Introduction
Cataracts are a typical age-correlated condition and
the leading cause of visual loss worldwide (1). Previous studies have demonstrated
that age-correlated cataracts cause approximately half of all cases
of visual loss (2–4). A previous epidemiological study
demonstrated that 96% of people aged >60 years have different
types or severities of lens opacity (5). According to estimates by the World
Health Organization, the number of people with cataract blindness
worldwide will reach 40 million by 2025 (6). Age-correlated cataracts cause severe
visual impairment, reduce quality of life, and become a burden on
social economy and health resources. As an important component of
the visual refraction system, human lenses are relatively isolated
tissues surrounded by the vitreous body and the aqueous humor.
Lenses, which are transparent in younger individuals, change with
age. These changes begin with the development of a compact, hard
nucleus and regional opacity, which result in the occurrence of
pathological cataracts.
MicroRNAs (miRNAs), a class of endogenous small
non-coding RNAs of 20–25 nucleotides, modulate the expression of
genes at the post-transcriptional level (7). miRNAs bind to complementary sequences
present in the 3′-untranslated regions (3′-UTR) of target gene
mRNAs and, thus, regulate translational interference or degradation
of mRNAs (8). Several miRNAs have
previously been detected in animal eyes (9,10);
miRNA-31 (miRNA-31), miR-184, miR-204, miR-125b, miR-26a, let-7b
and others were identified in the lens; miR-26a, miR-24, miR-31
miR-184 and miR-205 were identified in the cornea; and miR-181a,
miR-124a, miR-182, miR-183, miR-125b and miR-30 were demonstrated
to be produced in the retina (9,10).
Accumulating evidence has demonstrated that abnormal expression of
miRNAs is closely correlated with the pathogenesis of a wide range
of age-associated conditions (11–14),
including as cataracts (15).
miRNA studies have provided novel insights to the treatment of such
diseases (16,17).
Variants in primary miRNAs or in the 3′-UTR of the
target gene may interfere with the production of the miRNA or the
binding between miRNA and mRNA of the target gene. miRNAs
negatively regulate the expression of target genes by binding to
the ʻseed sequence' in the 3′-UTR of the genes, and the variants
within or nearby the seed sequence may inhibit or enhance
miRNA/mRNA interaction leading to either ʻloss-of-function' or
ʻgain-of-function'. The present study investigated the potential
effect of the rs78378222 polymorphism (18,19)
in the 3′-UTR of tumor protein p53 (TP53) on the miR-125b-induced
apoptosis of lens epithelial cells, and the association between the
rs78378222 polymorphism and the risk of age-associated
cataracts.
Materials and methods
Study population
Lens epithelial cells were collected as previously
described (20) from 12 patients
with cataracts and 12 normal controls receiving eye surgery at
Beijing Tongren Hospital (Beijing, China). The study protocol
followed the guidelines of the Declaration of Helsinki (21) and was approved by the institutional
review board of Beijing Tongren Hospital. Following a full
explanation of the surgical procedures and possible complications,
all patients provided written informed consent. Patients were
selected based on clinically observable nuclear cataracts of grade
2 or 3 according to the Lens Opacities Classification System III
(22). Exclusion criteria included
cataract hardness greater than grade 3. Patients with type 1
diabetes mellitus (DM), rheumatologic disease and other systemic
diseases, with the exception of type 2 DM, were also excluded.
SRA01/04 cells were obtained from the American Type Culture
Collection (Manassas, VA, USA) and cultured in RPMI-1640 medium
(Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 100
mg/ml streptomycin, 100 U/ml penicillin and 10% fetal calf serum
(Invitrogen; Thermo Fisher Scientific, Inc.) in a humidified
atmosphere of 5% CO2 at 37°C.
DNA sequencing and cell biology
A DNA extraction kit (Qiagen, Inc., Valencia, CA,
USA) was used to extract genomic DNA from patient lens cells. The
Sanger method was used to bidirectionally sequence the TP53 3′-UTRs
(Beijing Tongren Hospital). A TP53 reference sequence (NM_000546.5;
genome.ucsc.edu) was compared with all sequences
and used to determine the genotype of the polymorphism.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) was used to extract total RNA from cataractous
lens samples and cultured SRA01/04 epithelial cells according to
the manufacturer's instructions. A UV-Vis spectrophotometer
(UV-1800; Shimadzu Corporation, Kyoto, Japan) was used to determine
the quality of RNA. Agarose gel (1.5%) electrophoresis with 260/280
values between 1.8 and 2.0 was used to confirm RNA integrity.
PrimerScript RT reagent kit (Takara Biotechnology Co., Ltd.,
Dalian, China) was used produce cDNA from 1 ng RNA for RT-qPCR
analysis. An ABI 7500 cycler (Applied Biosystems; Thermo Fisher
Scientific, Inc.), RT primer and TaqMan probes (Guangzhou RiboBio
Co., Ltd., Guangzhou, China) were used to determine the expression
of miR-125b and TP53. RNA U6 small nuclear 2 was used as the
endogenous reference control. The conditions for the RT reaction
were 65°C for 5 min, 25°C for 10 min, 42°C for 1 h and 75°C for 10
min. The PCR cycling conditions were as follows: 95°C for 5 min; 40
cycles of 95°C for 10 sec, and 60°C for 1 min. RT-qPCR analysis was
performed twice on three independent specimens. The primer
(Shanghai Jieli Biotechnology Co., Ltd., Shanghai, China) sequences
were as follows: Forward, 5′-TCAGTTTGCTGTTCTGGGTG-3′ and reverse,
5′-CGGTTGGCTGGAAAGGAG-3′ for GAPDH; forward,
5′-CCCCTCTGAGTCAGGAAACA-3′ and reverse, 5′-AGACAGAAGGGCCTGACTCA-3′
for TP53; forward, 5′-TCAGTTTGCTGTTCTGGGTG-3′ and reverse,
5′-CGGTTGGCTGGAAAGGAG-3′ for U6; and forward,
5′-UCCCUGAGACCCUAACUUGUGA-3′ and reverse,
5′-ACAAGUUAGGGUCUCAGGCACU-3′ for miR-125b. The equation
RQ=2−ΔΔCq was used to calculate the relative abundance
of miR-125b and TP53 mRNA in cell lines and tissues (23). GraphPad Prism 5 software (GraphPad
Software, Inc., La Jolla, CA, USA) was used to produce graphs and
perform analyses.
Luciferase assay
Human genomic DNA was used for amplification of the
3′-UTR of the TP53 gene containing conserved binding sites for
miR-125b. Following amplification, the fragments were implanted
into the pmiR-RB-REPORT vector (Guangzhou RiboBio Co., Ltd.). A
mutant 3′-UTR fragment containing mutations in seed binding sites
was obtained using a QuikChange Site-Directed Mutagenesis kit
(Agilent Technologies, Inc., Santa Clara, CA, USA), according to
the manufacturer's protocols, to introduce deletions in the 3′-UTR
of TP53. Similarly, the TP53-3′-UTR mutant fragment was inserted
into the same sites of the pmiR-RB-REPORT control vector. For
reporter assays, SRA01/04 cells were plated in 24-well plates and
cultured overnight in Iscove's modified Dulbecco's medium
(Invitrogen; Thermo Fisher Scientific, Inc.). Following incubation,
the cells were cotransfected with miR-125b mimic/mimic control and
wild-type/mutant reporter plasmid (Guangzhou RiboBio Co., Ltd.)
using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.). At 48 h post-transfection, the Dual-Luciferase Reporter
assay system (Promega Corporation, Madison, WI, USA) was used to
determine luciferase activity according to the manufacturer's
protocol. The GloMax™ 96 Microplate Luminometer (Promega
Corporation) was used to measure luciferase activity. The
endogenous control was Renilla luciferase plasmid. Each
experiment was conducted with three independent specimens and
repeated twice.
Apoptosis analysis
At 48 h following transfection, an apoptosis assay
was performed on SRA01/04 lens epithelial cells (1.5×105
cells/well). An apoptosis detection kit (Nanjing KeyGen Biotech.
Co. Ltd., Nanjing, China) was used to determine apoptosis using
Annexin V/propidium iodide staining. The specimens were cultured at
room temperature for 15 min in darkness and analyzed by flow
cytometry (BD Influx™ Cell Sorter; BD Biosciences, Franklin Lakes,
NJ, USA). CellQuest software (BD Biosciences) was used to analyze
the results.
Western blot analysis
Protein extraction from SRA01/04 and primary cells
was performed by incubation with lysis buffer containing 1% NP-40,
0.1% sodium dodecyl sulfate (SDS), 2 mg/ml aprotinin, 1 mM
phenylmethane sulfonyl fluoride and 150 mM NaCl at 4°C for 30 min.
A BCA Protein assay kit (Bio-Rad Laboratories, Inc., Hercules, CA,
USA) was used to quantify the protein levels. The separation of
protein extracts (20 μg) was performed via 10%
SDS-polyacrylamide gel electrophoresis (Roche Applied Science,
Penzberg, Germany) and transferred onto polyvinylidene difluoride
membranes (PerkinElmer, Inc., Waltham, MA, USA). Membranes were
blocked in Tris-buffered saline containing 0.05% Tween 20 (TBST;
BioSharp, Hefei, China) with 5% non-fat milk, 2.7 mmol/l KCl, 137
mmol/l NaCl and 25 mmol/l Tris-HCl (pH 7.5) at 37°C for 1 h. The
membranes were then incubated with primary antibodies, mouse
monoclonal against β-actin (1:8,000; Santa Cruz Biotechnology,
Inc., Dallas, TX, USA; cat. no. sc-47778) or rabbit polyclonal
against TP53 (1:3,000; Santa Cruz Biotechnology, Inc.; cat. no.
6243) in TBST with 5% non-fat milk at 4°C overnight. Following
washes with TBST three times, membranes were cultured with
horseradish peroxidase-conjugated goat anti-rabbit polyclonal
(1:15,000; Invitrogen; Thermo Fisher Scientific, Inc.; cat. no.
Q11402MP) and goat anti-mouse (1:10,000; Santa Cruz Biotechnology,
Inc.; cat. no. sc-2005) secondary antibodies for 1 h at room
temperature. Membranes were washed again with TBST, then observed
using a SuperSignal West Pico Chemilumunescent Substrate enhanced
chemiluminescence kit (Thermo Fisher Scientific, Inc.) and analyzed
with ImageJ 1.48 (imagej.nih.gov/ij/).
Statistical analysis
The differences in microarray data were compared
between the two groups using the P-values corrected by false
discovery rate, which were below 0.05 obtained by significant
analysis of microarrays software (statweb.stanford.edu/~tibs/SAM/). SPSS software
version 16.0 (SPSS, Inc., Chicago, IL, USA) and an independent
samples t-test were used to estimate differences between two
groups. P<0.05 was considered to indicate a statistically
significant difference.
Results
Effect of rs78378222 on the binding site
in the 3-UTR of TP53
miRNAs negatively regulate the expression of target
genes by binding to the ʻseed sequence' in the gene 3′-UTR.
Variants within or nearby the ʻseed sequences' may compromise or
enhance miRNA/mRNA interaction leading to either ʻloss-of-function'
or ʻgain-of-function' effects. The present study evaluated the
potential effect of the rs78378222 polymorphism in the 3′-UTR of
TP53 on miRNA-125b-induced apoptosis of lens epithelial cells, and
the association between the rs78378222 polymorphism and the risk of
age-associated cataracts. Initially, sequences of mature miR-125b
and TP53 (wild-type and polymorphic) were analyzed and compared. As
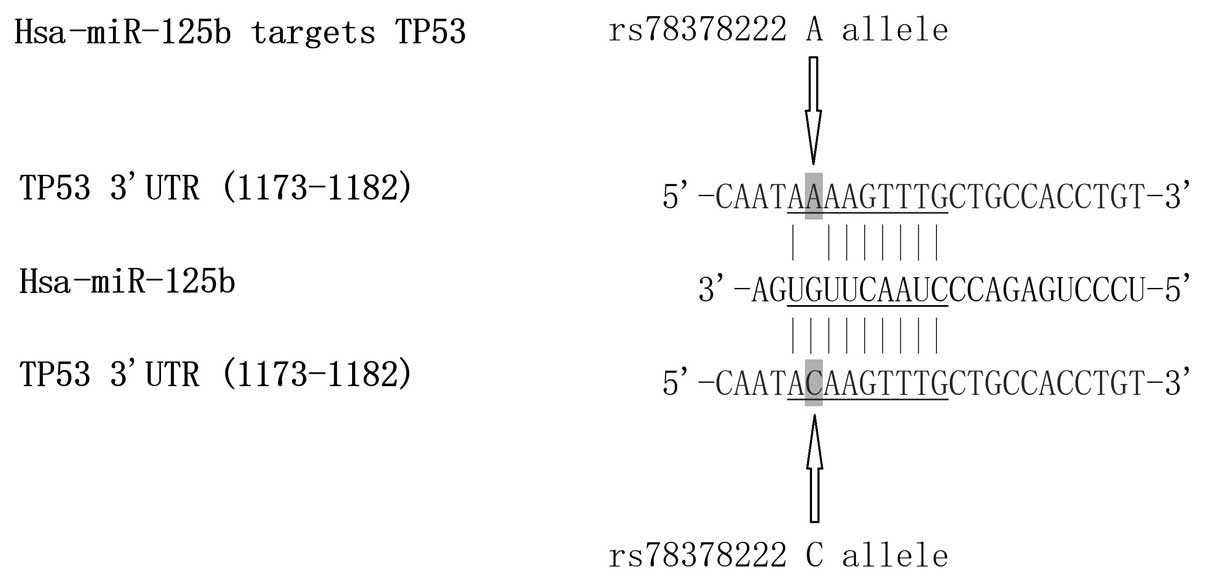
presented in Fig. 1, replacement
of A with C (at position 1177) introduces a novel potential binding
site in the 3′-UTR of TP53 with consecutive 8-bp perfect match,
indicating that the rs78378222 polymorphism results in a
ʻgain-of-function' to regulate TP53 expression.
Effect of rs78378222 on TP53
expression
To examine the effect of the rs78378222 polymorphism
on TP53 expression, the full length TP53 3′-UTR was subcloned and
inserted into a pcDNA3 vector containing a firefly luciferase gene
downstream, and the minor allele of the rs78378222 polymorphism (C)
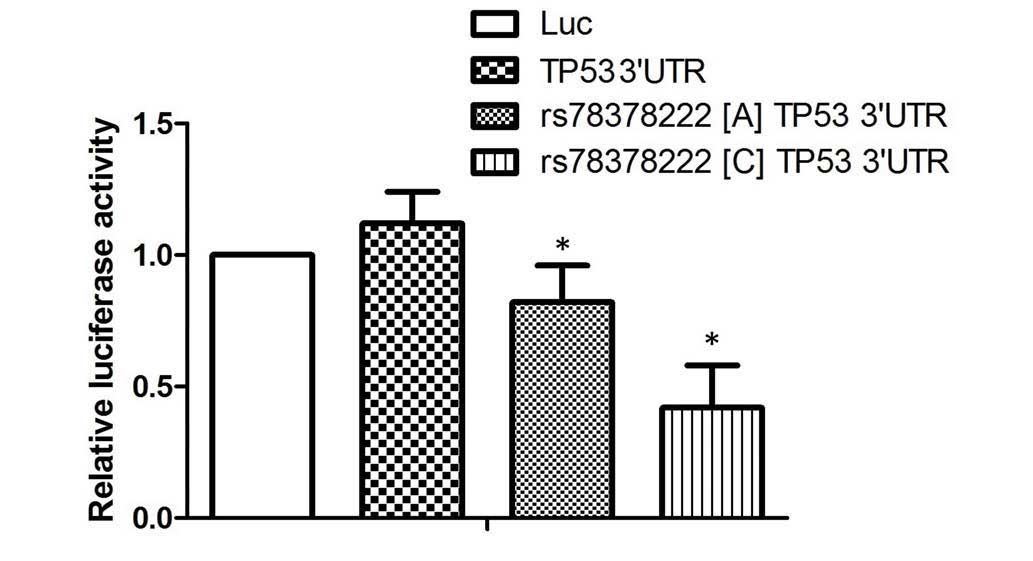
was introduced. The results of the luciferase assay demonstrated
that transfection with wild type TP53 3′-UTR (A) significantly
reduced the luciferase activity of the miR-125b overexpressing
cells, compared with the scramble controls. Furthermore, the
miR-125b overexpressing cells transfected with the construct
containing the minor allele of rs78378222 polymorphism (C) was
reduced further than those transfected with wild type 3′-UTR,
suggesting miR-125 negatively regulates the expression of TP53 by
targeting the 3′-UTR of the gene (P<0.01; Fig. 2), and introduction of a novel
binding site caused by the rs78378222 polymorphism further promoted
the negative regulatory association between miR-125 and TP53.
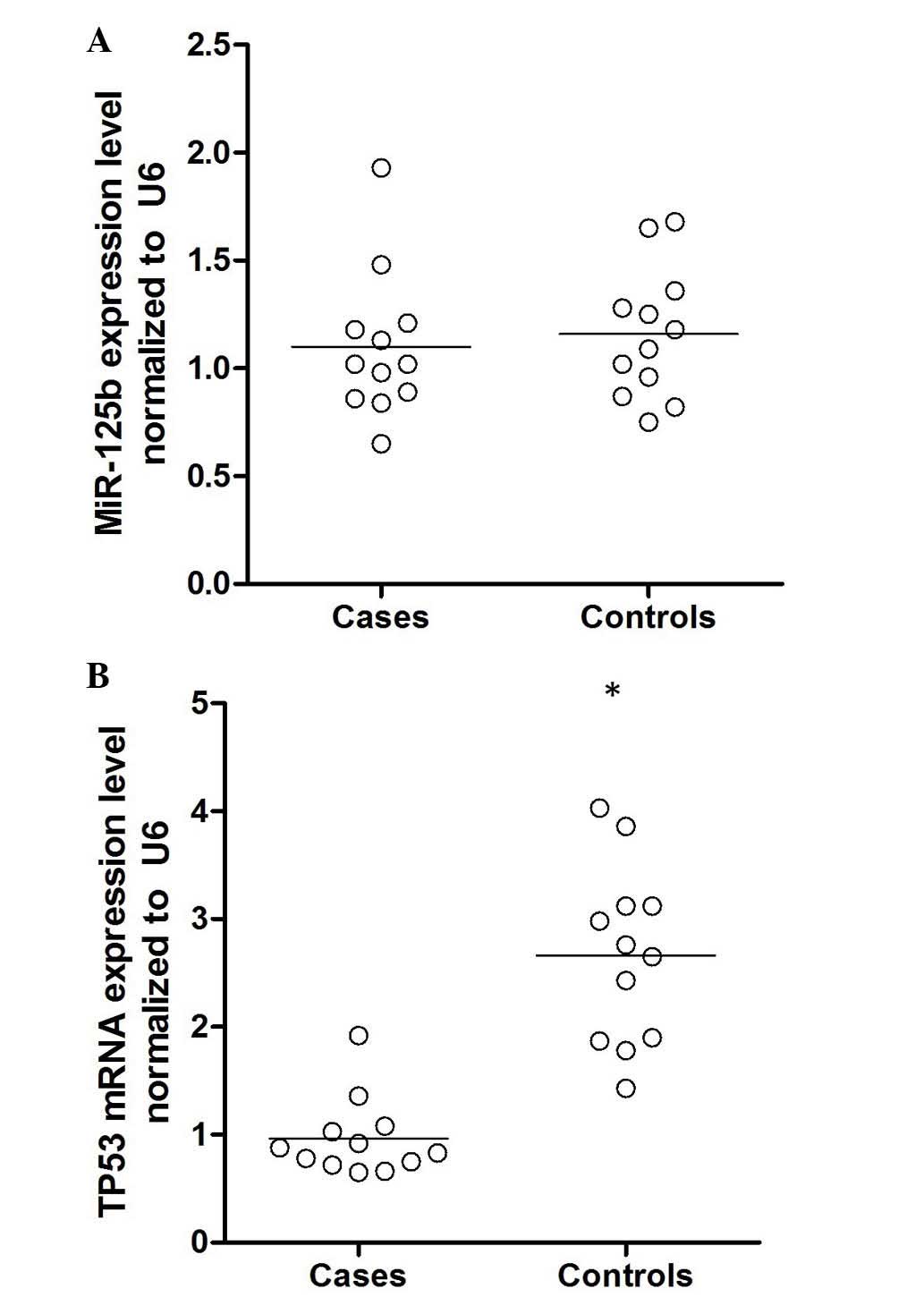
mRNA levels of miR-125b and TP53
To further confirm this hypothesis, epithelial cells
were collected from patients with age-associated cataracts and
controls, and the mRNA expression levels of miR-125b and TP53 were
determined. As demonstrated in Fig.
3, the expression level of miR-125b was comparable between the
two groups, whereas the mRNA expression level of TP53 was
significantly higher in the cataract group compared with the
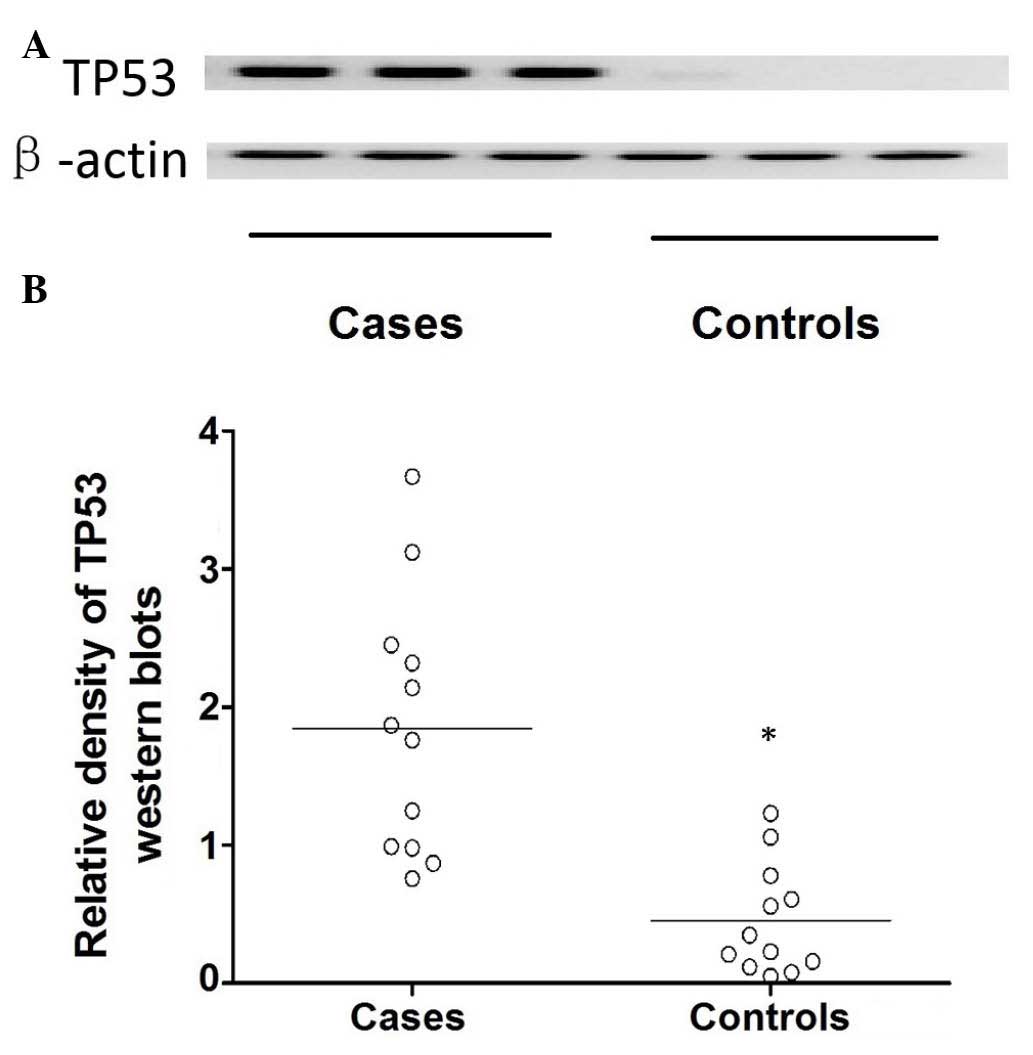
control (P<0.01). Additionally, the expression of TP53 protein
was determined using western blot analysis (Fig. 4). The target bands were quantified
by analyzing their relative density, which demonstrated that the
protein expression level of TP53 was significantly higher in the
cataract cases compared with the control group (P<0.01; Fig. 4).
Regulatory association between miR-125b
and TP53
To further investigate the regulatory association
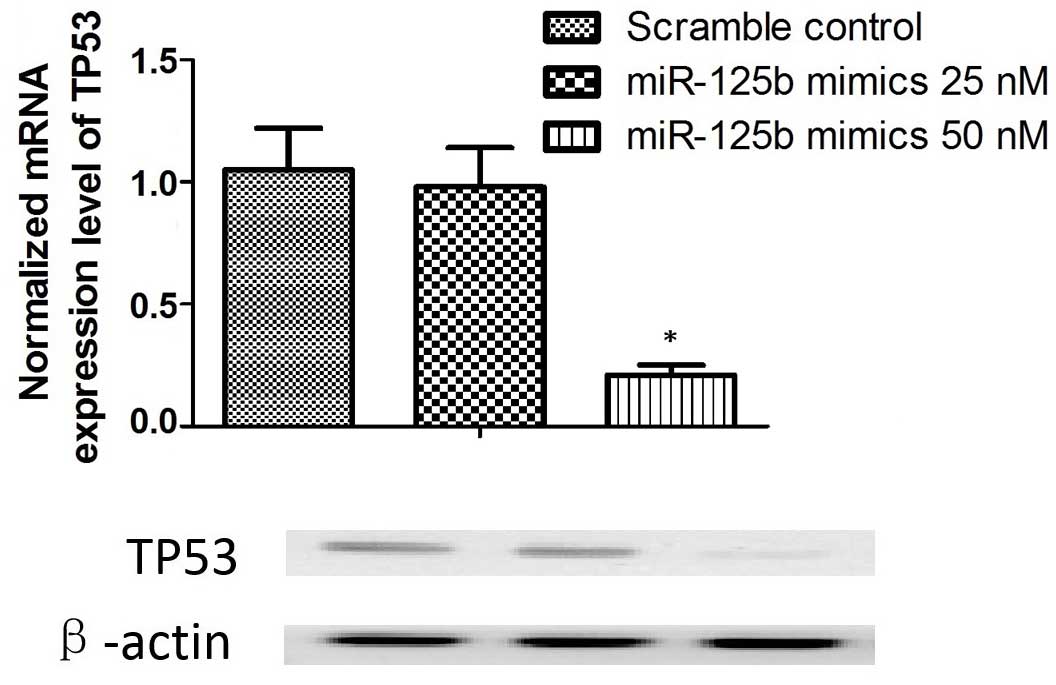
between miR-125b and TP53, miR-125b mimics were transfected into
cultured SRA01/04 epithelial cells. Transfection with 25 nM mimics
was not identified to significantly alter the expression of TP53;
however, 50 nM miR-125b mimics substantially reduced the mRNA and
protein expression levels of TP53 in the lens epithelial cells
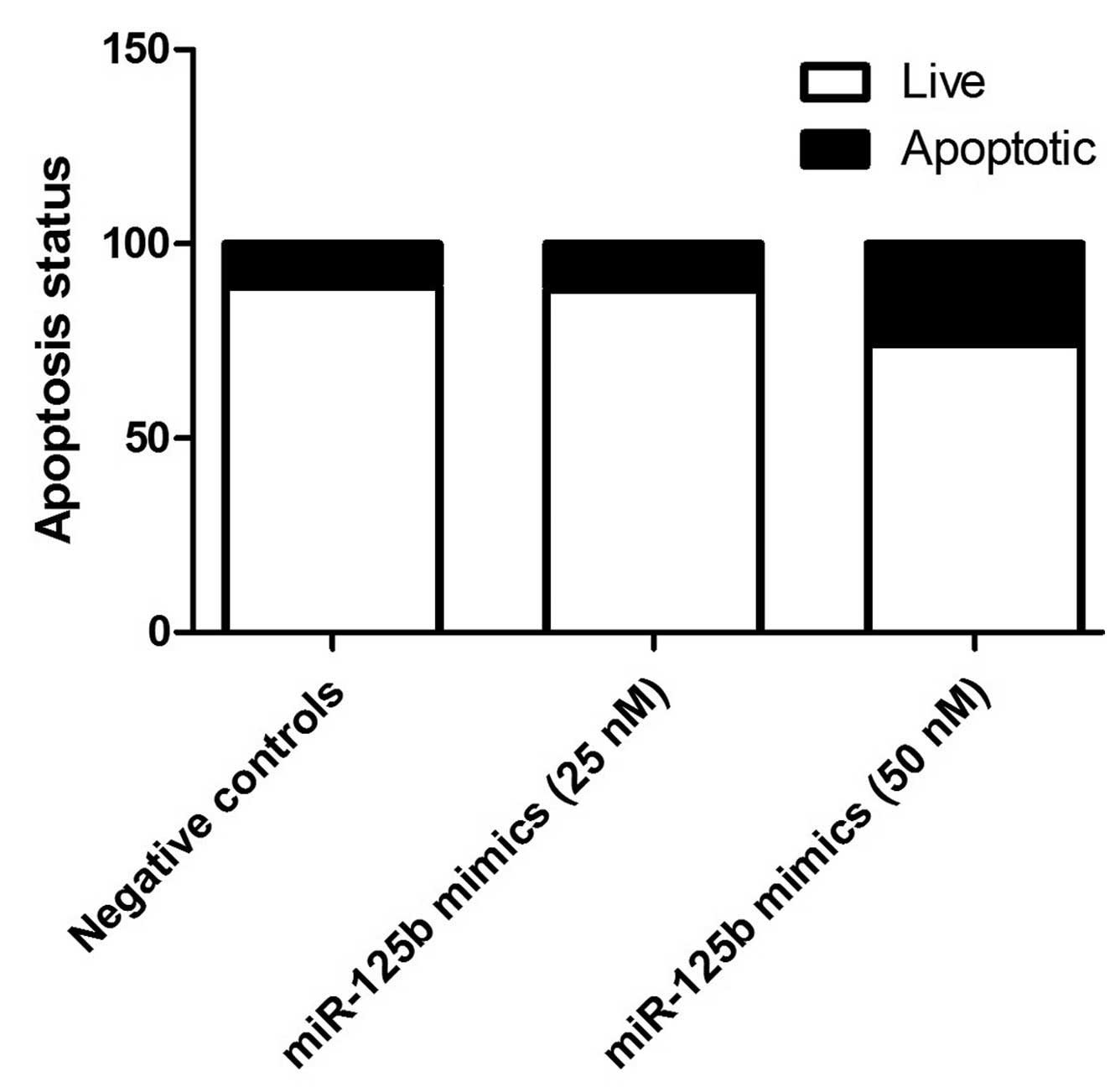
(P<0.01; Fig. 5). Additionally,
compared with negative controls, miR-125b (50 nM) significantly
induced apoptosis in the epithelial cells (Fig. 6).
Discussion
Previous investigation demonstrated that miRNAs are
an important group of gene regulators. The complete miRNA
transcriptome in human lens cells is unclear, although miRNAs have
been detected in a variety of mammalian organs and tissues,
including the eyes. To the best of our knowledge, the present study
is the first to investigate miRNAs in the central epithelium of
cataractous and transparent human lens. Determining the mutations
that lead to the development of age-associated cataracts may
improve understanding of the mechanisms that mediate
cataractogenesis, and provide novel insights into the development
and physiology of the normal lens. Furthermore, functional analysis
of candidate variants is a critical measure to understand the
molecular defects involved in age-associated cataracts, which has a
strong genetic component to its etiology. This may aid in
elucidating novel therapeutic strategies to delay lens
opacification. The present study evaluated the potential effect of
TP53 3′-UTR rs78378222 on the miR-125b-induced apoptosis of lens
epithelial cells, and the association between the rs78378222
polymorphism and the risk of the development of age-associated
cataracts. Initially, the sequences of mature miR-125b and TP53
(wild-type and polymorphic) were analyzed and compared. Replacement
of A with C introduces a novel potential binding site in the 3′-UTR
of TP53 with a consecutive 8-bp perfect match, indicating that the
rs78378222 polymorphism creates a ʻgain-of-function' effect to
regulate TP53 expression via miR-125b.
As a highly conserved miRNA expressed throughout an
array of species ranging from nematode to human, miR-125b acts as
either a repressor or promoter of gene expression, and is
associated with various diseases (24). By acting on a range of different
transcription factors (25),
growth factors (26) and matrix
metalloproteinases (27,28), miR-125b is important for various
cellular processes, including cell proliferation, differentiation
and apoptosis. The current study identified TP53 as a target of
miR-125b, and replacement with the rs78378222 polymorphism minor
allele (C) significantly promoted interaction between the miR-125b
and TP53 mRNA, as demonstrated by a luciferase assay. Furthermore,
the present study demonstrated that transfection of lens epithelial
cells with 25 nM mimics did not significantly alter the expression
of TP53, whereas 50 nM miR-125b mimics significantly reduced the
mRNA and protein expression level of TP53 in the lens epithelial
cells compared with negative control miRNA.
TP53 is a well-studied pro-apoptotic protein in
vivo. Prior to 2011, no genome-wide association study had
demonstrated a significant correlation between any cancer and a
TP53 polymorphism germline (other than variants of Li-Fraumeni
syndrome) (29). The rs78378222
TP53 polymorphism was previously demonstrated to be significantly
associated with breast cancer (30). An independent study confirmed that
rs78378222 was correlated with a 3.5-fold higher risk of glioma
development (31). Stacey et
al (32) demonstrated that the
transcript levels of TP53 expressed by rs78378222 [A/C]
heterozygotes was lower than that in wild type homozygotes in human
blood samples. rs78378222 is located in the fifth nucleotide of the
TP53 polyadenylation signal and was initially hypothesized to be
required for a wide range of processes, including polyadenylation
machinery recognition, cleavage, polyadenylation and transport of
mature mRNAs to the cytoplasm. The current study demonstrated that
replacement with C in the sequence introduces a novel potential
miRNA binding site in the TP53 3′-UTR, with a consecutive 8-bp
perfect match, indicating that rs78378222 creates a
ʻgain-of-function' effect. miR-125 is another important regulator
of TP53, and the minor allele (C) of the rs78378222 polymorphism
ʻgenerates' a novel perfect match binding site for miR-125b in the
3′-UTR of TP53. Additionally, the present study demonstrated that
the expression level of miR-125b was comparable in epithelial cells
from patients with cataracts and controls, whereas the mRNA
expression level of TP53 was significantly higher in the cataract
group compared with the controls, suggesting that the polymorphism
significantly promotes interaction between miR-125b and TP53.
In conclusion, the current study demonstrated that
the minor allele (C) of the rs78378222 polymorphism introduces a
potential novel miR-125b binding site in the 3′-UTR of TP53, with a
consecutive 8-bp perfect match, indicating that the rs78378222
polymorphism induces a ʻgain-of-function' effect to regulate of
TP53 expression via miR-125b. Furthermore, the present study
demonstrated that miR-125b is a novel negative regulator of TP53
in vivo, which may be a mechanism of age-associated cataract
development. miR-125b may be a potential therapeutic target for the
management of age-associated cataracts. Further research in
transgenic animals may be able to increase understanding of the
importance of miR-125b and confirm the results of the present
study.
Acknowledgments
The present study was supported by the National
Natural Science Foundation of Youth Science Foundation (grant no.
51302176).
References
|
1
|
Pascolini D, Mariotti SP, Pokharel GP,
Pararajasegaram R, Etya'ale D, Négrel AD and Resnikoff S: 2002
global update of available data on visual impairment: A compilation
of population-based prevalence studies. Ophthalmic Epidemiol.
11:67–115. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Congdon N, O'Colmain B, Klaver CC, Klein
R, Muñoz B, Friedman DS, Kempen J, Taylor HR and Mitchell P; Eye
Diseases Prevalence Research Group: Causes and prevalence of visual
impairment among adults in the United States. Arch Ophthalmol.
122:477–485. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
West S: Epidemiology of cataract:
Accomplishments over 25 years and future directions. Ophthalmic
Epidemiol. 14:173–178. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Resnikoff S, Pascolini D, Etya'ale D,
Kocur I, Pararajasegaram R, Pokharel GP and Mariotti SP: Global
data on visual impairment in the year 2002. Bull World Health
Organ. 82:844–851. 2004.
|
|
5
|
Cruciani F, Amore F, Albanese G and
Anzidei R: Investigation about causes of blindness and low vision
among members of Blind and Visually Impaired Italian Union (UICI).
Clin Ter. 162:e35–e42. 2011.PubMed/NCBI
|
|
6
|
WHO releases the new global estimates on
visual impairment. Accessed on, http://www.who.int/blindness/en/2011.
|
|
7
|
Chen K and Rajewsky N: The evolution of
gene regulation by transcription factors and microRNAs. Nat Rev
Genet. 8:93–103. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Peters L and Meister G: Argonaute
proteins: Mediators of RNA silencing. Mol Cell. 26:611–623. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Karali M, Peluso I, Marigo V and Banfi S:
Identification and characterization of microRNAs expressed in the
mouse eye. Invest Ophthalmol Vis Sci. 48:509–515. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ryan DG, Oliveira-Fernandes M and Lavker
RM: MicroRNAs of the mammalian eye display distinct and overlapping
tissue specificity. Mol Vis. 12:1175–1184. 2006.PubMed/NCBI
|
|
11
|
Maegdefessel L: The emerging role of
microRNAs in cardiovascular disease. J Intern Med. 276:633–644.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lee YH, Kim SY and Bae YS: Upregulation of
miR-760 and miR-186 is associated with replicative senescence in
human lung fibroblast cells. Mol Cells. 37:620–627. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mimura S, Iwama H, Kato K, Nomura K,
Kobayashi M, Yoneyama H, Miyoshi H, Tani J, Morishita A, Himoto T,
et al: Profile of microRNAs associated with aging in rat liver. Int
J Mol Med. 34:1065–1072. 2014.PubMed/NCBI
|
|
14
|
Khee SG, Yusof YA and Makpol S: Expression
of senescence-associated microRNAs and target genes in cellular
aging and modulation by tocotrienol-rich fraction. Oxid Med Cell
Longev. 2014:7259292014.PubMed/NCBI
|
|
15
|
Hughes AE, Bradley DT, Campbell M, Lechner
J, Dash DP, Simpson DA and Willoughby CE: Mutation altering the
miR-184 seed region causes familial keratoconus with cataract. Am J
Hum Genet. 89:628–633. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kota J, Chivukula RR, O'Donnell KA,
Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P,
Torbenson M, Clark KR, et al: Therapeutic microRNA delivery
suppresses tumorigenesis in a murine liver cancer model. Cell.
137:1005–1017. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lanford RE, Hildebrandt-Eriksen ES, Petri
A, Persson R, Lindow M, Munk ME, Kauppinen S and Ørum H:
Therapeutic silencing of microRNA-122 in primates with chronic
hepatitis C virus infection. Science. 327:198–201. 2010. View Article : Google Scholar
|
|
18
|
Wang Z, Rajaraman P, Melin BS, Chung CC,
Zhang W, McKean-Cowdin R, Michaud D, Yeager M, Ahlbom A, Albanes D,
et al: Further confirmation of germline glioma risk variant
rs78378222 in TP53 and its implication in tumor tissues via
integrative analysis of TCGA data. Hum Mutat. 36:684–688.
PubMed/NCBI
|
|
19
|
Guan X, Wang LE, Liu Z, Sturgis EM and Wei
Q: Association between a rate novel TP53 variant (rs78378222) and
melanoma, squamous cell carcinoma of the head and neck and lung
cancer susceptibility in non-Hispanic Whites. J Cell Mol Med.
17:873–878. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Long AC, Bomser JA, Grzybowski DM and
Chandler HL: All-trans retinoic acid regulates cx43 expression, gap
junction communication and differentiation in primary lens
epithelial cells. Curr Eye Res. 35:670–679. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Carlson RV, Boyd KM and Webb DJ: The
revision of the Declaration of Helsinki: Past, present and future.
Br J Clin Pharmacol. 57:695–713. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chylack LT Jr, Wolfe JK, Singer DM, Leske
MC, Bullimore MA, Bailey IL, Friend J, McCarthy D and Wu SY: The
lens opacities classification system III. The longitudinal study of
cataract study group. Arch Ophthalmol. 111:831–836. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ferracin M, Bassi C, Pedriali M, Pagotto
S, D'Abundo L, Zagatti B, Corrà F, Musa G, Callegari E, Lupini L,
et al: miR-125b targets erythropoietin and its receptor and their
expression correlates with metastatic potential and ERBB2/HER2
expression. Mol Cancer. 12:1302013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun YM, Lin KY and Chen YQ: Diverse
functions of miR-125 family in different cell contexts. J Hematol
Oncol. 6:62013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bousquet M, Nguyen D, Chen C, Shields L
and Lodish HF: MicroRNA-125b transforms myeloid cell lines by
repressing multiple mRNA. Haematologica. 97:1713–1721. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ge Y, Sun Y and Chen J: IGF-II is
regulated by microRNA-125b in skeletal myogenesis. J Cell Biol.
192:69–81. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bi Q, Tang S, Xia L, Du R, Fan R, Gao L,
Jin J, Liang S, Chen Z, Xu G, et al: Ectopic expression of MiR-125a
inhibits the proliferation and metastasis of hepatocellular
carcinoma by targeting MMP11 and VEGF. PLoS One. 7:e401692012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xu N, Zhang L, Meisgen F, Harada M,
Heilborn J, Homey B, Grandér D, Ståhle M, Sonkoly E and Pivarcsi A:
MicroRNA-125b down-regulates matrix metallopeptidase 13 and
inhibits cutaneous squamous cell carcinoma cell proliferation,
migration, and invasion. J Biol Chem. 287:29899–29908. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Thomas DC, Haile RW and Duggan D: Recent
developments in genomewide association scans: A workshop summary
and review. Am J Hum Genet. 77:337–345. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Rao AK, Vinothkumar V, Revathidevi S,
Arunkumar G, Manikandan M, Arun K, Rajkumar KS, Ramani R,
Ramamurthy R and Munirajan AK: Abscence of the TP53 poly-A signal
sequence variant rs78378222 in oral, cervical and breast cancers in
South India. Asian Pac J Cancer Prev. 15:9555–9556. 2014.
View Article : Google Scholar
|
|
31
|
Egan KM, Nabors LB, Olson JJ, Monteiro AN,
Browning JE, Madden MH and Thompson RC: Rare TP53 genetic variant
associated with glioma risk and outcome. J Med Genet. 49:420–421.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Stacey SN, Sulem P, Jonasdottir A, Masson
G, Gudmundsson J, Gudbjartsson DF, Magnusson OT, Gudjonsson SA,
Sigurgeirsson B, Thorisdottir K, et al: A germline variant in the
TP53 polyadenylation signal confers cancer susceptibility. Nat
Genet. 43:1098–1103. 2011. View
Article : Google Scholar : PubMed/NCBI
|