Introduction
Deafness is the partial or total inability to hear
(1). In adults, it causes problems
in communication (2). In children,
as well as problems in communication, deafness affects the
development of language. Several factors may cause deafness
(3,4), including genetic mutations, noise
exposure, illness, toxic chemicals and aging. A substantial
proportion of cases of deafness are caused by genetic mutations
(4–7). Genetic deafness can be divided into
two categories: Syndromic and nonsyndromic deafness. The clinical
phenotypes of genetic deafness remain to be fully elucidated and
the same mutation may result in different clinical phenotypes in
different individuals.
Mutations in genes, including the GJB3 (8), GJB2 (9), SLC26A4 (10) and 12S RNA genes (11), have been previously associated with
genetic deafness. Large vestibular aqueduct syndrome (LVAS) is one
type of genetic deafness that can be caused by genetic mutations in
the SLC26A4 gene (12).
Conversely, mutations in the GJB3 gene have been associated with
nonsyndromic deafness (DFNA2B), which is inherited in an autosomal
dominant pattern (13,14). The clinical phenotypes, including
age of onset and the hearing loss severity, of mutations in the
GJB3 gene vary. Whether there is a correlation between genetic
deafness and mutations in the GJB3 and SLC26A4 genes remains to be
elucidated.
In the present study, mutations in the GJB3, GJB2,
SLC26A4 and 12S RNA genes were screened in a family with genetic
deafness. The screening identified mutations in the GJB3 and
SLC26A4 genes of the deaf patient, from which the relative clinical
information were examined and discussed. The results of the present
study may enable the establishment of genetic counseling specific
to deaf patients harboring mutations in the GJB3 and SLC26A4
genes.
Materials and methods
Patients and genomic DNA isolation
Whole blood samples were obtained from the study
family at the First People's Hospital of Yunnan Province (Kunming,
China) during their visit to the clinical center on 13th August
2013. The affected individual exhibited hearing loss, whereas
neither the father nor the mother of the affected child showed
signs of hearing loss or impairment. The present study was approved
by the Ethics Committee of the First People's Hospital of Yunnan
Province (Affiliated Hospital of Kunming University of Science and
Technology). Written informed consent was signed and obtained from
the participants. Genomic DNA from the whole blood samples were
extracted using a DNA extraction kit (cat. no. ZTLYQ-F50; Xi'an
Tianlong Science & Technology Co., Ltd., China), according to
the manufacturer's protocol.
Polymerase chain reaction (PCR)
amplification and sequencing
The GJB2, GJB3, SLC26A4 and 12S rRNA gene fragments
were amplified using PCR. The primers used for amplification and
sequencing of the fragments are shown in Table I and were obtained from the Beijing
Genomics Institute (Shenzhen, China). The amplification of the
fragments in the 12S rRNA gene referred to the descriptions by Wang
et al (15), and was
performed in a 25 µl reaction mixture containing 10X LA PCR
Buffer II (Mg2+ Plus), 2.5 units of Takara LA Taq
(Takara Biotechnology Co., Ltd., Dalian, China), 400 µM of
each dNTP, 0.2 µM of each primer and 50 ng DNA. The
following conditions were used for PCR amplification: Denaturation
cycle at 94°C for 1 min, followed by 30 cycles of denaturation at
94°C for 30 sec and 65.6°C for 5 min, and ended with a final
extension at 72°C for 10 min. Amplification of the GJB2, GJB3 and
SLC26A4 gene fragments was performed in a volume of 25 µl
containing 30 ng genomic DNA, 50 µM dNTP, 10X LA TaqTM PCR
buffer, 2.5 units of Takara LA TaqTM and 0.2 µM of each
forward and reverse primer. The PCR amplification for these gene
fragments followed a denaturation cycle of 94°C for 5 min, followed
by 35 cycles of denaturation at 94°C for 30 sec, annealing at 55°C
for SLC26A4 exon10, 58°C for GJB3, 59°C for SLC26A4 exon19, 63°C
for SLC26A4 intron7 and exon7+8, or 60°C for GJB2, and extension at
72°C for 30 sec for SLC26A4 exon10, 35 sec for SLC26A4 exon 19, 40
sec for SLC26A4 intron 7 and exon 7+8, 55 sec for GJB3 or 60 sec
for GJB2, and ended with a final extension step at 72°C for 7
min.
 | Table IPrimers used for amplification of
GJB2, GJB3, SLC26A4 and 12S RNA gene fragments. |
Table I
Primers used for amplification of
GJB2, GJB3, SLC26A4 and 12S RNA gene fragments.
| Author (year) | Gene | Primer | Primer sequence
(5′-3′) | Size | Use | Refs. |
|---|
| Wang et al
(2008) | 12S RNA | mt-H2187 |
TGTTGAGCTTGAACGCTTTCTTAATTGGTG | >4 kb | PCR | (15) |
| Wang et al
(2008) | | mt-L13894 |
ACTTAAAATAAAATCCCCACTATGCACAT | | PCR | (15) |
| Wang et al
(2008) | | mt-L1156 |
GAACACTACGAGCCACAGC | | Sequencing | (15) |
| Bicego et al
(2006) | GJB2 | GJB2F |
TGCTTACCCAGACTCAGAGAA | 864 bp | PCR; Sequencing | (7) |
| Bicego et al
(2006) | | GJB2R |
GACTGAGCCTTGACAGCTGAG | | PCR; Sequencing | (7) |
| Liu et al
(2009) | GJB3 | GJB3F |
TACGATGGTTTTTCCTCTAATTCT | 986 bp | PCR | (14) |
| Liu et al
(2009) | | GJB3R |
TTGCATAACTTAGTGAACTCAGAG | | PCR | (14) |
| Li and Zhu | | GJB3S |
ACACCGCCCTGCATGTCCCAT | | Sequencing | Present |
| Li and Zhu | SLC26A4 intron 7 and
exon 7+8 | SLC78F |
GAGTGTTGTTTGATGCTGAT | 670 bp | PCR | Present |
| Li and Zhu | SLC78S |
GCTGCTTTTAAACAAATGGC | | PCR | Present |
| Li and Zhu | | SLC78R1 |
GTTTCTTCCAGATCACACAC | | Sequencing | Present |
| Li and Zhu | SLC26A4 exon 10 | SLC10F |
GTCCAAACTCCTGATGTCGT | 447 bp | PCR | Present |
| Li and Zhu | | SLC10R2 |
CCACAGCAGGTAAGTGTAGC | | PCR | Present |
| Li and Zhu | | SLC10R1 |
GGGAGTGGAACAAGAGGAAT | | Sequencing | Present |
| Li and Zhu | SLC26A4 exon 19 | SLC19F |
GCTAATTGGGAGGGTGAGGT | 551 bp | PCR | Present |
| Li and Zhu | | SLC19R2 |
CAGTACTGGGTACTACCAGG | | PCR; Sequencing | Present |
The PCR products were purified using a Genomic DNA
Purification kit (cat. no. DP204-02; Tiangen Biotech Co., Ltd.,
Tiangen, China) and were sequenced using the sequencing primers
(Table I) and Big Dye Terminator
v.3.1 Cycle Sequencing kit (cat. no. 4337456; Applied Biosystems;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) on an ABI Prism
3730 DNA sequencer (Applied Biosystems; Thermo Fisher Scientific,
Inc.).
Evolutionary conservation analysis
Evolutionary conservation analysis was performed by
aligning amino acid sequences of the GJB3 and SLC26A4 proteins of
eight vertebrate species from the GenBank database (http://www.ncbi.nlm.nih.gov/genbank/),
as follows: Homo sapiens (GJB3, GenBank accession no.
NP_001005752.1; SLC26A4 GenBank accession no. NP_000432.1),
Macaca mulatta (GJB3, GenBank accession no.
XP_001108150.1;SLC26A4 GenBank accession no. XP_001094049.1),
Canis familiaris (GJB3, GenBank accession no.
XP_005629030.1;SLC26A4 GenBank accession no. XP_005631003.1),
Bos taurus (GJB3, GenBank accession no.
NP_001098465.1;SLC26A4 GenBank accession no, XP_002686849.2),
Mus musculus (GJB3, GenBank accession no.
NP_001153484.1;SLC26A4 GenBank accessio no. NP_035997.1), Rattus
norvegicus (GJB3, GenBank accession no. NP_062113.1;SLC26A4
GenBank accession no. NP_062087.1), Gallus gallus (GJB3,
GenBank accession no. XP_004947838.1; SLC26A4 GenBank accession no.
XP_425419.3), and Danio rerio (GJB3, GenBank accession no.
NP_001017685.1; SLC26A4 GenBank accession no. NP_001159387.1).
Results and Discussion
In the present study, the 12S rRNA gene, exons 7, 8,
10 and 19, and intron 7 of the SLC26A4 gene, and the entire coding
region of the GJB3 and GJB2 genes, were examined by direct
sequencing in a Chinese family comprising a child with LVAS
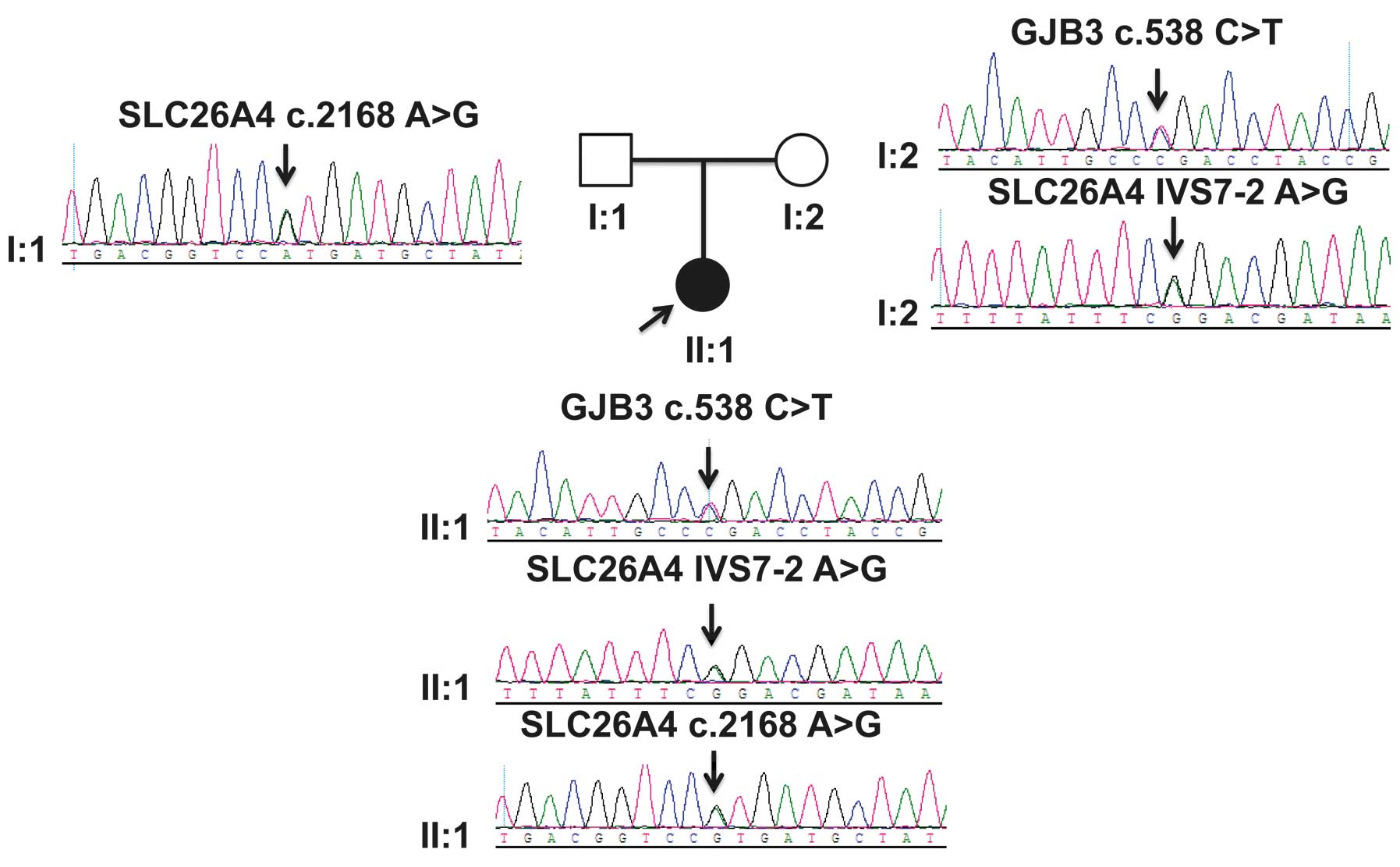
(Fig. 1). The family was from the
Yunnan province of southwestern China. Chromosome examination of
the affected individual (subject II:1) showed no chromosome
aberration (data not shown). The heterozygous mutations of the GJB3
and SLC26A4 genes in the family were identified (Fig. 1). The affected individual (subject
II:1) harbored the following heterozygous mutations of SLC26A4:
IVS7-2 A>G and c.2168 C>T, and the GJB3 mutation, c.538
C>T. The age at onset of deafness was 4 years old. The mutations
confirmed the clinical diagnosis following a computed tomography
scan, since LVAS has been reported to be caused by compound
mutations in SLC26A4 (IVS7-2 A>G and c.2168 C>T) (10,12).
The mother of the affected individual (subject I:2) showed no
hearing loss, but harbored heterozygous mutations of SLC26A4
(IVS7-2 A>G) and GJB3 (c.538 C>T). The GJB3 c.538 C>T
mutation has been shown to be correlated with a form of
nonsyndromic deafness, termed DFNA2B, which is inherited in an
autosomal dominant pattern (13,14).
The time of onset and clinical phenotype of deafness caused by the
GJB3 c.538 C>T mutation varies in different individuals, and
this may explain why subject I:2, who harbored the GJB3 c.538
C>T mutation, did not exhibit any hearing loss. The father of
the affected individual (subject I:1) harbored heterozygous
mutations of SLC26A4 (c.2168 C>T) and did not exhibit any
hearing loss. These findings are consistent with reports that
heterozygous mutations of SLC26A4 c.2168 C>T alone are unable to
cause deafness.
In addition to the three mutations described above,
no other common mutations associated with inheritable nonsyndromic
deafness were identified in the fragments sequenced in the present
study, including c.35delG, c.167delT, c.176_191del16, c.235delC and
c.299_300delAT in the GJB2 gene; c.547 G>A in the GJB3 gene;
IVS7-2 A>G, c.1174 A>T, c.1226 G>A, c.1229 C>T and
c.2162 C>T in the SLC26A4 gene, and m.1494 C>T and m.1555
A>G in the 12S rRNA gene.
The present study also found a number of reported
genetic variants of the GJB3 and GJB2 genes in the family examined,
which were listed in the dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP/). These
variants included rs41310442 in the GJB3 gene (subject I:1, CC;
subject I:2, CT; subject II:1, CT) and rs138547875 in the GJB2 gene
(subject I:1. AG; subject I:2, GG; subject II:1, AG). The SNPs did
not result in the alteration of amino acids. The m.1736 A>G
mutation in the 16S rRNA gene was found in subject I:1, whereas A
was identified at this position in the codon of subjects I:1 and
II:1.
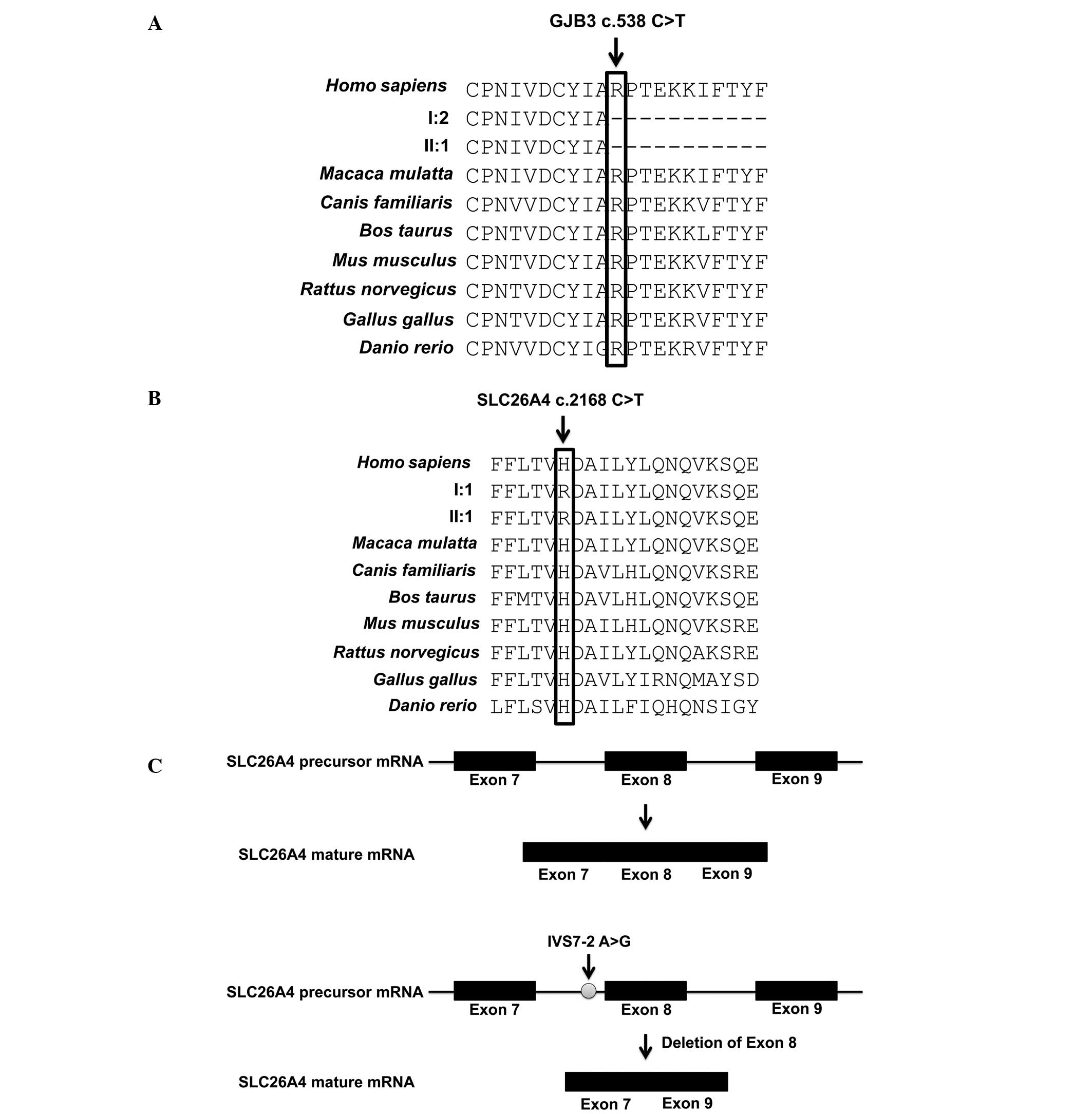
Evolutionary conservation analysis showed that GJB3
180R and SLC26A4 723H were highly conserved in different vertebrate
species (Fig. 2A and B). These
results indicated that mutations in GJB3 180R and SLC26A4 723H may
result in significant functional changes of these proteins. The
splicing site mutation SLC26A4 IVS7-2 A>G can alter the normal
splicing of SLC26A4 precursor mRNA (Fig. 2C). SLC26A4 IVS7-2 A>G results in
the deletion of exon 8 during splicing of the SLC26A4 precursor
mRNA, as shown in Fig. 2C. Taken
together, SLC26A4 IVS7-2 A>G and c.2168 C>T, and GJB3 c.538
C>T disrupt the normal function of SLC26A4 and GJB3
respectively.
In the present study, the affected individual with
LVAS harbored three heterozygous mutations of SLC26A4 IVS7-2
A>G, SLC26A4 c.2168 C>T and GJB3 c.538 C>T. LVAS can be
caused by the combined heterozygous mutations of SLC26A4 IVS7-2
A>G and c.2168 C>T. Whether the GJB3 c.538 C>T mutation
can increase or decrease the hearing loss caused by SLC26A4 IVS7-2
A>G and c.2168 C>T mutations remains to be elucidated. The
affected individual (subject II:1) was examined at the onset time
by multiple auditory steady-state evoked responses (ASRR),
otoacoustic emission (OAE) and auditory brainstem response (ABR)
assessments. In the ASRR test, the right ear passed at 500 and
1,000 HZ, and the auditory thresholds were 80 and 120 dBHL
respectively. The right ear showed no response to 2,000 and 4,000
HZ. The left ear only passed at 4,000 HZ, with an auditory
threshold of 110 dBHL. The left ear showed no response to the tests
at 500, 1,000 or 2,000 HZ. In the ABR assessment, a 95 dBHL click
evoked no ABR waveforms in the left ear and normal ABR waveforms in
the right ear. The auditory threshold of the right ear was 85 dBHL.
Neither the right or left ear passed the transient OAE (TEOAE) or
distortion product OAE assessments. These results indicated that
the combined mutations in the SLC26A4 and GJB3 genes may have
resulted in a severe hearing loss.
Deafness, or hearing loss, is one of the most common
birth defects worldwide. Statistically, 1/500 newborns has
bilateral permanent sensorineural hearing loss (>40 dBHL)
(1). The exact pathologic
mechanism of deafness remains to be fully elucidated. The GJB2,
GJB3, SLC26A4 and 12S RNA genes are considered to be closely
associated with inheritable deafness (16–18).
By examining the nucleotide sequence of these genes in a Chinese
family, a deaf patient carrying combined heterozygous mutations in
the SLC26A4 and GJB3 genes was identified in the present study. The
clinical data of this patient showed severe hearing loss.
Although patients with deafness have been shown to
harbor the same mutations, the clinical phenotype of inheritable
nonsydromic deafness varies (14,19,20).
The results of the present study indicated that combined
heterozygous mutations of the SLC264 and GJB3 genes may result in
severe hearing loss. These results contribute to the understanding
of clinical phenotype of deaf patients carrying combined mutations
in the SLC26A4 and GJB3 genes. These conclusions require further
confirmation in other patients carrying similar mutations. Further
functional investigations are also required to reveal the molecular
mechanism underlying deafness associated with these three
heterozygous mutations in the SLC26A4 and GJB3 genes.
Acknowledgments
The present study was supported by the Education
Commission of Yunnan Province (grant no. 2014Z036), the National
Natural Science Foundation of China (grant nos. 81460424 and
31060155) and the Kunming University of Science and Technology
(grant no. KKZ3201360025). The authors would like to thank the
patients who participated in this study.
References
|
1
|
Lasak JM, Allen P, McVay T and Lewis D:
Hearing loss: Diagnosis and management. Prim Care. 41:19–31. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Oishi N and Schacht J: Emerging treatments
for noise-induced hearing loss. Expert Opin Emerg Drugs.
16:235–245. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Roizen NJ: Nongenetic causes of hearing
loss. Ment Retard Dev Disabil Res Rev. 9:120–127. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Willems PJ: Genetic causes of hearing
loss. N Engl J Med. 342:1101–1109. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Levenson D: New testing guidelines for
hearing loss support next-generation sequencing: Testing method may
help determine genetic causes of hearing loss among patients whose
phenotypes are not easily distinguished clinically. Am J Med Genet
A. 164A:vii–viii. 2014.
|
|
6
|
Wei Q, Wang S, Yao J, Lu Y, Chen Z, Xing G
and Cao X: Genetic mutations of GJB2 and mitochondrial 12S rRNA in
nonsyndromic hearing loss in Jiangsu Province of China. J Transl
Med. 11:1632013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bicego M, Beltramello M, Melchionda S,
Carella M, Piazza V, Zelante L, Bukauskas FF, Arslan E, Cama E,
Pantano S, et al: Pathogenetic role of the deafness-related M34T
mutation of Cx26. Hum Mol Genet. 15:2569–2587. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Frei K, Ramsebner R, Hamader G, Lucas T,
Schoefer C, Baumgartner WD, Wachtler FJ and Kirschhofer K: Lack of
association between Connexin 31 (GJB3) alterations and
sensorineural deafness in Austria. Hear Res. 194:81–86. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kokotas H, Grigoriadou M, Korres GS,
Ferekidou E, Giannoulia-Karantana A, Kandiloros D, Korres S and
Petersen MB: Are GJB2 mutations an aggravating factor in the
phenotypic expression of mitochondrial non-syndromic deafness? J
Hum Genet. 55:265–269. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Smith RJ and Robin NH: Genetic testing for
deafness-GJB2 and SLC26A4 as causes of deafness. J Commun Disord.
35:367–377. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Estivill X, Govea N, Barceló E, Badenas C,
Romero E, Moral L, Scozzri R, D'Urbano L, Zeviani M and Torroni A:
Familial progressive sensorineural deafness is mainly due to the
mtDNA A1555G mutation and is enhanced by treatment of
aminoglycosides. Am J Hum Genet. 62:27–35. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Azaiez H, Yang T, Prasad S, Sorensen JL,
Nishimura CJ, Kimberling WJ and Smith RJ: Genotype-phenotype
correlations for SLC26A4-related deafness. Hum Genet. 122:451–457.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Oh SK, Choi SY, Yu SH, Lee KY, Hong JH,
Hur SW, Kim SJ, Jeon CJ and Kim UK: Evaluation of the pathogenicity
of GJB3 and GJB6 variants associated with nonsyndromic hearing
loss. Biochim Biophys Acta. 1832:285–291. 2013. View Article : Google Scholar
|
|
14
|
Liu XZ, Yuan Y, Yan D, Ding EH, Ouyang XM,
Fei Y, Tang W, Yuan H, Chang Q, Du LL, et al: Digenic inheritance
of non-syndromic deafness caused by mutations at the gap junction
proteins Cx26 and Cx31. Hum Genet. 125:53–62. 2009. View Article : Google Scholar :
|
|
15
|
Wang HW, Jia X, Ji Y, Kong QP, Zhang Q,
Yao YG and Zhang YP: Strikingly different penetrance of LHON in two
Chinese families with primary mutation G11778A is independent of
mtDNA haplogroup background and secondary mutation G13708A. Mutat
Res. 643:48–53. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang S, Wang G, Jiang Y, Yuan Y, Han D,
Song Y and Dai P: Phenotype and genotype of deaf patients with
combined genomic and mitochondrial inheritance models.
Mitochondrion. 13:791–794. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sagong B, Baek JI, Oh SK, Na KJ, Bae JW,
Choi SY, Jeong JY, Choi JY, Lee SH, Lee KY and Kim UK: A rapid
method for simultaneous screening of multi-gene mutations
associated with hearing loss in the Korean population. PLoS One.
8:e572372013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Du W, Cheng J, Ding H, Jiang Z, Guo Y and
Yuan H: A rapid method for simultaneous multi-gene mutation
screening in children with nonsyndromic hearing loss. Genomics.
104:264–270. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Smith RJH, Shearer AE, Hildebrand MS and
Van Camp G: Deafness and hereditary hearing loss overview.
GeneReviews® (Internet). Pagon RA, Adam MP, Ardinger HH, Wallace
SE, Amemiya A, Bean LJH, Bird TD, Fong CT, Mefford HC, Smith RJH
and Stephens K: University of Washington; Seattle, WA: pp.
1993–2016. 1993
|
|
20
|
Bitner-Glindzicz M: Hereditary deafness
and phenotyping in humans. Br Med Bull. 63:73–94. 2002. View Article : Google Scholar : PubMed/NCBI
|