Introduction
Gastric cancer (GC) is the second leading cause of
cancer-associated mortality, despite its steady declining trend in
most parts of the world. A total of 989,600 newly diagnosed cases
of GC and 738,000 mortalities have been estimated for 2008,
accounting for 8% of all newly diagnosed cancer and 10% of all
cancer-associated mortalities (1).
>70% of new diagnoses and mortalities occur in developing
countries. Eastern Asia, Eastern Europe and South America have the
highest incidence rates of GC, while rates are low in North America
and most parts of Africa (1). As
early GC is typically asymptomatic and features a small tumor size,
it is usually detected at advanced stages (2,3).
Therapeutic interventions to treat such late-stage tumors are often
restricted to non-curative gastrectomy, lymphadenectomy and
post-operative chemoradiotherapy (4). Thus, the prognosis is poor and the
five-year relative survival rate for GC is <30% in most
countries (5). It is therefore of
great clinical importance to identify novel biomarkers for early
diagnosis and targeted treatment of GC.
Small ubiquitin-like modifiers (SUMOs) are highly
conserved 11-kDa proteins that are detected in nearly all tissues
of eukaryotic organisms, covalently attach and detach from target
proteins to regulate their functions. SUMOylation, which is highly
dynamic and reversible, is a type of post-translational
modification and regulates numerous cellular functions and
processes, including protein stability, protein localization,
protein-protein interaction, cell cycle progression, DNA
replication and repair, chromatin organization, transcription and
RNA metabolism (6–8). Numerous diseases are associated with
SUMO conjugation, including brain ischemia, heart failure,
arthritis, degenerative diseases and cancer (9). DeSUMOylation enzymes are proteases
that remove SUMOs from their substrate proteins and reverse the
SUMOylation modification of target proteins, which counteracts
ubiquitination (10,11). Ubiquitin-specific peptidase 39
(USP39), which is a 65-kDa SR-related protein of the U4/U6·U5
triple small nuclear ribonucleoprotein, is involved in the assembly
of the mature spliceosome (12).
USP39 is required to maintain the spindle checkpoint and sustain
successful cytokinesis during the splicing of Aurora B (13). Mutation of USP39 in zebrafish has
been shown to cause Rb1 mRNA splicing defects and pituitary lineage
expansion (14). In addition, a
previous study indicated that USP39 is overexpressed in human
breast cancer tissues compared with that in normal breast tissues,
and lentivirus-mediated suppression of USP39 inhibited the growth
of breast cancer cells in vitro (15). Furthermore, USP39 was found to be a
target protein of SUMO and mutation of its SUMOylation sites (K6,
K16, K29, K51 and K73) was observed to promote the effects of USP39
on the proliferation of prostate cancer cells (16). However, the role of USP39 in GC has
remained largely elusive.
In the present study, the expression of USP39 in the
MGC80-3 GC cell line was knocked down using a lentivirus stably
expressing small hairpin (sh)RNA targeting USP39 and the resulting
effects on the proliferation, colony formation capacity and cell
cycle were investigated. Knockdown of USP39 was found to markedly
reduce the proliferation and colony formation capacity of
MGC80-3-cells and to induce G2/M-phase arrest through regulating
the cleavage of poly(adenosine diphosphate ribose) polymerase
(PARP).
Materials and methods
Cell culture
MGC80-3, SGC-7901 and AGS human gastric cancer cell
lines, and the 293T human embryonic kidney cell line were purchased
from the Cell Bank of the Chinese Academy of Science (Shanghai,
China). The MGC80-3 and SGC-7901 cell lines were cultured in 1640
medium (Hyclone; GE Healthcare Life Sciences, Logan, UT, USA), and
AGS were cultured in F12 medium (Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum
(FBS; Biological Industries, Kibbutz Beit-Haemek, Israel) and the
293T cell line was cultured in Dulbecco's modified Eagle's medium
(Hyclone) containing 10% FBS. Cells were kept in a humidified
atmosphere containing 5% CO2 at 37°C.
Lentiviral vector construction and
transfection
The construction of vectors was performed previously
described (17) shRNA targeting
the USP39 sequence and a scrambled control shRNA were cloned into
the pFH-L lentiviral vector (Shanghai Hollybio, Shanghai, China).
Briefly, DNA oligonucleotides were synthesized, annealed and
inserted into the pFH-L vector by double digestion sites
NheI and PacI (Takara Biotechnology Co., Ltd.,
Dalian, China), and ligated with T4 DNA ligase (Takara
Biotechnology Co., Ltd.) according to the manufacturer's protocol.
The ligation products were transformed in E. coli competent
cells (Beijing Transgen Biotech Co., Ltd., Beijing, China), which
were then cultured on coated plates and selected for monoclones.
Recombination of lentiviral expression vectors were confirmed by
DNA sequencing. Their target sequences, synthesized by Genewiz,
Inc. (South Plainfield, NJ, USA) were as follows: shUSP39,
5′-GATTTGGAAGAGGCGAGATAACTCGAGTTATCTCGCCTCTTCCAAATCTTTTT-3′; and
shCon,
5′-GCGGAGGGTTTGAAAGAATATCTCGAGATATTCTTTCAAACCCTCCGCTTTTTT-3′.
For lentivirus production, 293T cells were
transfected with 10 µg pFH-L lentiviral vectors along with
7.5 µg envelope plasmid pVSVG-I (Shanghai Hollybio) and 5
µg packaging plasmid pCMVΔR8.92 (Shanghai Hollybio) via
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.).
After 48 h of incubation, the culture medium containing the
lentivirus was collected.
To perform USP39 knockdown in MGC80-3 cells,
5×104 cells were cultured in six-well plates and the
USP39 shRNA-expressing lentivirus (shUSP39) or scrambled control
shRNA-expressing lentivirus (shCon) was added with a multiplicity
of infection of 60. After 96 h of transfection, cells were observed
by fluorescence microscopy (#CKX41; Olympus, Tokyo, Japan).
Positive cells were identified based on the expression of green
fluorescence protein (GFP).
Reverse-transcription quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells using TRIzol
reagent (Invitrogen). RT reaction was performed using 2 µg
total RNA in a reaction mixture containing 2 µl oligo dT
primers (50 µM), 4 µl 5X Moloney-Murine Leukemia
Virus (M-MLV) buffer, 1 µl dNTPs (10 mM), 0.5 µl
RNasin, 0.5 µl M-MLV RT (RNase H-) and nuclease-free water
in a total volume of 20 µl. The reaction mixture was
incubated at 42°C for 1 h, 75°C for 15 min, then refrigerated on
ice according to the M-MLV RT protocol. RT reagents was obtained
from Promega Corporation (Madison, WI, USA).
PCR was performed using a CFX96 Touch Real-Time PCR
detection system (Bio-Rad Laboratories, Inc., Hercules, CA, USA)
according to the manufacturer's instructions. The primers used
(synthesized by Genewiz, Inc.) were as follows: USP39 forward,
5′-GCCAGCAGAAGAAAAAGAGC-3′ and reverse, 5′-GCCATTGAACTTAGCCAGGA-3′;
ACTB forward, 5′-GTGGACATCCGCAAAGAC-3′ and reverse,
5′-AAAGGGTGTAACGCAACTA-3′. Forward and reverse primers were mixed
and diluted to 2.5 µM. The PCR reaction mixture contained
0.8 µl primers, 5 µl cDNA (30 ng/µl), 10
µl 2X SYBR Premix Ex Taq (Takara Biotechnology Co., Ltd.)
and 4.2 µl RNA-free water in a total volume of 20 µl.
All reactions were performed in triplicate. Thermal cycling
conditions comprised initial denaturation at 95°C for 1 min,
followed by 40 cycles of denaturation at 95°C for 5 sec and
annealing extension at 60°C for 20 sec. Expression levels were
normalized to the internal control ACTB. Relative quantification
was performed using the 2−ΔΔCq method (18).
Western blot analysis
MGC80-3 cells were harvested after lentiviral
transfection for five days. Cells were washed with
phosphate-buffered saline (PBS; Sangon Biotech Co., Ltd., Shanghai,
China) and lysed with ice-cold 2X sodium dodecyl sulfate (SDS)
lysis buffer containing 100 mM Tris-HCl, pH 6.8, 10 mM
ethylenediaminetetraacetic acid, 4% sodium dodecyl sulfate (SDS)
and 10% glycine (all from Sangon Biotech Co., Ltd.), followed by 30
min of incubation on ice and centrifugation at 10,800 × g for 5 min
at 4°C. The protein concentration was determined using the
bicinchoninic acid protein assay (Beyotime Institute of
Biotechnology Inc., Haimen, China). Protein extracts (30
µg/lane) were separated on a 10% SDS-polyacrylamide gel
(Genscript, Nanjing, China) and transferred onto a polyvinylidene
difluoride membrane (EMD Millipore, Billerica, MA, USA). After
blocking with 5% non-fat milk in Tris-buffered saline containing
Tween 20 (TBS-T; Sigma-Aldrich, St. Louis, MO, USA) for 1 h at room
temperature, the membranes were incubated with rabbit anti-USP39
antibody (cat. no. 23865-1-AP; 1:1,000 dilution; Proteintech Group,
Inc., Chicago, IL, USA) or rabbit anti-glyceraldehyde-3-phosphate
dehydrogenase antibody (cat. no. 10494-1-AP; 1:100,000 dilution;
Proteintech Group, Inc.) at 4°C overnight and washed 3 times with
TBS-T. Subsequently, membranes were incubated with horseradish
peroxidase (HRP)-conjugated goat anti-rabbit immunoglobulin G (cat.
no. SC-2054; 1:5,000 dilution; Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA) and blots were developed with enhanced
chemiluminescence reagent (RPN2132; GE Healthcare, Little Chalfont,
UK) and exposed to X-ray film (Kodak, Rochester, NY, USA).
Cell proliferation assay
Cell proliferation was assessed using a
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay. After transfection with lentivirus for four days, MGC80-3
cells stably transfected with shCon or shUSP39 (2,000 per well)
were cultured in 96-well plates with five parallel wells per
condition. On days 1, 2, 3, 4 and 5, 20 µl MTT (5 mg/ml in
PBS) was added, followed by incubation for 4 h. The reaction was
terminated by addition of acidic isopropanol [10% SDS, 5%
isopropanol (Sangon Biotech Co., Ltd.) and 0.01 mol/l HCl (Sangon
Biotech Co., Ltd.)], and the absorbance was measured at a
wavelength of 595 nm using an Epoch microplate reader (BioTek
Instruments, Inc., Winooski, VT, USA).
Colony formation assay
Following four days of lentiviral transfection,
MGC80-3 cells were trypsinized, re-suspended and seeded into
six-well plates at a concentration of 400 cells/well. Cells were
cultured for seven days with replacement of the media every two
days. Subsequently, the cells were washed with PBS twice and fixed
with 4% paraformaldehyde (Sigma-Aldrich). Following two further
washes with PBS, cells were stained with 700 µl crystal
violet (Beyotime Institute of Biotechnology) for 5 min and washed
three times with PBS. The colonies were counted under an Olympus
CH2 microscope (Olympus Corporation, Tokyo, Japan). Three
independent assays were performed.
Cell cycle analysis
Cell cycle analysis was performed using flow
cytometry. After lentiviral infection for four days, MGC80-3 cells
(2×105 per dish) were seeded into 6-cm dishes and
incubated for three days until a confluency of 70% was reached.
Following two washes with cold PBS, cells were fixed in ice-cold
75% ethanol (Sinopharm Chemical Reagent Co., Ltd., Shanghai, China)
overnight at 4°C. Following washing in pre-cooled PBS, cells were
stained with propidium iodide (Sigma-Aldrich) for 1 h at 37°C in
the dark. Cell cycle distributions were determined using a Gallios
flow cytometer (Beckman Coulter, Inc., Brea, CA, USA). All
experiments were performed in triplicate.
Intracellular signaling array
Following lentiviral transfection for five days,
MGC80-3 cells were lysed in Cell Lysis Buffer (cat. no. 7018; Cell
Signaling Technology, Inc., Danvers, MA, USA). Cell lysates were
assayed using the PathScan Intracellular Signaling Array kit (cat.
no. 7323; Cell Signaling Technology, Inc.) for simultaneous
detection of phosphorylation or cleavage of 18 important and
well-characterized signaling molecules according to the
manufacturer's instructions. In brief, the lysate was diluted to
0.2 mg/ml in array diluent buffer (Cell Signaling Technology,
Inc.), and 75 µl lysate was added onto a
nitrocellulose-coated glass slide pre-coated with capture
antibodies. Following incubation of the slide overnight at 4°C,
biotinylated detection antibody cocktail was added, following
incubation for 1 h at room temperature. The slide was then washed 3
times with 1X array wash buffer (Cell Signaling Technology, Inc.)
for 5 min at room temperature. Subsequently, HRP-linked
streptavidin was added and the plate was incubated for 30 min at
room temperature. Finally, chemiluminescent substrate was added and
the signals were detected using X-ray film.
Statistical analysis
Values are expressed as the mean ± standard
deviation. Statistical analysis was performed using GraphPad Prism
version 5 (GraphPad Software, Inc., La Jolla, CA, USA) Statistical
significance was determined using the two-tailed Student's
t-test. P<0.05 was considered to indicate a statistically
significant difference between values.
Results
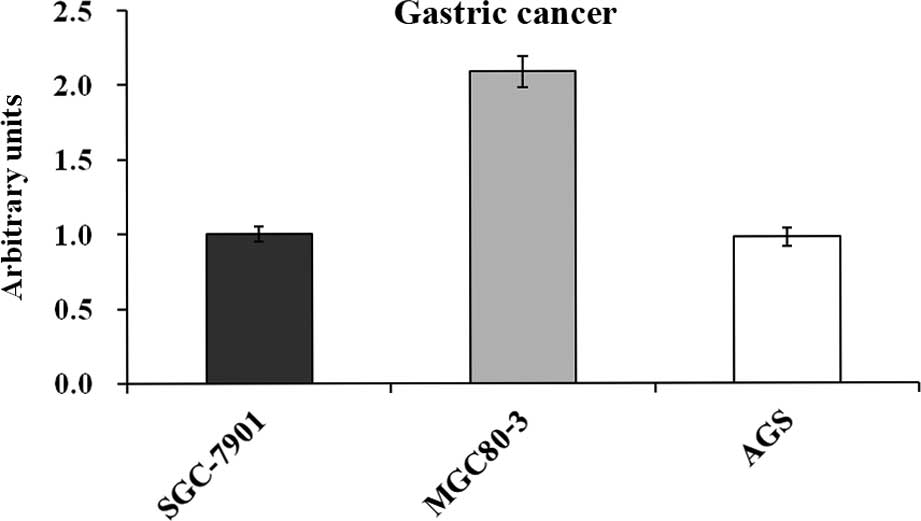
MGC80-3 cells express USP39 mRNA at high
levels
Since USP39 levels had not previously been assessed
in gastric cancers, the present study used RT-qPCR to determine
USP39 expression in three human GC cell lines. As shown in Fig. 1, USP39 was expressed in the
SGC-7901, MGC80-3 and AGS cell lines, with MGC80-3 cells expressing
USP39 at markedly higher levels than the other two cell lines.
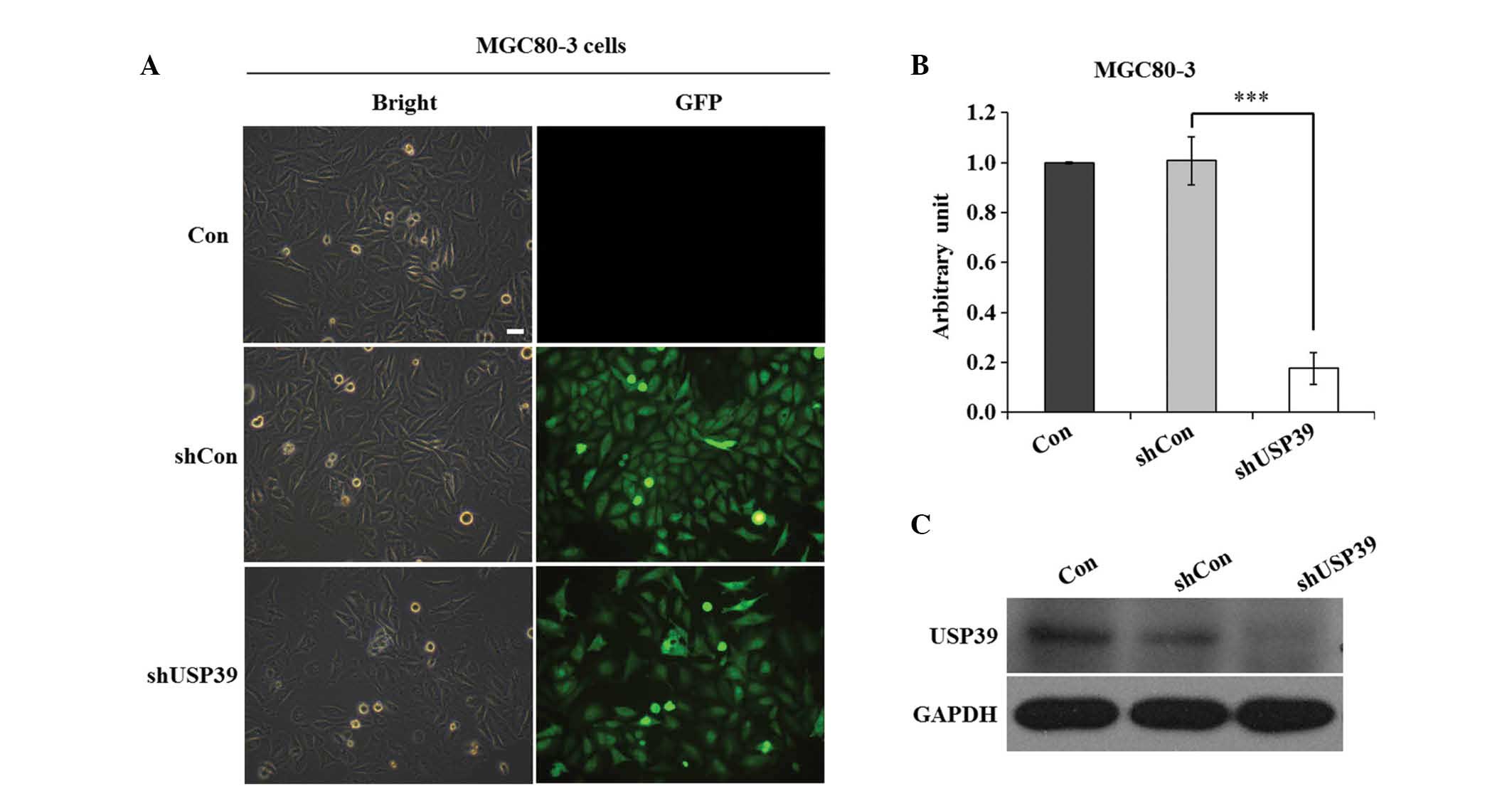
Knockdown of USP39 in MGC80-3 cells with
the shRNA lentivirus system
To elucidate the functional role of USP39 in gastric
cancer, sequences encoding for the expression of shUSP39 or shCon
were cloned into the pFH-L lentiviral vector. The shUSP39 and shCon
lentiviruses expressing GFP were generated and individually
transfected into MGC80-3 cells. The transfection efficiency of the
lentivirus was 95% after four days of transfection (Fig. 2A). RT-qPCR analysis revealed that
transfection with shUSP39 reduced the mRNA levels of USP39 by 83%
(Fig. 2B). In addition, western
blot analysis revealed that in MGC80-3 cells transfected with the
shUSP39 construct, the protein levels of USP39 were obviously
reduced (Fig. 2C). These results
suggested that the lentivirus stably expressing shRNA targeting
USP39 was successfully constructed and transfected, and that it
efficiently suppressed the expression of USP39.
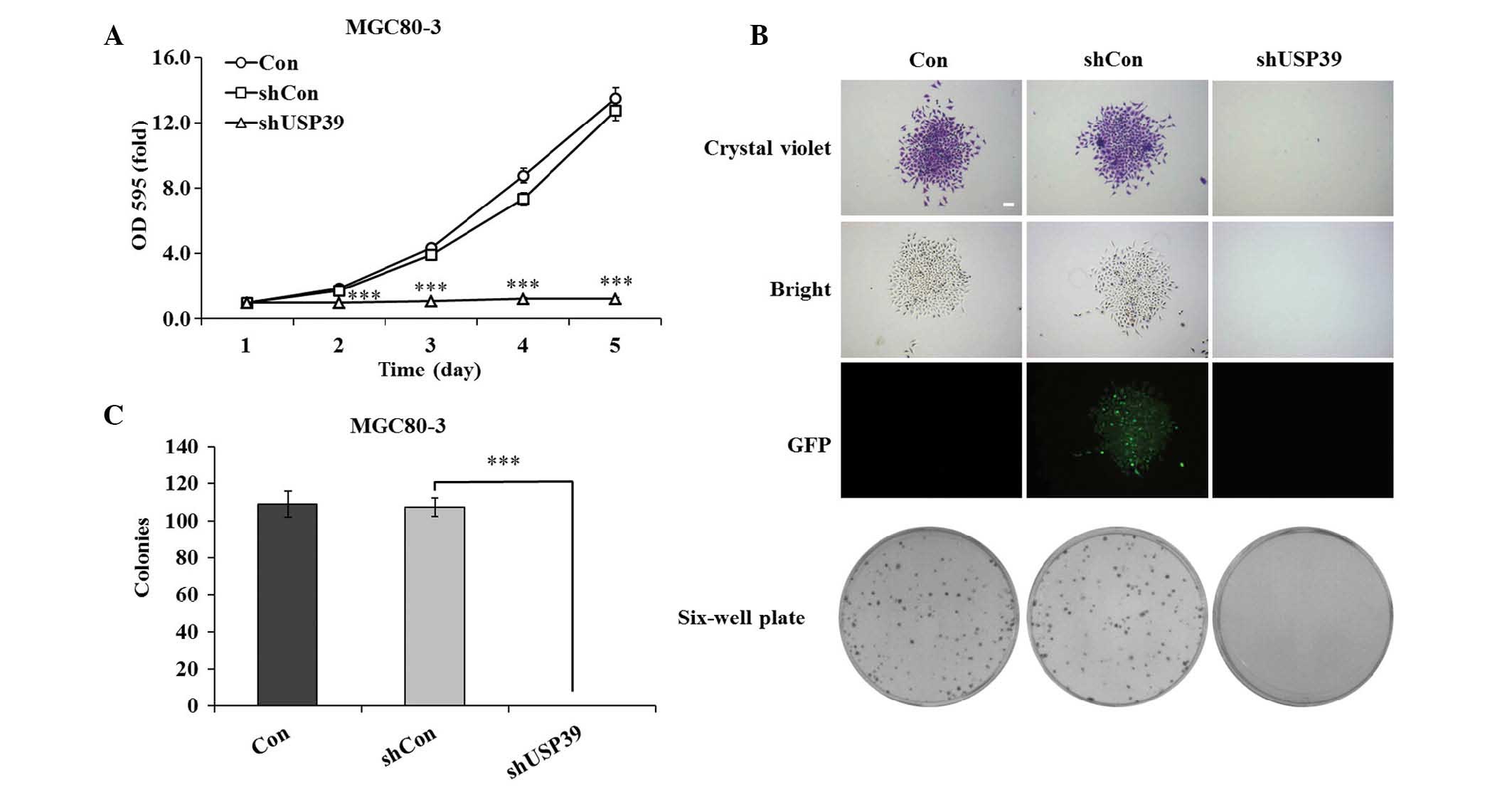
USP39 knockdown inhibits GC-cell
proliferation and colony formation
To investigate the effects of lentivirus-mediated
downregulation of USP39 on the growth of GC cells, MGC80-3 cell
proliferation was assessed using an MTT assay. As shown in Fig. 3A, USP39 knockdown completely
inhibited the proliferation of MGC80-3 cells at days 2–5, while
proliferation in the shCon group was not significantly different
from that in the Con group (P<0.001). On days four and five,
cell growth in the USP30 knockdown group was reduces by 83.2 and
90.5%, respectively, compared with that in the control groups.
Furthermore, a clonogenic assay showed that in the shUSP39 group,
colony formation was completely inhibited, as no colonies were
formed over the assay period of seven days, while colony size in
the shCon group was equal to that in the Con group (Fig. 3B). In addition, the number of
colonies in the shCon and Con groups was similar, while no colonies
were formed in the shUSP39 group (Fig.
3C). These findings indicated that the knockdown of USP39
completely inhibited the cell proliferative ability and colony
formation capacity of GC cells.
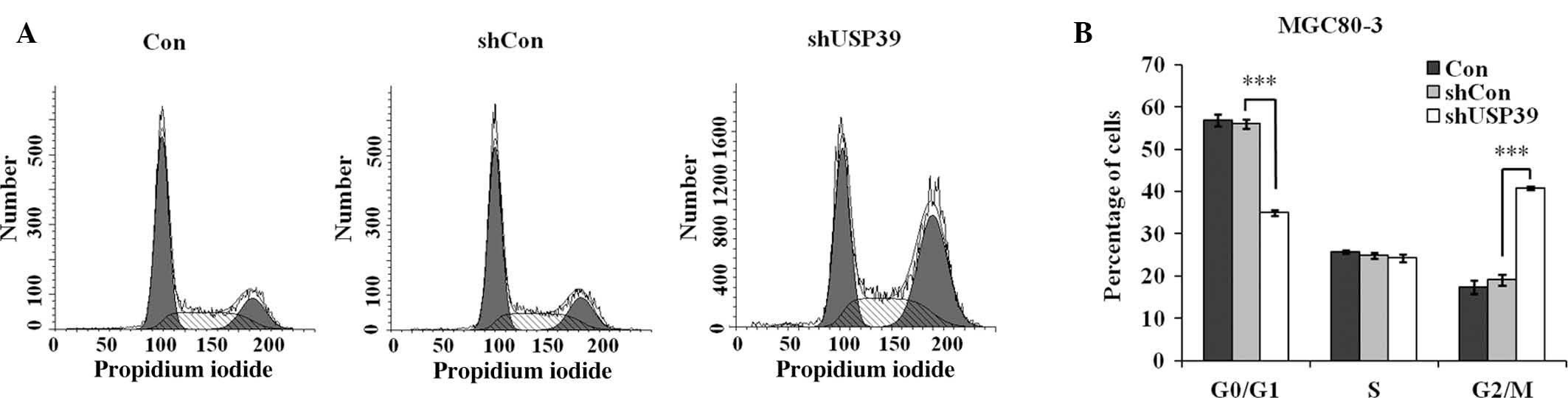
Suppression of USP39 causes cell cycle
arrest in G2/M phase
Based on the observed repression of MGC80-3-cell
proliferation and colony formation by shUSP39, flow cytometric
analysis was performed to observe the effects of USP39 on cell
cycle progression (Fig. 4A). The
results demonstrated that after transfection with shUSP39
lentivirus, the G0/G1-phase population accounted for 35.0%, which
was obviously decreased compared to that in the shCon group (56.1%;
P<0.001), while the G2/M-phase population in the shUSP39 group
was increased (40.8 vs. 19.1% in shCon group; P<0.001) (Fig. 4B). This result suggested that USP39
depletion causes G2/M-phase arrest in GC cells.
USP39 silencing induces the cleavage of
PARP
To reveal the mechanisms governing the inhibitory
effects of shUSP39 on cell growth, the PathScan Intracellular
Signaling Array kit was used to detect 18 important and
well-characterized signaling molecules in the shUSP39- or
shCon-transfected MGC80-3 cells. The results showed that the
cleavage of PARP at Asp214 was markedly enhanced in the
shUSP39-transfected MGC80-3 cells, indicating that the cleavage of
PARP may be involved in shUSP39-mediated growth suppression in GC
cells.
Discussion
Understanding the molecular aberrations behind the
initiation and progression of GC is important in finding novel
molecular markers for early diagnosis, targeted treatment and
prognosis evaluation. Overexpression of USP39 has been reported in
human breast cancers, and inhibition of USP39 repressed the growth
of breast cancer cells (15).
However, to date, the functional role of USP39 in GC has remained
elusive.
To the best of our knowledge, the present study was
the first to shed light on the functional role of USP39 in GC. A
lentiviral shRNA system was used to effectively inhibit the
expression of USP39 at the mRNA and protein level. RT-qPCR and
western blot analysis showed efficient silencing of USP39. An MTT
assay and a colony formation assay were then used to identify the
effects of USP39 knockdown on GC-cell proliferation. USP39 deletion
was shown to completely inhibit the proliferation and colony
formation ability of MGC80-3 cells. The present study also revealed
that knockdown of USP39 caused G2/M-phase arrest in MGC80-3 cells.
Furthermore, the PathScan Intracellular Signaling Array indicated
that inhibition of USP39 increased the cleavage of PARP.
USP39, containing a central zinc finger and two
ubiquitin C-terminal hydrolase domains, belongs to the
ubiquitin-specific protease family (14). Previous studies have shown that
USP39 is overexpressed in breast cancer tissues and that knockdown
of USP39 obviously inhibits the proliferative and colony formation
ability of MCF-7 breast cancer cells. In addition, USP39 knockdown
was revealed to induce G0/G1-phase arrest and apoptosis of breast
cancer cells (15). However, the
present study showed that shUSP39 effectively inhibited GC-cell
growth and colony formation, possibly by causing cell cycle arrest
in G2/M phase. Recently, Wen et al (16) indicated that the overexpression of
USP39 increased the proliferation of the androgen-independent PC3
and androgen-dependent LNCaP prostate cancer cells. Of note,
mutation of the SUMOylation sites of USP39 was demonstrated to
further strengthen its ability to promote the proliferation of
prostate cancer cells. These results suggested that SUMO
modifications have a significant role in the function of USP39.
To reveal the underlying mechanisms of the effects
of shUSP39 on GC-cell growth, the present study screened for
intracellular signaling target proteins by using a high-throughput
proteomics method. The cleavage of PARP (Asp214) was found to be
obviously increased in shUSP39-transfected MGC80-3 cells. PARP1,
also known as PARP, is involved in DNA repair. As USP39 is required
to maintain the spindle checkpoint and sustain successful
cytokinesis during splicing (13,14),
it is likely that loss of USP39 led to a halt or dysregulation of
mitosis, which explains for the observed G2/M-phase arrest. PARP
may have been activated due to DNA damage or faulty splicing in the
absence of USP39. Indeed, PARP, which is involved in DNA repair, is
cleaved during apoptosis. The cleavage of PARP often occurs between
Asp-214 and Gly-215 and has been demonstrated to be an early marker
of apoptosis (19). PARP-1
762Ala/Ala has been indicated to be a risk factor for GC in a Han
Chinese population (20), and the
PARP1 rs1136410 genotype has been correlated with the risk of lymph
node metastasis and tumor invasion in GC (21). It was previously reported that PARP
expression in yeast was increased during the G2/M phase of the cell
cycle (22). Furthermore, G2/M
arrest was enhanced by PARP inhibitors in mammalian cells (23,24).
However, the underlying mechanism of PARP activity in the
regulation of G2/M phase arrest remains unclear and requires
clarification in future studies. Taken together, the results of the
present study indicated that USP39 knockdown caused a de-regulation
of mitosis, resulting in G2/M-phase arrest and cleavage of PARP
(Asp214) for DNA damage repair, leading to total inhibition of GC
cell proliferation. However, it remains to be clarified whether
inhibition of USP39 affects normal cells in patients, leading to
considerable side effects (13,14).
In conclusion, the present study was the first to
reveal that shRNA-mediated knockdown of USP39 inhibited the growth
and colony formation ability of GC cells. Furthermore, suppression
of USP39 induced G2/M-phase arrest and increased the cleavage of
PARP (Asp214). The present study suggested that USP39 is crucial
for the proliferation of GC cells. Due to its upregulated
expression in certain cancer types and with overproliferation being
a hallmark of cancer, USP39 may be targeted for the treatment of
cancer. Future studies assessing USP39 in normal and cancer cells
will reveal whether USP39 is a feasible and cancer-specific
molecular target.
Acknowledgments
The present study was supported by grants from the
Nature & Science Foundation of Zhejiang Province (no. Y205757),
the Medical Research Fund of Zhejiang Province (no. 2009A029) and
the Outstanding Research Personnel Training Funds of Zhejiang
Cancer Hospital.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nishida T, Tsutsui S, Kato M, Inoue T,
Yamamoto S, Hayashi Y, Akasaka T, Yamada T, Shinzaki S, Iijima H,
et al: Treatment strategy for gastric non-invasive intraepithelial
neoplasia diagnosed by endoscopic biopsy. J Gastrointest
Pathophysiol. 2:93–99. 2011. View Article : Google Scholar
|
|
3
|
Layke JC and Lopez PP: Gastric cancer:
Diagnosis and treatment options. Am Fam Physician. 69:1133–1140.
2004.PubMed/NCBI
|
|
4
|
Cheng L, Wang P, Yang S, Yang Y and Zhang
Q, Zhang W, Xiao H, Gao H and Zhang Q: Identification of genes with
a correlation between copy number and expression in gastric cancer.
BMC Med Genomics. 5:142012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Brenner H, Rothenbacher D and Arndt V:
Epidemiology of stomach cancer. Methods Mol Biol. 472:467–477.
2009. View Article : Google Scholar
|
|
6
|
Yeh ET: SUMOylation and De-SUMOylation:
Wrestling with life's processes. J Biol Chem. 284:8223–8227. 2009.
View Article : Google Scholar :
|
|
7
|
Nie M, Xie Y, Loo JA and Courey AJ:
Genetic and proteomic evidence for roles of Drosophila SUMO in cell
cycle control, Ras signaling and early pattern formation. PLoS One.
4:e59052009. View Article : Google Scholar
|
|
8
|
Johnson ES: Protein modification by SUMO.
Annu Rev Biochem. 73:355–382. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang W and Paschen W: SUMO proteomics to
decipher the SUMO-modified proteome regulated by various diseases.
Proteomics. 15:1181–1191. 2015. View Article : Google Scholar :
|
|
10
|
Nijman SM, Luna-Vargas MP, Velds A,
Brummelkamp TR, Dirac AM, Sixma TK and Bernards R: A genomic and
functional inventory of deubiquitinating enzymes. Cell.
123:773–786. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wilkinson KD: Regulation of
ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J.
11:1245–1256. 1997.PubMed/NCBI
|
|
12
|
Makarova OV, Makarov EM and Lührmann R:
The 65 and 110 kDa SR-related proteins of the U4/U6.U5 tri-snRNP
are essential for the assembly of mature spliceosomes. EMBO J.
20:2553–2563. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
van Leuken RJ, Luna-Vargas MP, Sixma TK,
Wolthuis RM and Medema RH: Usp39 is essential for mitotic spindle
checkpoint integrity and controls mRNA-levels of aurora B. Cell
Cycle. 7:2710–2719. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rios Y, Melmed S, Lin S and Liu NA:
Zebrafish usp39 mutation leads to rb1 mRNA splicing defect and
pituitary lineage expansion. PLoS Genet. 7:e10012712011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang H, Ji X, Liu X, Yao R, Chi J, Liu S,
Wang Y, Cao W and Zhou Q: Lentivirus-mediated inhibition of USP39
suppresses the growth of breast cancer cells in vitro. Oncol Rep.
30:2871–2847. 2013.PubMed/NCBI
|
|
16
|
Wen D, Xu Z, Xia L, Liu X, Tu Y, Lei H,
Wang W, Wang T, Song L, Ma C, et al: Important role of SUMOylation
of Spliceosome factors in prostate cancer cells. J Proteome Res.
13:3571–3582. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang X, Yu Q, Zhang Y, Ling Z and Yu P:
Tectonic 1 accelerates gastric cancer cell proliferation and cell
cycle progression in vitro. Mol Med Rep. 12:5897–5902.
2015.PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
19
|
Sallmann FR, Bourassa S, Saint-Cyr J and
Poirier GG: Characterization of antibodies specific for the caspase
cleavage site on poly(ADP-ribose) polymerase: specific detection of
apoptotic fragments and mapping of the necrotic fragments of
poly(ADP-ribose) polymerase. Biochem Cell Biol. 75:451–456. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang Q, Li Y, Li X, Zhou W, Shi B, Chen H
and Yuan W: PARP-1 Val762Ala polymorphism, CagA+ H. pylori
infection and risk for gastric cancer in Han Chinese population.
Mol Biol Rep. 36:1461–1467. 2009. View Article : Google Scholar
|
|
21
|
Kim J, Pyun JA, Cho SW, Lee K and Kwack K:
Lymph node metastasis of gastric cancer is associated with the
interaction between poly (ADP-ribose) polymerase 1 and matrix
metallopeptidase 2. DNA Cell Biol. 30:1011–1017. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tao Z, Gao P and Liu HW: Studies of the
expression of human poly(ADP-ribose) polymerase-1 in Saccharomyces
cerevisiae and identification of PARP-1 substrates by yeast
proteome microarray screening. Biochemistry. 48:11745–11754. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nozaki T, Masutani M, Akagawa T, Sugimura
T and Esumi H: Suppression of G1 arrest and enhancement of G2
arrest by inhibitors of poly(ADP-ribose) polymerase: Possible
involvement of poly(ADP-ribosyl)ation in cell cycle arrest
following gamma-irradiation. Jpn J Cancer Res. 85:1094–1098. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Piao L, Nakagawa H, Ueda K, Chung S,
Kashiwaya K, Eguchi H, Ohigashi H, Ishikawa O, Daigo Y, Matsuda K
and Nakamura Y: C12orf48, termed PARP-1 binding protein, enhances
poly(ADP-ribose) polymerase-1 (PARP-1) activity and protects
pancreatic cancer cells from DNA damage. Genes Chromosomes Cancer.
50:13–24. 2011. View Article : Google Scholar
|