR and P=0.02869). The computational data obtained
in the current study suggested that the miR-335-5p binding site may
be strengthened in T allele carriers; however, miR-28-5p, miR-665,
and miR-708-5p were presumed to have weaker binding site in the T
allele variant (Fig. 1).
Therefore, downregulation of ErbB4 expression levels in
individuals carrying the T allele in rs1836724 may result in
greater susceptibility to breast cancer.
Conversely, the present study demonstrated an
association between the C/T and perhaps C/C genotypes of rs1836724
and an ER-positive phenotype. As was predicted, the C allele was
associated with more post-translational suppression corresponding
to stronger mRNA binding sites within the 3′-UTR ErbB4.
Consequently, further bioinformatics analysis was executed to
reveal identical regulatory miRNAs of ErbB4 and ESR1.
As shown in Table IV, the miRNAs
that regulate ErbB4 and ESR1 were similar. According
to previously demonstrated association between ErbB4 and
ESR1 expression and breast cancer outcome (25), and the identical miRNAs,
association between the C allele and an ER-positive phenotype can
be explained. The present study hypothesized that stronger target
binding sites at the 3′-UTR ErbB4 due to the presence of the
C allele may reduce the possibility of ESR1 being targeted by the
same miRNAs. In other words, the C allele leads to more preferable
sequences at 3′-UTR ErbB4, which may in turn recruit miRNAs
able to target 3′-UTR ESR1. Therefore, observation of ER
positive phenotype in the state of C/T genotype 6 times (OR=6,
P=0.028) more than T/T genotype seemed completely logical. It is
notable that, further discussion of miR-335-5p in the ER phenotype
has not been conducted as it was not listed as one of the
ESR1/ErbB4 common regulatory miRNAs.
In conclusion, to the best of our knowledge, this
population based case-control study demonstrated for the first time
a correlation between the SNP rs1836724 located at the 3′-UTR
ErbB4 and susceptibility to breast cancer. The data
suggested a significant association between the T allele and a risk
of breast cancer. Computational data correlated the experimental
observations with altered target binding sites of three
ErbB4 regulatory miRNAs, hsa-miR-28-5p, hsa-miR-7085p and
hsa-miR-665. Furthermore, the ER-positive phenotype was shown to be
associated with the C carriers of rs1836724. miRNAs that regulate
both ESR1 and ErbB4 explained a competitive
correlation between these genes, which leads to less ESR1
downregulation in the C allele state due to stronger target binding
in the 3′-UTR of ErbB4. Together, this investigation
suggests that the rs1836724T/T SNP is a potential risk factor for
the development of breast cancer.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Collaborative Group on Hormonal Factors in
Breast Cancer: Familial breast cancer: Collaborative reanalysis of
individual data from 52 epidemiological studies including 58,209
women with breast cancer and 101,986 women without the disease.
Lancet. 358:1389–1399. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lalloo F and Evans DG: Familial breast
cancer. Clin Genet. 82:105–114. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hynes NE and MacDonald G: ErbB receptors
and signaling pathways in cancer. Curr Opin Cell Biol. 21:177–184.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gullick WJ: c-erbB-4/HER4: Friend or foe?
J Pathol. 200:279–281. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
McCubrey JA, Abrams SL, Fitzgerald TL,
Cocco L, Martelli M, Montalto G, Cervello M, Scalisi A, Candido S,
Libra M and Steelman LS: Roles of signaling pathways in drug
resistance, cancer initiating cells and cancer progression and
metastasis. Adv Biol Regul. 57:75–101. 2015. View Article : Google Scholar
|
|
7
|
Junttila TT, Sundvall M, Lundin M, Lundin
J, Tanner M, Härkönen P, Joensuu H, Isola J and Elenius K:
Cleavable ErbB4 isoform in estrogen receptor-regulated growth of
breast cancer cells. Cancer Res. 65:1384–1393. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Naresh A, Long W, Vidal GA, Wimley WC,
Marrero L, Sartor CI, Tovey S, Cooke TG, Bartlett JM and Jones FE:
The ERBB4/HER4 intracellular domain 4ICD is a BH3-only protein
promoting apoptosis of breast cancer cells. Cancer Res.
66:6412–6420. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liang H, Liu M, Yan X, Zhou Y, Wang W,
Wang X, Fu Z, Wang N, Zhang S, Wang Y, et al: miR-193a-3p functions
as a tumor suppressor in lung cancer by down-regulating ERBB4. J
Biol Chem. 290:926–940. 2015. View Article : Google Scholar :
|
|
10
|
Rokavec M, Justenhoven C, Schroth W,
Istrate MA, Haas S, Fischer HP, Vollmert C, Illig T, Hamann U, Ko
YD, et al: A novel polymorphism in the promoter region of ERBB4 is
associated with breast and colorectal cancer risk. Clin Cancer Res.
13:7506–7514. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Murabito JM, Rosenberg CL, Finger D,
Kreger BE, Levy D, Splansky GL, Antman K and Hwang SJ: A
genome-wide association study of breast and prostate cancer in the
NHLBI's Framingham heart study. BMC Med Genet. 8(8 Suppl 1):
S62007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Marjan MN, Hamzeh MT, Rahman E and Sadeq
V: A computational prospect to aspirin side effects: Aspirin and
COX-1 interaction analysis based on non-synonymous SNPs. Comput
Biol Chem. 51:57–62. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gong J, Tong Y, Zhang HM, Wang K, Hu T,
Shan G, Sun J and Guo AY: Genome-wide identification of SNPs in
microRNA genes and the SNP effects on microRNA target binding and
biogenesis. Hum Mutation. 33:254–263. 2012. View Article : Google Scholar
|
|
14
|
Gong J, Liu C, Liu W, Wu Y, Ma Z, Chen H
and Guo AY: An update of miRNASNP database for better SNP selection
by GWAS data, miRNA expression and online tools. Database.
2015:1–8. 2015. View Article : Google Scholar
|
|
15
|
Bao L, Zhou M, Wu L, Lu L, Goldowitz D,
Williams RW and Cui Y: PolymiRTS database: Linking polymorphisms in
microRNA target sites with complex traits. Nucleic Acids Res.
35(Database issue): D51–D54. 2007. View Article : Google Scholar
|
|
16
|
Solé X, Guinó E, Valls J, Iniesta R and
Moreno V: SNPStats: A web tool for the analysis of association
studies. Bioinformatics. 22:1928–1929. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mesrian Tanha H, Mojtabavi Naeini M,
Rahgozar S, Rasa SM and Vallian S: Modified tetra-primer ARMS PCR
as a single-nucleotide polymorphism genotyping tool. Genetic Test
Mol Biomarkers. 19:156–161. 2015. View Article : Google Scholar
|
|
18
|
Su XZ, Wu Y, Sifri CD and Wellems TE:
Reduced extension temperatures required for PCR amplification of
extremely A+T-rich DNA. Nucleic Acids Res. 24:1574–1575. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dweep H, Sticht C, Pandey P and Gretz N:
miRWalk-database: Prediction of possible miRNA binding sites by
'walking' the genes of three genomes. J Biomed Inform. 44:839–847.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim HC, Lee JY, Sung H, Choi JY, Park SK,
Lee KM, Kim YJ, Go MJ, Li L, Cho YS, et al: A genome-wide
association study identifies a breast cancer risk variant in ERBB4
at 2q34: Results from the Seoul breast cancer study. Breast Cancer
Res. 14:R562012. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ni Y and Siyuan Z: Erbb4 signaling: An
overlooked backup system? Cell Cycle. 14:16232015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang M, Yang Q, Zhang L, Zhou S, Ye W,
Yao Q, Li Z, Huang C, Wen Q and Wang J: miR-302b is a potential
molecular marker of esophageal squamous cell carcinoma and
functions as a tumor suppressor by targeting ErbB4. J Exp Clin
Cancer Res. 33:102014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bandyopadhyay S, Mitra R, Maulik U and
Zhang MQ: Development of the human cancer microRNA network.
Silence. 1:62010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yu T, Li J, Yan M, Liu L, Lin H, Zhao F,
Sun L, Zhang Y, Cui Y, Zhang F, et al: MicroRNA-193a-3p and -5p
suppress the metastasis of human non-small-cell lung cancer by
down-regulating the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway.
Oncogene. 34:413–423. 2015. View Article : Google Scholar
|
|
25
|
Zhu Y, Sullivan LL, Nair SS, Williams CC,
Pandey AK, Marrero L, Vadlamudi RK and Jones FE: Coregulation of
estrogen receptor by ERBB4/HER4 establishes a growth-promoting
autocrine signal in breast tumor cells. Cancer Res. 66:7991–7998.
2006. View Article : Google Scholar : PubMed/NCBI
|