Introduction
Although the signal transduction cascade of the
Notch signaling pathway is simple, it is able to precisely control
multiple binary cell fate decisions, cell proliferation and
differentiation, and stem cell maintenance during embryogenesis and
postnatal development (1,2). The dual activity of its nuclear
effector, recombination signal binding protein for immunoglobulin κ
J region (RBP-Jκ) is crucial to the dynamics of Notch signaling
responses (3). RBP-Jκ activates
the expression of target genes in cells receiving the Notch signal
and represses target expression in the non-receiving cells. This
dual role of RBP-Jκ allows a fine spatial and temporal control of
Notch-regulated transcription; however, the underlying mechanism is
not fully understood (1).
The G protein-coupled receptor 4 (GPR4) family of
proton-sensing G protein-coupled receptors (GPCRs) has recently
been identified to be novel pH sensors (4–6).
GPR4, originally cloned as an orphan GPCR, is expressed in a wide
range of tissues, such as the lung, kidney, heart and liver
(7). GPR4 is highly conserved
during evolution, with >90% amino acid sequence homology among
mammalian orthologs and >70% homology between human and
zebrafish orthologs. GPR4 was previously reported as a receptor for
sphingosylphosphorylcholine and lysophosphatidylcholine, however,
this observation has not been consistently confirmed and the
original publication was withdrawn (8,9). Our
previous in vitro study indicated that GPR4 is capable of
mediating the tube formation of blood vessels by regulating the
function of endothelial cells (ECs) (10,11).
When GPR4 was knocked down in ECs, the growth, migration and tube
formation of ECs were significantly inhibited. In addition, the
GPR4 expression levels appear to be associated with EC survival.
When GPR4 was restored in ECs with GPR4 knocked down, the growth,
migration, and tube formation of ECs fully recovered, confirming
the critical role of this protein for healthy EC function. In
vivo studies have provided further evidence in support of the
GPR4 functions in angiogenesis (4,12).
Dilated and tortuous subcutaneous blood vessels, spontaneous
hemorrhages, and defective vascular smooth muscle cell coverage
were found in ~17% of GPR4-null embryos and neonates (4). These observations indicated that GPR4
is required for the normal vascular development of multiple
tissues/organs. However, the mechanism by which GPR4 regulates the
angiogenesis of ECs has not been clearly defined to the best of our
knowledge.
In the current study, the human (h)GPR4 protein was
associated with the Notch1 protein in ECs and was observed to be
essential in tube formation in the Notch signaling pathway in
vitro.
Materials and methods
Cell culture and reagents
Human HMEC-1 cells were purchased from the Centers
for Disease Control and Prevention (Atlanta, GA, USA). The HMEC-1
cells were cultured at 37°C in 5% CO2 in Gibco
Dulbecco's modified Eagle's medium (DMEM) supplemented with Gibco
10% (v/v) fetal bovine serum (FBS), 100 µg ml−1
penicillin and 10 µg ml−1 streptomycin (all
purchased from Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Cells were passaged every 2–3 days. Suspensions of HMEC-1 cells
were produced from confluent cultures using trypsin/EDTA solution
and the cell concentration determined using a Burker hemocytometer
(Neubauer, Darmstadt, Germany). HMEC-1 cells were seeded at
1×106 either directly into wells of a standard 6-well
plate or into modified well-inserts, which were mounted with the
polymer and located in the wells of a 6-well plate. The growth
medium was replaced every day or as required. The Notch receptor
inhibitor, γ-secretase inhibitor I (GSI-I; Z-LLNle-CHO) was
obtained from EMD Millipore (Billerica, MA, USA).
GSI-I (100 µM) was dissolved in distilled
water and stored at −20°C as a stock solution. Prior to treatment
with GSI-I, the cells were starved with low-serum medium
(containing 0.5% FBS) for 24 h. To block the Notch signaling
pathway, 1 µM GSI-I was added to the medium for 24 h.
Luciferase reporter assay
The HMEC-1 cells were transfected with the
pcDNA3-hGPR4 plasmid for 24 h and infected with the Lenti-RBP-Jκ
Reporter lentiviruses [a section of the RBP-Jκ target sequence,
CGTGGGAA (repeated four times), with the luciferase gene], which
was obtained from Qiagen China Co., Ltd., (Shanghai, China).
Following a 48-h transfection, the cells were lysed in
chemiluminescence lysis buffer [18.3% 1 m
K2HPO4, 1.7% 1 m
KH2PO4, 1 mM phenylmethylsulfonyl fluoride
(PMSF), and 1 mM dithiothreitol] and luciferase activity was
assayed using a luciferase assay kit (cat. no. E1910; Promega
Corporation, Madison, WI, USA). The results were presented as the
mean ± standard deviation (SD) of three independent
experiments.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the HMEC-1 cells using
Invitrogen TRIzol reagent (Thermo Fisher Scientific, Inc.) and RNA
aliquots (200 ng) were reverse transcribed using Random Rochez
(Roche Diagnostics GmbH, Mannheim, Germany) and Reverse
Transcriptase (Takara Biotechnology Co., Ltd., Dalian, China),
according to the manufacturer's protocol. qPCR was performed with
the LightCycler® 480 (Roche Diagnostics) and the
Real-Time detection system (Roche Diagnostics) was used according
to the following conditions: 95°C, 1 min; and 40 cycles of 98°C for
5 sec and 60°C for 20 sec. qPCR of the core genes, Notch,
GAPDH, β-2-microglobulin (B2M), actin,
β (ACTB), hypoxanthine phosphoribosyltransferase
(HPRT1) and ribosomal protein, large, P0
(RPLP0) was performed using the RT2 Profiler™ PCR
Array (Qiagen China Co., Ltd.,) for the human Notch signaling
pathway. Reactions were conducted, and gene expression levels were
calculated relative to GAPDH, B2M, ACTB,
HPRT1 and RPLP0 mRNA levels, which served as
endogenous controls. Relative expression was calculated as
2(Cq gene under investigation − Cq GAPDH).
Tube formation assay
Growth factor-reduced Matrigel (BD Biosciences, San
Jose, CA, USA) was dissolved at 4°C overnight, and 50 µl was
pipetted onto the 96-well culture plates and allowed to polymerize
for 2 h at 37°C. Three quaters of the HMEC-1 cells were transfected
with pcDNA3-hGPR4. After 24 h, 3×104 cells were plated
on 96-well plates coated with the Matrigel and incubated for 8 h
with or without 1 µM GSI-I. Images of the cells were
obtained using an Olympus BX-60 digital camera (Olympus
Corporation, Tokyo, Japan). Three randomly selected fields of view
were photographed per well, and the average was calculated, using
ImageJ software, version 1.47 (National Institutes of Health,
Bethesda, MD, USA), to analyze the length of the total capillary
structure.
Gene silencing using small interfering
(si)RNA
siRNAs against Notch1 (siNotch1) or the non-specific
RNAi (siCon) were purchased from Dharmacon, Inc., (Lafayette, CO,
USA). The Notch1 sequence: 5′-AAGTGTCTGAGGCCAGCAAGA-3′ was as
reported by Rizzo et al (13). Cells (2×105) were plated
in a 6-well plate and cultured for 24 h prior to transfection to
reach ~50% confluence. The cells were transfected with 50 nM siRNA
against Notch1 in the presence of 2.5 µl Invitrogen
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.), in
a final volume of 1 ml serum-free DMEM/HIGH glucose medium. The
reaction was stopped following 6 h of treatment and the medium was
replaced with fresh 10% FBS supplemented medium.
Preparation of cell extracts and western
blotting analysis
The cells were lysed in lysis buffer [50 mM Tris (pH
7.5), 100 mM NaCl, 1 mM EDTA, 0.5% NP-40, 0.5% Triton X-100, 2.5 mM
sodium orthovanadate, 10 mM protease inhibitor cocktail and 1 mM
PMSF) by incubating for 20 min at 4°C. The protein concentration
was determined using the Bio-Rad assay system (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). Total proteins (20
µg) were fractionated using 12% SDS-PAGE (Sigma-Aldrich, St.
Louis, MO, USA) and transferred onto a polyvinylidene fluoride
membrane (EMD Millipore). The membranes were blocked with 5% nonfat
dried milk in 1X Tris-buffered saline buffer containing 0.1%
Tween-20 and subsequently incubated with the following primary
antibodies: Polyclonal rabbit anti-Notch1 (1:1,000; cat. no.
ab27526; Abcam, Cambridge, MA, USA), monoclonal mouse anti-HIF1α
(1:1,000; cat. no. ab113642; Abcam), poly-clonal rabbit anti-VEGF
(1:500; cat. no. ab46154; Abcam) and monoclonal mouse anti-GAPDH
(1:1,000; cat. no. ab8245; Abcam). Horseradish peroxidase
(HRP)-conjugated goat anti-rabbit IgG (1:10,000 dilution; ab6721;
Abcam) or HRP-conjugated goat anti-mouse IgG (1:10,000 dilution;
ab97023; Abcam) was used as the secondary antibodies, and the
protein bands were detected using an enhanced chemiluminescence
detection system (Santa Cruz Biotechnology, Inc., Dallas, TX, USA).
The western blot products were imaged using the Fusion FX6 system
(Vilber Lourmat Deutschland GmbH, Eberhardzell, Germany).
Quantification of the western blots was performed using laser
densitometry, and the relative protein expression was normalized to
the GAPDH levels in each sample. The results are presented as the
mean of three independent experiments with error bars representing
SDs.
Transendothelial migration
Migration assays were performed in Transwell plates
(Corning Incorporated, Corning, NY, USA) with a 6.5-mm diameter and
3-µm pore filters. The ECs were plated at 3×104
cells/well on gel-coated filters. The nonadherent cells were
removed after 18 h. The adherent cells were cultured for 2–3 days
to obtain 100% confluence. Freshly isolated lymphocytes
(1×105) were added to the upper compartment in 0.1 ml
serum-free medium with or without hGPR4/GSI-I, and 0.6 ml
serum-free medium was added to the lower compartment. The Transwell
plates were incubated at 37°C and at 5% CO2 for 6 h. The
cells that did not migrate were removed using a cotton swab,
whereas the cells that migrated were fixed with 4% paraformaldehyde
and stained with 1% crystal violet. The cells that migrated to the
lower compartment were collected and counted. The membrane was then
fixed in formalin for 10 min at room temperature prior to staining
with 0.1% crystal violet for 5 min. The number of HMEC-1 cells that
migrated to the lower surface of the membrane were counted in 10
random fields at maginification, ×100 using a light microscope
(Leica Microsystems GmbH, Wetzlar, Germany). A chemotactic index
(CI) was calculated to express stimulated migration: CI = treated
migration (number of HMEC-1 per field)/random migration (number of
HMEC-1 per field). Each assay was performed in triplicate
wells.
Statistical analysis
Data were analyzed using the two-tailed Student's
t-test using SPSS software, version 19.0 (IBM SPSS, Armonk, NY,
USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
hGPR4 induces the expression of Notch1 in
HMEC-1
As Notch signaling has a profound effect on
angiogenesis, the involvement of Notch signaling in hGPR4-induced
angiogenesis was examined. HMEC-1 cells were transfected with
pcDNA3-hGPR4 for 24 h. Then the cells were infected with the
lentivirus-mediated RBP-Jκ reporter for 24 h. As shown in Fig. 1A, following treatment with hGPR4
for 48 h, the level of luciferase was upregulated significantly.
Using validated Notch cDNA microarray datasets, the mRNA expression
of Notch-related genes in HMEC-1 with or without pcDNA3-hGPR4
transfection were compared. Notch1 was identified to be the
predominant Notch receptor expressed, although Notch2, 3 and 4 were
detected at low levels in the HMEC-1 cells. A higher level of
expression of Notch1, CCND1 and LFNG were observed in HMEC-1 with
pcDNA3-hGPR4 transfection (Fig.
1B).
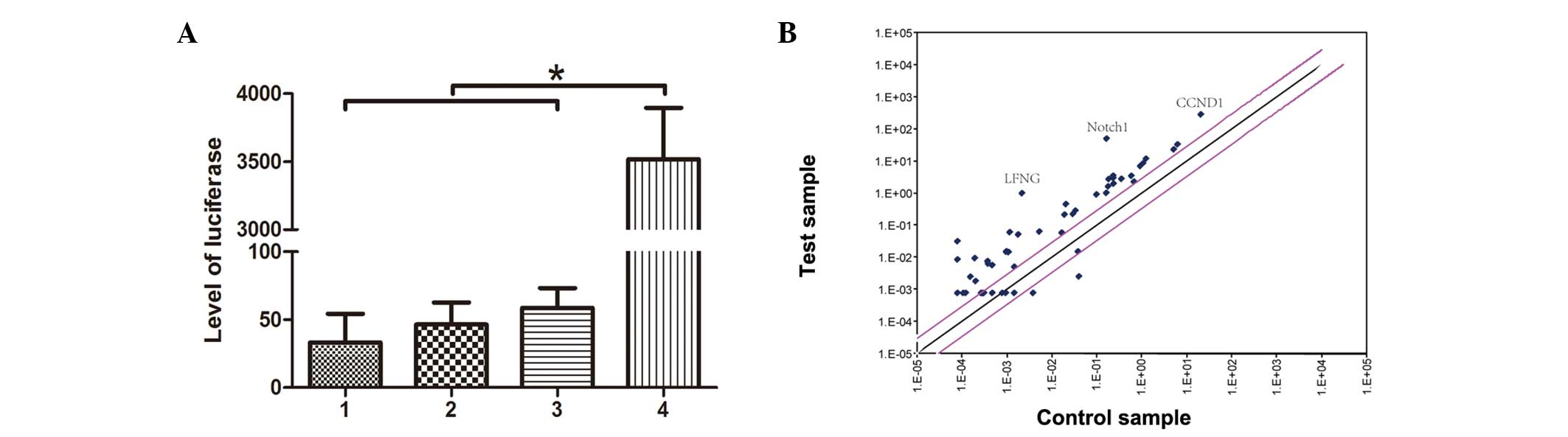 | Figure 1Transcriptional regulation of Notch
target genes following pcDNA3-hGPR4 transfection in HMEC-1 cells.
(A) HMEC-1 cells were transfected with pcDNA3-hGPR4 for 24 h. Then
the cells were infected with the lentivirus-mediated RBP-Jκ
reporter for 24 h. The luciferase activity was detected using a
luciferase assay kit. 1, HMEC-1 blank cells; 2, HMEC-1 cells
transfected with pcDNA3-hGPR4; 3, HMEC-1 cells infected with
lentivirus without RBP-Jκ reporter; 4, HMEC-1 cells infected with
lentivirus-mediated RBP-Jκ reporter. *P<0.0001. (B)
Transcriptional regulation of Notch target genes following
pcDNA3-hGPR4 treatment in HMEC-1 cells. cDNA was synthesized from
HMEC-1 cells before and after treatment with pcDNA3-hGPR4. A
mini-array of Notch-relevant genes demonstrates that few genes were
downregulated upon pcDNA3-hGPR4 treatment; however, certain
significant genes including LFNG, Notch1 and CCND1 were
upregulated. The gray lines indicate a two-fold change. hGPR4,
human G protein-coupled receptor 4; RBP-Jκ, recombination signal
sequence binding protein-Jκ; LFNG, LFNG O-fucosylpeptide
3-β-N-acetylglucosaminyltransferase; CCND1, cyclin D1. |
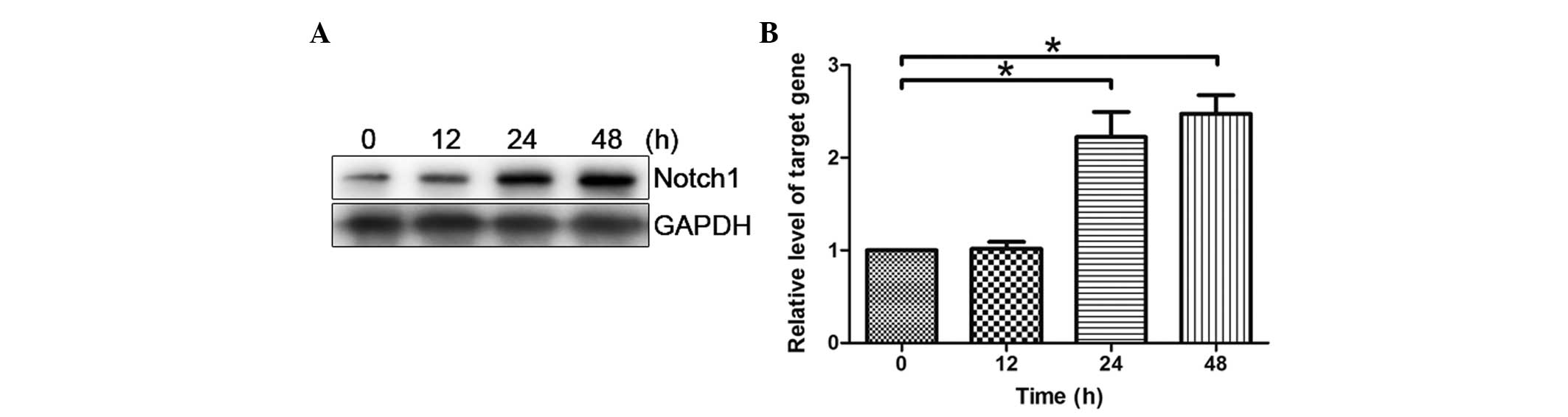
HMEC-1 cells were transfected with pcDNA3-hGPR4 for
12, 24 and 48 h. As shown in Fig. 2A
and B, after treatment with pcDNA3-hGPR4 for 12 h, the
expression of Notch1 was demonstrated to be markedly enhanced and
reached its maximum at 48 h. Notch1 demonstrated a time-dependent
response to hGPR4.
Notch1 participates in hGPR4-induced
HMEC-1 tube formation
In a previous study, the function of hGPR4 in
angiogenesis was demonstrated in the tube formation test (14,15),
and the present study showed similar results; as hGPR4
significantly enhanced HMEC-1 tube formation when compared with a
control group without hGPR4. To further estimate whether Notch
signaling is involved in hGPR4-induced tube formation, the Notch
inhibitor, GSI-I was used. GSI-I (1 µM) markedly inhibited
hGPR4-induced tube formation. Tube lengths were significantly
decreased from 430.67±148.41 in the control cells to 150.0±89.27
µm in the HMEC-1 cells treated with pcDNA3-hGPR4 and
siNotch1 (P=0.0485; Fig. 3A and
B).
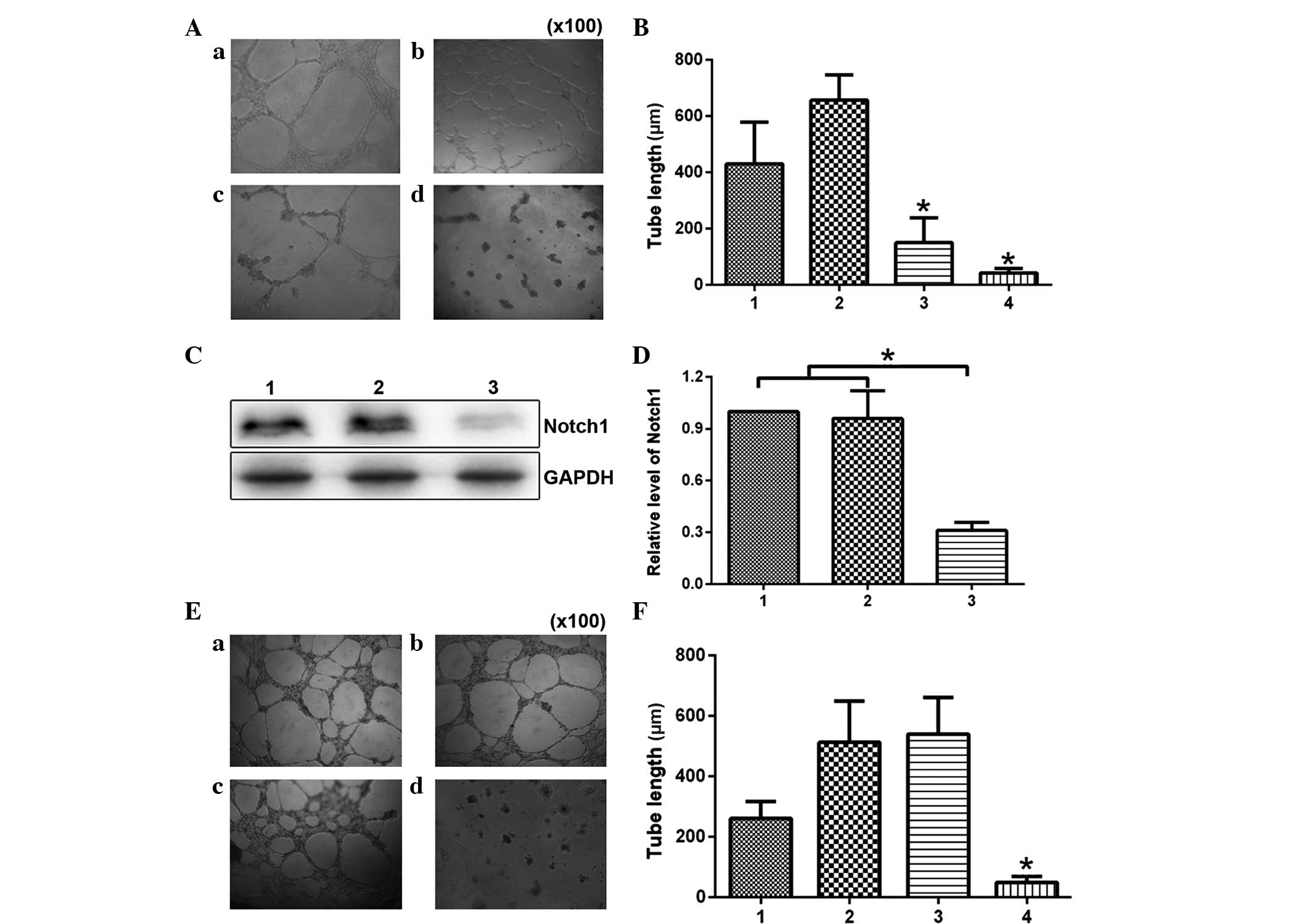 | Figure 3Notch1 participates in HMEC-1 tube
formation induced by hGPR4. (A) HMEC-1 cells treated with
pcDNA3-hGPR4, 1 µM GSI-I and a combination of the two
(magnification, ×100). (a) Blank HMEC-1 cells served as a control;
(b) HMEC-1 cells transfected with pcDNA3-hGPR4; (c) HMEC-1 cells
treated with pcDNA3-hGPR4 and siNotch1; (d) HMEC-1 cells
transfected with pcDNA3-hGPR4 and treated with GSI-I. (B) The tube
length was quantified in eight fields after the corresponding
treatment. 1, Blank HMEC-1 cells served as a control; 2, HMEC-1
cells transfected with pcDNA3-hGPR4; 3, HMEC-1 cells treated with
pcDNA3-hGPR4 and siNotch1; 4, HMEC-1 cells transfected with
pcDNA3-hGPR4 and then treated with GSI-I. (C) HMEC-1 cells
transfected with pcDNA3-hGPR4 and then treated with GSI-I. The cell
lysates were also subjected to western blot analysis with
anti-Notch1 antibody. The antibody to GAPDH served as a loading
control. 1, HMEC-1 cells; 2, HMEC-1 cells transfected by
pcDNA3-hGPR4; 3, HMEC-1 cells transfected with pcDNA3-hGPR4 and
GSI-I. (D) Densitometric analysis of western blot assay to quantify
target protein levels. Results are expressed as mean ± standard
deviation of three independent experiments, *P<0.05
vs. the control. (E) HMEC-1 cells treated with pcDNA3-hGPR4, siCon
or siNotch1 (magnification, ×100); (a) Blank HMEC-1 cells served as
a control; (b) HMEC-1 cells transfected with pcDNA3-hGPR4 and
siCon; (c) HMEC-1 cells transfected with pcDNA3-hGPR4; (d) HMEC-1
cells transfected with siNotch1. (d) HMEC-1 cells transfected with
pcDNA3-hGPR4 and siNotch1. (F) The tube length was quantified in
eight fields after the corresponding treatment. 1, Blank HMEC-1
cells served as a control; 2, HMEC-1 cells transfected with
pcDNA3-hGPR4 and siCon; 3, HMEC-1 cells transfected with
pcDNA3-hGPR4; 4, HMEC-1 cells transfected with pcDNA3-hGPR4 and
siNotch1. *P<0.05. hGPR4, human G protein-coupled
receptor 4; GSI-I, γ-secretase inhibitor I; si, small interfering;
siCon, non-specific RNAi. |
As it has previously been demonstrated that GSIs are
able to act through different biochemical pathways (16), the present study investigated
whether the effects induced by GSI-I were associated with the
specific inhibition of Notch signaling, which is induced by GSI-I.
The Notch gene was silenced and the effects of hGPR4 addition were
evaluated. An siRNA sequence was obtained for the knockdown of
Notch1. After confirming the reduction in Notch1 expression level
(Fig. 3C and D), the effect of
hGPR4 on HMEC-1 tube formation activity was analyzed. As shown in
Fig. 3E and F, Notch1 siRNA
significantly inhibited hGPR4-induced tube formation from
540.0±121.45 to 49.67±19.50 µm (P=0.0143), while siCon did
not exert an effect hGPR4-induced tube formation.
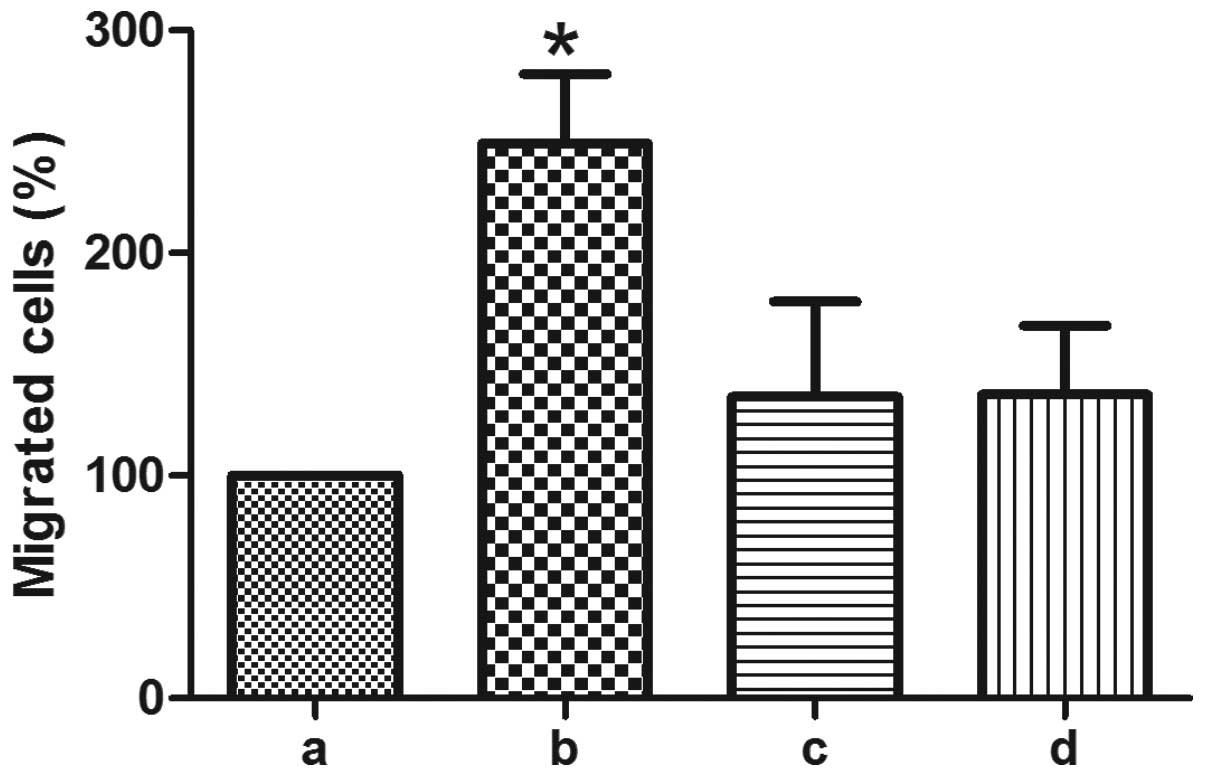
HGPR4 induces lymphocyte transendothelial
migration through Notch1
Vascular leakage and lymphocyte transendothelial
migration are the critical, initial steps in angiogenesis. To
detect if Notch signaling is involved in hGPR4-induced lymphocyte
transendothelial migration, GSI-I and siNotch were used. As shown
in Fig. 4, 1 µM GSI-I significantly decreased
hGPR4-induced lymphocyte transendothelial migration. hGPR4-induced
lymphocyte transendothelial migration was also significantly
attenuated from 249.02±31.45 to 135.29±42.72 with Notch1 knockdown
(P=0.0116).
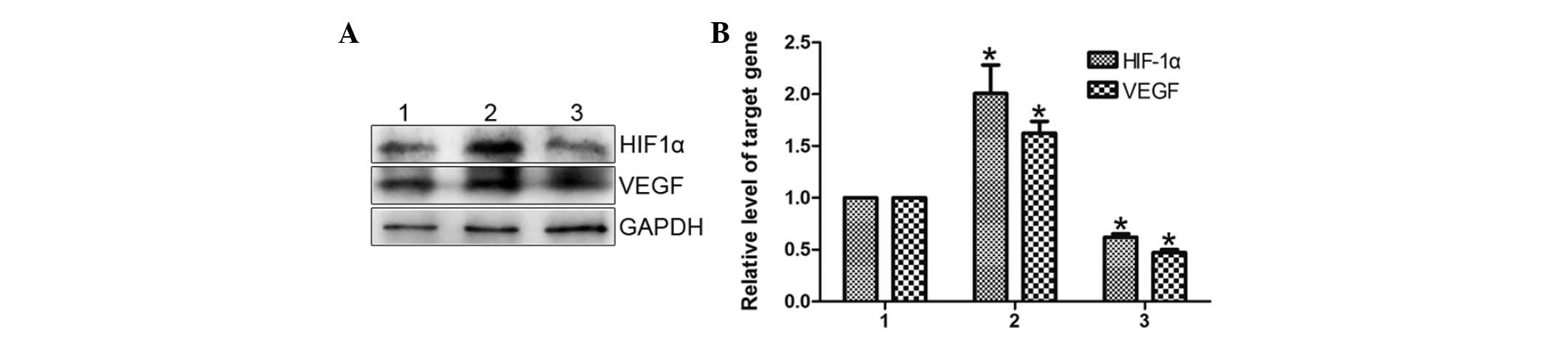
Notch1 mediates hGPR4 regulation of VEGF
and HIF-1α
VEGF is considered to be the most important gene in
angiogenesis. The present study aimed to determine whether hGPR4
also regulates VEGF via Notch signaling. It was found that hGPR4
overexpression significantly induced VEGF expression (Fig. 5A). VEGF expression was
significantly attenuated by the knockdown of Notch1, when
compared with cells transfected with control cells (Fig. 5A).
Subsequently, whether HGPR4 regulates HIF-1α and
whether this regulation is also through Notch signaling was
investigated, as HIF-1α is the main mediator of VEGF and controls
the upregulation of VEGF. As demonstrated in Fig. 5, hGPR4 significantly augmented
HIF-1α expression when compared with the control, while siNotch1
significantly inhibited hGPR4-induced HIF-1α expression.
Discussion
The results of the present study demonstrated the
Notch involvement in EC activation, apoptosis and proliferation.
Although the effects of various mediators of cell fate on the Notch
signaling pathway in ECs have been characterized, little is known
about the role of hGPR4 upon Notch in ECs (13,17).
The requirement for Notch signaling in
vasculogenesis and angiogenesis is well documented in a number of
studies. Hellstrom et al (18) report that delta-like 4
(DLL4)–Notch1 signalling regulates the formation of appropriate
numbers of tip cells to control vessel sprouting and branching in
the mouse retina. Yang et al (19) reported that differential activation
of the hypoxia/HIF1-VEGF-Notch pathway may serve a role in
epicardial cell interactions that promote epicardial
epithelial/mesenchymal transition and coronary progenitor cell
differentiation during epicardial development and coronary
vasculogenesis, particularly in hypoxic sulcus regions. Notch1, the
key regulator of vasculogenesis and embryonic differentiation, has
shown a correlation with a poor prognosis in hepatocellular
carcinoma (HCC). Notch1 may serve as a potential target for
vasculogenic mimicry development in HCC (20). Notch signaling is reported to
regulate angiogenesis by interacting with VEGF signaling.
Increasing evidence indicates that Notch signaling promotes
angio/arteriogenesis not only in developmental states but also in
ischemia-induced angiogenesis in adults (21,22).
Additionally, Notch signal activation regulates VEGF receptor
expression and angiogenic activity in endothelial cells in a
ligand-dependent manner. DLL4-mediated Notch signaling suppresses
tip cells sprouting in the retina, which is antagonized by Notch
signal activation by Jagged-1 (23). Thus, negative and positive roles
for Notch signaling in endothelial sprouting and angiogenetic
activity have been reported in a number of previous studies. In the
present study, the relationship between hGPR4 and Notch1 was
investigated. These findings suggest that Notch1 is an important
downstream target of hGPR4 in vascular endothelial cells. Although
it remains largely unknown why in some endothelial cells had to
develop into mature endothelium while vasculogenic mimicry may
already serve the same purpose, it has been demonstrated that
Notch1 might contribute to these processes.
In the current study, the results showed that the
RBP-Jκ-mediated Notch signaling may be critical for HMEC-1 tube
formation. The Notch signaling pathway is important in cell-cell
communication, and the self-renewal, migration and differentiation
of cells (1,24). The present study identified a
positive role for Notch signaling in endothelial morphogenesis via
the induction of cellular extensions mediated by Notch1. This
finding was supported by the observation that the Notch signaling
pathway is involved in the regulation of VEGF and HIF-1α levels,
and the increase of VEGF and HIF-1α levels correlated with
increased endothelial responsiveness to the Notch1. It was found
that Notch signaling increased angiogenesis by inducing Notch1
expression.
The present study demonstrated that Notch1 is
upregulated by Notch signaling in ECs following hGPR4
overexpression. A positive role for Notch signaling was identified
in endothelial morphogenesis via the induction of cellular
extensions, which were mediated by hGPR4. This was demonstrated by
the observation that overexpression of hGPR4 increased Notch1
expression levels and this increase correlated with increased
endothelial form of HMEC-1 in vitro. Using a protein-based
Notch inhibitor, GSI-I, the current study demonstrated that the
perturbation of endogenous Notch signaling resulted in reduced VEGF
and HIF-1α expression levels. Thus, loss- and gain-of-function
studies reveal that hGPR4 regulates Notch signaling expression in
HMEC-1 cells.
Acknowledgments
The present study was supported by the National
Natural Science Foundations of China (grant nos. 31201060/C0709,
30973175/H1621 and 81172490/H1621); the Program for New Century
Excellent Talents in University (grant no. NCET-12-0440); the
Scientific and Technological Research Foundation of Shaanxi
Province (grant nos. 2012K13-01-06 and 2007K09-09); the project was
sponsored by the Scientific Research Foundation for the Returned
overseas Chinese Scholars of State Education Ministry, (grant no.
0601-18920006); the Research Foundation of the Health Department of
Shaan'xi Province (grant no. 2010D41); Qing Nian Jiao Shi Gen Zong
Ji Hua of Xi'an Jiaotong University ('The Fundamental Research
Funds for the Central Universities'; grant no. 2012-FRFCU-121); and
supported by the Program for Changjiang Scholars and Innovative
Research Team in University (grant no. PCSIRT:1171) and the
Research Foundation of Xi'an Jiao Tong University of China (grant
no. RFXJTU:1231).
References
|
1
|
Kopan R and Ilagan MX: The canonical Notch
signaling pathway: Unfolding the activation mechanism. Cell.
137:216–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray SJ: Notch signalling: A simple
pathway becomes complex. Nat Rev Mol Cell Biol. 7:678–689. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Borggrefe T and Oswald F: The Notch
signaling pathway: transcriptional regulation at Notch target
genes. Cell Mol Life Sci. 66:1631–1646. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang LV, Radu CG, Roy M, Lee S, McLaughlin
J, Teitell MA, Iruela-Arispe ML and Witte ON: Vascular
abnormalities in mice deficient for the G protein-coupled receptor
GPR4 that functions as a pH sensor. Mol Cell Biol. 27:1334–1347.
2007. View Article : Google Scholar :
|
|
5
|
Ludwig MG, Vanek M, Guerini D, Gasser JA,
Jones CE, Junker U, Hofstetter H, Wolf RM and Seuwen K:
Proton-sensing G-protein-coupled receptors. Nature. 425:93–98.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Murakami N, Yokomizo T, Okuno T and
Shimizu T: G2A is a proton-sensing G-protein-coupled receptor
antagonized by lysophosphatidylcholine. J Biol Chem.
279:42484–42491. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mahadevan MS, Baird S, Bailly JE, Shutler
GG, Sabourin LA, Tsilfidis C, Neville CE, Narang M and Korneluk RG:
Isolation of a novel G protein-coupled receptor (GPR4) localized to
chromosome 19q13.3. Genomics. 30:84–88. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bektas M, Barak LS, Jolly PS, Liu H, Lynch
KR, Lacana E, Suhr KB, Milstien S and Spiegel S: The G
protein-coupled receptor GPR4 suppresses ERK activation in a
ligand-independent manner. Biochemistry. 42:12181–12191. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhu K, Baudhuin LM, Hong G, Williams FS,
Cristina KL, Kabarowski JH, Witte ON and Xu Y:
Sphingosylphosphorylcholine and lysophosphatidylcholine are ligands
for the G protein-coupled receptor GPR4. J Biol Chem.
276:41325–41335. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kim KS, Ren J, Jiang Y, Ebrahem Q, Tipps
R, Cristina K, Xiao YJ, Qiao J, Taylor KL, Lum H, et al: GPR4 plays
a critical role in endothelial cell function and mediates the
effects of sphingosylphosphorylcholine. FASEB J. 19:819–821.
2005.PubMed/NCBI
|
|
11
|
Afrasiabi E, Blom T, Ekokoski E, Tuominen
RK and Törnquist K: Sphingosylphosphorylcholine enhances calcium
entry in thyroid FRO cells by a mechanism dependent on protein
kinase C. Cell Signal. 18:1671–1678. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wyder L, Suply T, Ricoux B, Billy E,
Schnell C, Baumgarten BU, Maira SM, Koelbing C, Ferretti M, Kinzel
B, et al: Reduced pathological angiogenesis and tumor growth in
mice lacking GPR4, a proton sensing receptor. Angiogenesis.
14:533–544. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rizzo P, Miao H, D'Souza G, Osipo C, Song
LL, Yun J, Zhao H, Mascarenhas J, Wyatt D, Antico G, et al:
Cross-talk between notch and the estrogen receptor in breast cancer
suggests novel therapeutic approaches. Cancer Res. 68:5226–5235.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim KS, Ren J, Jiang Y, Ebrahem Q, Tipps
R, Cristina K, Xiao YJ, Qiao J, Taylor KL, Lum H, et al: GPR4 plays
a critical role in endothelial cell function and mediates the
effects of sphingosylphosphorylcholine. FASEB J. 19:819–821.
2005.PubMed/NCBI
|
|
15
|
Ren J, Jin W, Gao YE, Zhang Y, Zhang X,
Zhao D, Ma H, Li Z, Wang J, Xiao L, et al: Relations between GPR4
expression, microvascular density (MVD) and clinical pathological
characteristics of patients with epithelial ovarian carcinoma
(EOC). Curr Pharm Des. 20:1904–1916. 2014. View Article : Google Scholar
|
|
16
|
Rasul S, Balasubramanian R, Filipović A,
Slade MJ, Yagüe E and Coombes RC: Inhibition of gamma-secretase
induces G2/M arrest and triggers apoptosis in breast cancer cells.
Br J Cancer. 100:1879–1888. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Funahashi Y, Shawber CJ, Vorontchikhina M,
Sharma A, Outtz HH and Kitajewski J: Notch regulates the angiogenic
response via induction of VEGFR-1. J Angiogenes Res. 2:32010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hellstrom M, Phng LK, Hofmann JJ, Wallgard
E, Coultas L, Lindblom P, Alva J, Nilsson AK, Karlsson L, Gaiano N,
et al: Dll4 signalling through Notch1 regulates formation of tip
cells during angiogenesis. Nature. 445:776–780. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang K, Doughman YQ, Karunamuni G, Gu S,
Yang YC, Bader DM and Watanabe M: Expression of active Notch1 in
avian coronary development. Dev Dyn. 238:162–170. 2009. View Article : Google Scholar
|
|
20
|
Zhu MS, Xu LB, Zeng H, Shi XD, Wu WR and
Liu C: Association of Notch1 with vasculogenic mimicry in human
hepatocellular carcinoma cell lines. Int J Clin Exp Pathol.
7:5782–5791. 2014.PubMed/NCBI
|
|
21
|
Takeshita K, Satoh M, Ii M, Silver M,
Limbourg FP, Mukai Y, Rikitake Y, Radtke F, Gridley T, Losordo DW
and Liao JK: Critical role of endothelial Notch1 signaling in
postnatal angiogenesis. Circ Res. 100:70–78. 2007. View Article : Google Scholar
|
|
22
|
Funahashi Y, Shawber CJ, Vorontchikhina M,
Sharma A, Outtz HH and Kitajewski J: Notch regulates the angiogenic
response via induction of VEGFR-1. J Angiogenes Res. 2:32010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Suchting S, Freitas C, le Noble F,
Benedito R, Bréant C, Duarte A and Eichmann A: The Notch ligand
Delta-like 4 negatively regulates endothelial tip cell formation
and vessel branching. Proc Natl Acad Sci U S A. 104:3225–3230.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang B, Tang Q, Post J, Zhou H, Huang XB,
Zhang XD, Wang Q, Sun YM and Fan FY: Effect of radiation on the
Notch signaling pathway in osteoblasts. Int J Mol Med. 31:698–706.
2013.PubMed/NCBI
|