Introduction
Lung cancer is a major contributor to mortality
rates worldwide, with a mortality rate of >160,000 in America
and a 5-year survival rate of ~16% (1). Lung squamous cell carcinoma (SCC) is
defined as a common type of non-small cell lung cancer (NSCLC),
which accounts for ~80% of all cases of lung cancer (2,3). SCC
is usually found in aged males and is correlated closely with
smoking (4). SCC is also
associated with high rates of metastasis and relapse (5). Therefore, further investigations on
prognostic markers are required to manage the clinical treatments.
Metastasis and angiogenesis are the hallmarks of cancer, and are
the most important factors leading to cancer-associated mortality.
Research has improved the understanding of metastasis (6), however, the precise molecular
mechanisms remain to be fully elucidated. Although it has been
revealed that mutations of several oncogenes, including Kirsten rat
sarcoma viral oncogene homolog, anaplastic lymphoma kinase and
epidermal growth factor receptor, or genome amplification
contribute to the promotion and advanced stages of SCC (7,8),
further investigations to identify novel targets and elucidate
molecular mechanisms are required.
MicroRNAs (miRNAs) are identified as a class of
short non-coding RNAs, which regulate gene expression through
binding to the 3′untranslated regions of the mRNAs of target genes,
mediating its stability and degradation (9). In previous years, studies on miRNAs
have shown that they are critical in cellular processes, including
proliferation, apoptosis and motility. The dysregulation of miRNAs
has been demonstrated in several types of tumor, including lung
cancer, breast cancer and colon cancer, promoting tumorigenesis
(10–12). Among these miRNAs, miR-588 has
previously been reported in colorectal cancer-associated miRNAs
(13), however, its role in lung
cancer, particularly in SCC, has not been reported.
GRN (progranulin), also known as PC cell-derived
growth factor, has been identified as a tissue and circulating
biomarker for NSCLC (14). The
deregulation of GRN has been found in several types of tumor,
including breast cancer, prostate cancer, chronic lymphocytic
leukemia and hepatocellular carcinoma (15). For example, in breast cancer, the
expression of GRN has been found to be associated with the
expression of estrogen receptor and the acquisition of resistance
to drugs (16). Various studies
have reported that the overexpression of GRN can activate multiple
important pathways, including the mitogen-activated protein kinase,
phosphoinositide 3-kinase/Akt and focal adhesion kinase pathways,
promoting cell proliferation, invasion and tumorgenesis (17,18).
However, the significance and molecular mechanisms accounting for
the upregulation of GRN remain to be fully elucidated.
The aims of the present study were to provide
evidence of the expression of miR-588 and its target gene, GRN, in
SCC. The results showed that the expression of miR-588 was higher
in non-tumor tissues, compared with SCC tissues, and that miR-588
suppressed SCC cell migration and invasion. In addition, GRN was
identified as a potential target of miR-588, which was upregulated
in SCC tissues, and the abnormal expression of GRN was involved in
the tumor-suppressive functions mediated by miR-588.
Materials and methods
Human tissues and cell culture
A total of 85 pairs of human SCC and adjacent
non-tumor tissues were collected between 2005 and 2014 at the
Department of Pathology, First Hospital of Shanxi Medical
University (Shanxi, China). The patients had no received systemic
chemotherapy or radiotherapy prior to surgery. The clinical data
collected consisted of age, gender, pathological tumor stage and
lymph node invasion. The detailed information is shown in Table I. All specimens were histologically
confirmed by two pathologists. Tumor subtype was examined based on
the World Health Organization (WHO) 2004 criteria, and the staging
was according to the TNM classification (UICC 2009) (19,20).
All human tissues were acquired with informed consent and the
approval of the Ethical Review Committee of the WHO of the
Collaborating Center for Research in Human Production.
 | Table IClinical characteristics of 85
patients with lung squamous cell carcinoma. |
Table I
Clinical characteristics of 85
patients with lung squamous cell carcinoma.
| Characteristic | Cases (n) |
|---|
| Age (years) | |
| <50 | 26 |
| ≥50 | 59 |
| Gender | |
| Male | 63 |
| Female | 22 |
| Stage | |
| I | 12 |
| II | 45 |
| III+IV | 28 |
| Lymph node
invasion | |
| Negative | 55 |
| Positive | 30 |
| Smoking
history | |
| Smoker | 48 |
| Non-smoker | 37 |
Human lung SCC cell lines (H226, H2170 and H1703)
were purchased form American Type Culture Collection (ATCC;
Manassas, VA, USA). All cell lines were maintained in RPMI 1640
medium (Gibco: Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% fetal bovine serum (Hyclone: GE Healthcare
Life Sciences, Logan, UT, USA) and antibiotics. The cells were
incubated under 5% CO2 and 37°C conditions according to
ATCC.
Oligonucleotide transfection
The miR-588 mimics and small interfering (si)RNA
against GRN (forward 5′-GCUUCCAAAGAUCAGGUAACATT-3′ and reverse
5′-UGUUACCUGAUCUUUGGAAGCTT-3′) were synthesized by GenePharma
(Shanghai, China). The miR-588 inhibitors were synthesized by
Thermo Fisher Scientific, Inc. Transfection was performed using
Lipofectamine 2000 reagents according to the manufacturer's
protocol.
Total RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis
Total RNA was extracted from the 85 clinical tissue
samples using TRIzol reagent (Invitrogen; Thermo Fisher Scientific,
Inc,) according to the manufacturer protocol. Tissues were
homogenized in the TRIzol reagent and then centrifuged at 13000 × g
at 4°C for 20 min. Reverse transcription was performed to convert
the extracted RNA (200 ng) into cDNA with oligo (dT) primer (25
pmol) and M-MLV reverse transcriptase (Takara Biotechnology Co.,
Ltd., Dalian, China) incubation at 42°C for 60 min. RT-qPCR was
performed using the SYBR-Green method (SYBR Premix Ex Taq II;
Takara Biotechnology Co., Ltd.) with GRN-specific and β-actin
primers (used at 0.4 µM) as an internal control. qPCR was
performed using an ABI7500 real-time PCR system (Applied
Biosystems: Thermo Fisher Scientific, Inc.) with incubation at 95°C
for 30 sec (predenaturation) followed by 40 cycles of 95°C for 30
sec and 60°C for 30 sec. The primers were as follows: GRN, forward
5′-ATCTTTACCGTCTCAGGGACTT-3′ and reverse
5′-CCATCGACCATAACACAGCAC-3′ and β-actin, forward
5′-ATGATGATATCGCCGCGCTC-3′ and reverse 5′-CATCACGCCCTGGTGCC-3′,
respectively. The relative expression levels of GRN were calculated
using the ΔΔCq method as described previously (21).
For measuring the expression of miR-588, TaqMan
miRNA assays (Applied Biosystems: Thermo Fisher Scientific, Inc.)
were performed according to the manufacturer's protocol, and U6
small nuclear RNA was used to normalize the relative gene
expression.
Western blot analysis
The tissues or cells were lyzed in RIPA protein
extraction reagent according to the manufacturer's protocol. The
proteins were quantified using a BCA protein assay kit (Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
Subsequently, 50 µg of total protein was loaded per lane of
a 10% SDS gel, and then transferred onto a nitrocellulose membrane
and blotted as described previously (22). The membrane was incubated with
primary antibody at 4°C overnight. Following washing with
phosphate-buffered saline, secondary antibody incubated with the
membrane for 1 h. Primary antibodies included anti-GRN (rabbit
anti-human monoclonal antibody; 1:1,000 dilution; cat. no.
ab108608; Abcam, Cambridge, UK), anti-vascular endothelial growth
factor (VEGF; mouse anti-human monoclonal antibody; 1:1,000
dilution; cat. no. ab69479; Abcam) and anti-GAPDH (mouse anti-human
monoclonal antibody; 1:2,000 dilution; cat. no. sc-365062; Santa
Cruz Biotechnology, Inc., Santa Cruz, CA, USA). Secondary
antibodies were goat horseradish peroxidase (HRP)-conjugated
anti-mouse IgG (1:5,000; cat. no. sc-2005) and goat anti-rabbit IgG
(1:5,000; cat. no. sc-2004) from Santa Cruz Biotechnology, Inc. The
protein bands were visualized by enhanced chemiluminescent
detection (Super Signal West Dura Extended Duration
Chemiluminescent Substrate, Thermo Fisher Scientific, Inc.) and
quantified by Quantity One software (version 4.4.036, Bio-Rad
Laboratories, Inc., Hercules, CA, USA).
Immunohistochemical staining
All tissue specimens were surgically resected with
adjacent normal lung tissues and fixed in 10% formalin followed by
being embedded in paraffin. Serial 4-µm-thick sections were
used for immunohistochemical analysis.
The formalin-fixed and paraffin-embedded sections
were heated at 60°C for 30 min and dewaxed twice in xylene,
followed by exposure to graded ethanol and finally water.
Endogenous peroxidase was blocked by incubation in 0.3% hydrogen
peroxide methanol for 10 min, followed by antigen retrieval, which
was performed via boiling in citrate buffer for 20 min. The
sections were incubated with a 1:200 dilution of anti-GRN primary
antibody overnight at 4°C. The following day, the sections were
washed and then incubated with HRP-labeled anti-IgG for 30 min.
Finally, the sections were stained using diaminobenzidine substrate
chromogen and counterstained with hematoxylin. All these steps were
performed under a light microscope to control the reactions, and
phosphate-buffered saline was performed instead of primary antibody
as a negative control.
Evaluation of immunohistochemical
staining
The immunostaining of GRN was quantified, blinded to
the patients' clinical data, as described previously (23). In brief, the percentage of staining
intensity was scored as follows: No staining=0; <15% staining=1
(weak); 15–50% staining =2 (moderate); >50% staining =3
(strong). Furthermore, according to the percentage staining of the
tumor cells and the sections, the scores were determined as
intensity x percentage, to produce a final score of 0–300. The
sections were distinguished into two groups, with either a high
expression level of GRN or a low expression level of GRN.
Migration and invasion assays
For the migration assays, 2×104 cells
were plated into the top chamber of cell culture inserts (BD
Biosciences, San Diego, CA, USA) with serum-free medium. For the
invasion assays, 2×104 cells were plated into the top
chamber of the insert, which was precoated with Matrigel (BD
Biosciences). The bottom chamber contained a 10% FBS medium.
Following 2 days of incubation 5% CO2 and 37°C
conditions, cells adhering to the lower membrane were fixed and
stained with 0.1% crystal violet. Images of the cells were
captured, and the cells were counted using a light microscope
(Olympus Corporation, Tokyo, Japan).
Target gene identification
TargetScan version 7.1 (www.targetscan.org/vert_71/) was used to predict the
potential binding target of miRNA and genes.
Luciferase activity assays
The 3′-UTR of GRN were cloned into the PGL3 basic
vector (Promega Corp., Madison, WI, USA). For the luciferase
assays, 200 ng of the PGL3-GRN-3′UTR vector were co-transfected
into 5×104 cells with 100 nM miR-588 mimics or control,
together with 40 ng Renilla luciferase vector (Promega
Corp,) as an internal control. The cells were harvested after 48 h
of incubation at 5% CO2 and 37°C conditions following
transfection and the luciferase activities were assayed according
to the manufacturer's protocol. The transfections were performed in
duplicate and repeated three times.
Statistical analysis
All statistical analysis was performed using SPSS
21.0 software (IBM SPSS, Armonk, NY, USA). The association between
miR-588 and GRN was determined using Spearman analysis. Other
experiments were repeated three times and assessed using Student's
t-test. Data are expressed as the mean ± standard error of
the mean. P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-588 is downregulated in SCC, and is
correlated with advanced clinical stage and lymph node
invasion
To determine the expression of miR-588 in SCC, the
mature miR-588 was detected in 85 pairs of SCC tissue samples and
non-tumor samples. The results showed that the expression of
miR-588 was significantly downregulated in the SCC tissues,
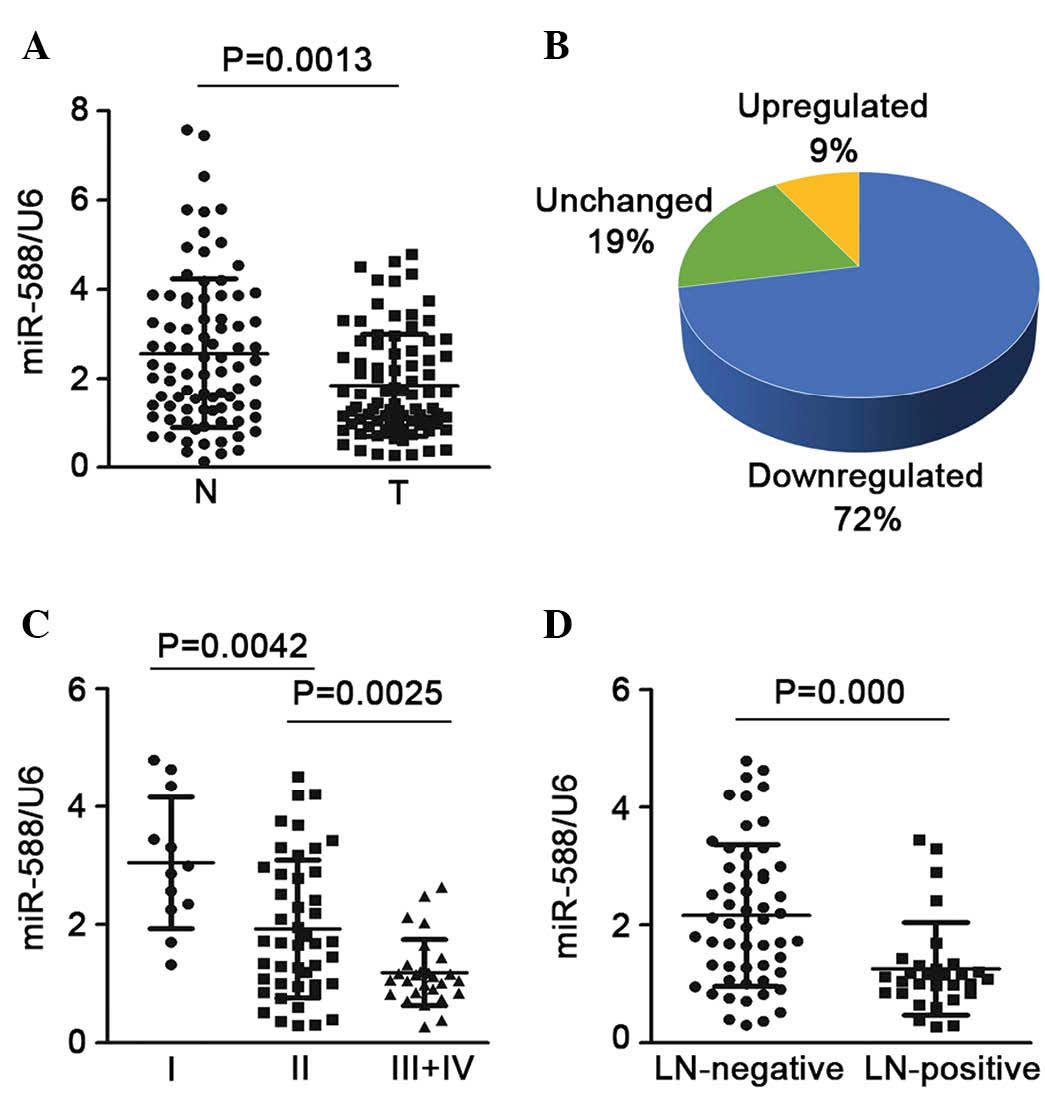
compared with the non-tumor tissues (Fig. 1A). In ~72% of the samples,
expression was downregulated >2-fold, whereas only 9% of the
samples showed upregulated expression (Fig. 1B). The present study also assessed
the association between the expression of miR-588 and advanced
stage or lymph node invasion states. It was found that SCC at an
advanced stage (stages III and IV) had a lower expression level of
miR-588, compared with SCC at an early stage (stage I or II;
Fig. 1C). In addition, when the 85
SCC samples were cataloged based on their status of lymph node
invasion, it was found that the expression of miR-588 was notably
downregulated in the SCC samples, which were positive for lymph
node invasion, compared with those without (Fig. 1D). Taken together, these finding
suggested that the downregulation of miR-588 may be critical in the
progression and metastasis of SCC.
miR-588 suppresses the migration and
invasion of SCC cells
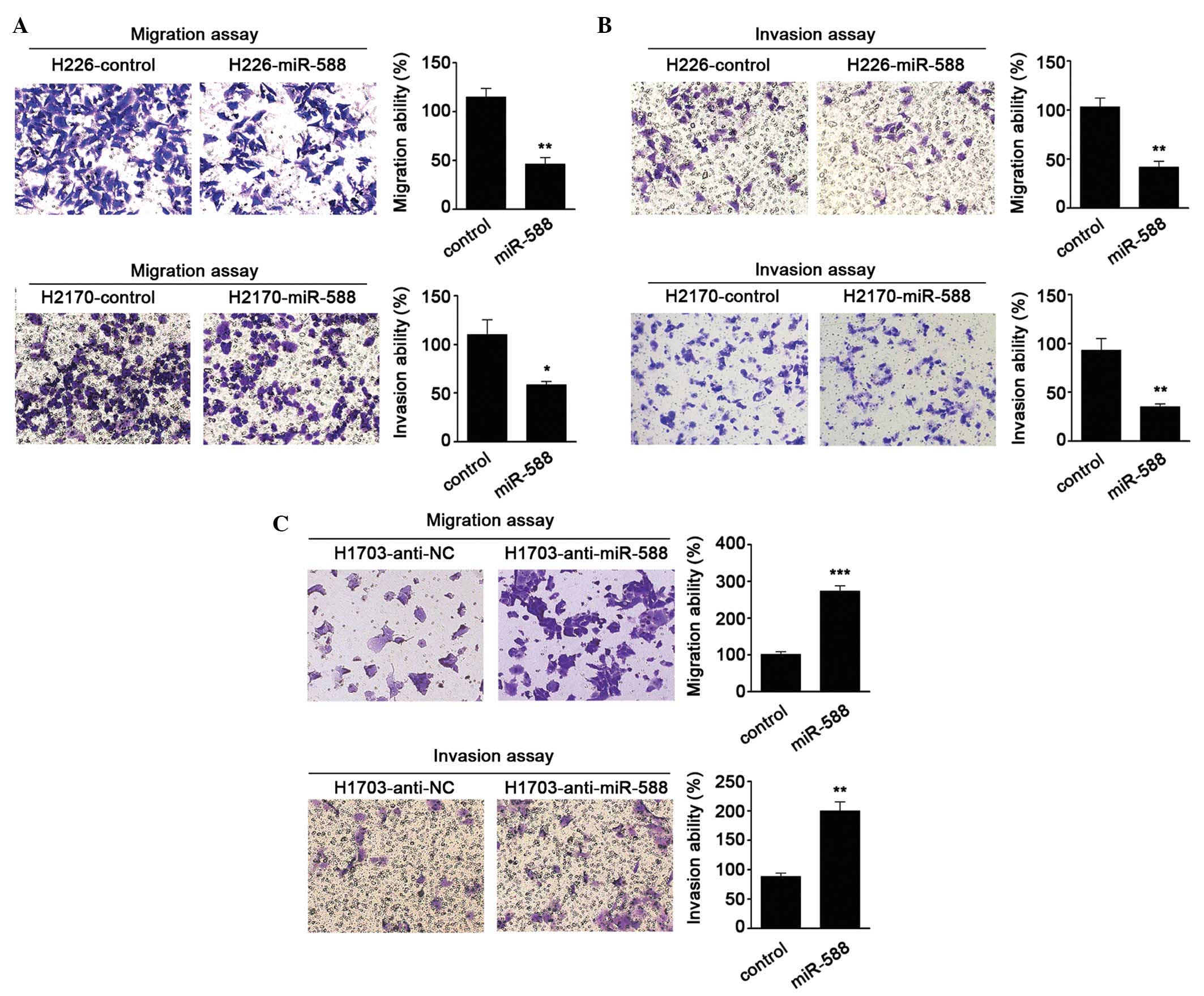
To further investigate the biological functions of
miR-588 in SCC, gain- and loss-of-function analyses were performed
in SCC cells. miR-588 mimics were transfected into H226 and H2170
cells, as these two cells had lower relative basal expression
levels of miR-588. It was found that miR-588 significantly
suppressed the migration and invasion abilities of the cells
(Fig. 2A and B). Antagonists were
then used to inhibit the expression of miR-588 in H1703 cell lines.
The results were confirmed the hypothesis that the knockdown of
miR-588 increases SCC cell migration and invasion (Fig. 2C). These data suggested that
miR-588 conferred negative effects on SCC metastasis.
GRN is a downstream target of miR-588 in
SCC
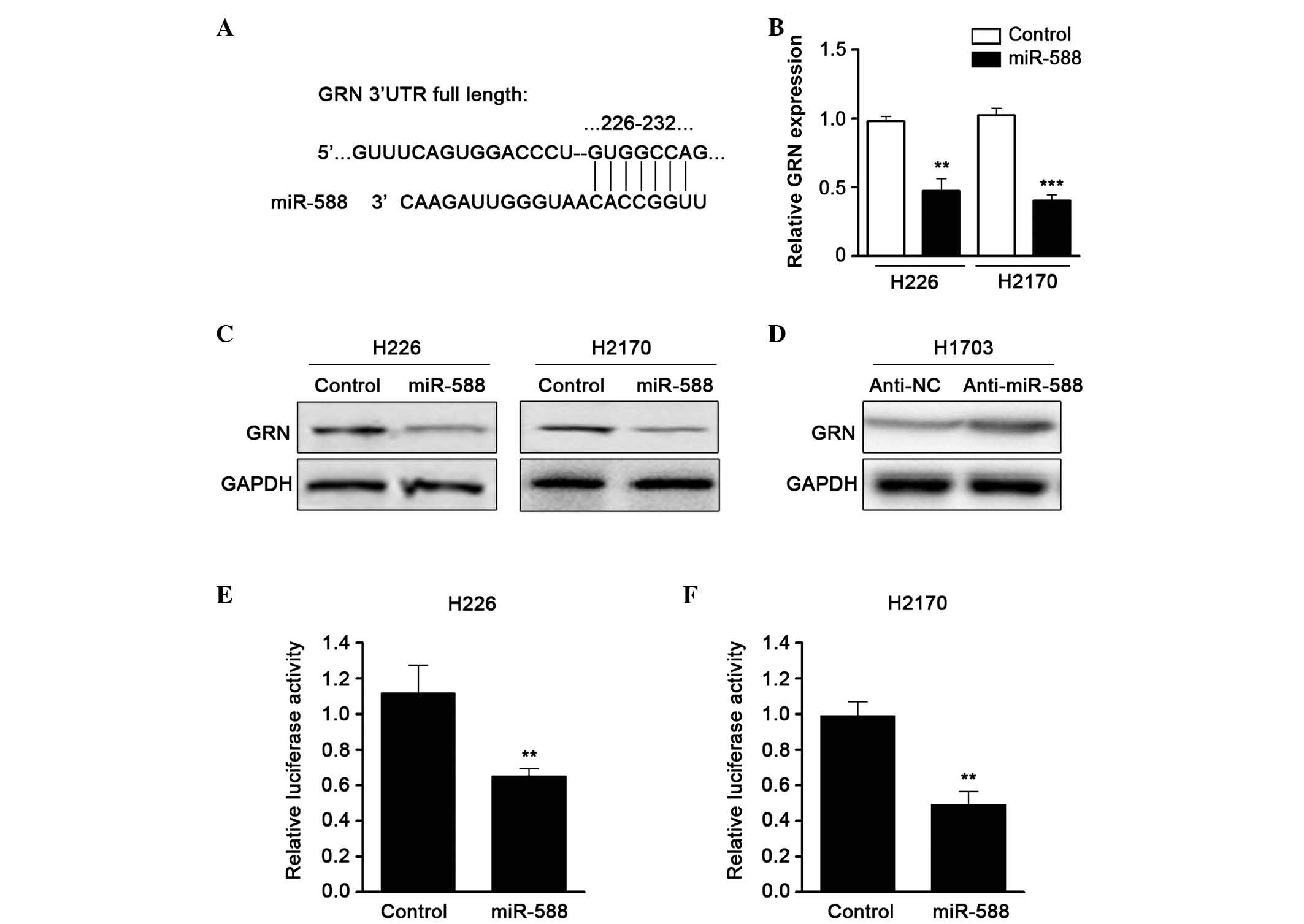
To further examine the molecular targets of miR-588
in SCC metastasis, the TargetScan algorithm was used to predict the
potential downstream gene of miR-588. GRN, a growth factor which
contributes to tumor invasion and metastasis, was one of the
candidates identified (Fig. 3A).
RT-qPCR was then performed to confirm the expression of GRN when
miR-588 was overexpressed by mimics. The results showed that the
mRNA expression of GRN was downregulated by miR-588 (Fig. 3B). In addition, western blot assays
revealed that the protein level of GRN was also reduced (Fig. 3C). The present study subsequently
used an antagonist of miR-588 and found that suppressing miR-588
significantly enhanced the expression of GRN in SCC cell lines
(Fig. 3D). To confirm that GRN was
a direct target of miR-588, a luciferase activity assay was
performed. A luciferase reporter vector containing the 3′UTR of GRN
was cloned and cotransfected with miR-588 mimics into the cells.
The relative luciferase activity was markedly reduced when the
cells were transfected with miR-588, compared with the control
(Fig. 3E and F). These data
suggested that the expression of GRN was regulated by miR-588 in
SCC.
Upregulation of GRN is frequent and
inversely associated with the expression of miR-588 in SCC
To determine the association between the expression
of GRN and miR-588 and the significance of the downregulation of
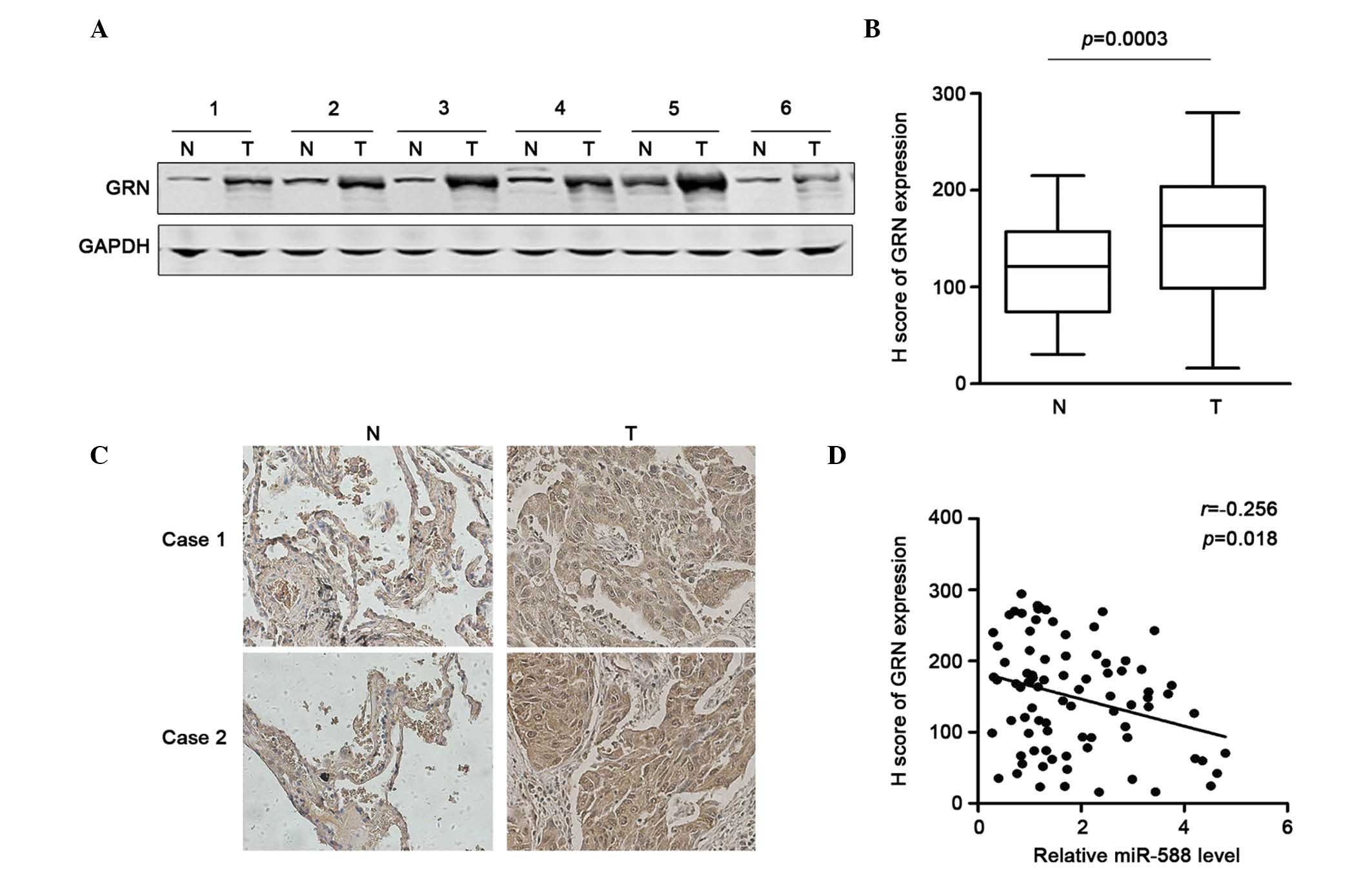
miR-588 in SCC, the present study first examined the expression of
GRN in six pairs of SCC and non-tumor tissues using western blot
analysis. The results showed that GRN was frequently overexpressed
in the SCC tissues (Fig. 4A). The
sample number was then increased and immunohistochemical staining
was performed to determine the protein expression of GRN in 85
pairs of SCC tissues and adjacent non-tumor tissues. Using H
scoring statistics, it was found that the expression of GRN was
significantly higher in the SCC tissues, compared with the
non-tumor tissues (Fig. 4B). The
representative results are shown in Fig. 4C. The correlation between the
expression of miR-588 and GRN in these samples were then analyzed.
Using Spearman analysis, it was found that the downregulation of
GRN was correlated with the upregulatied expression of miR-588
(Fig. 4D). Taken together, these
data suggested that the upregulation of GRN may be due to the
repression of miR-588 in SCC.
GRN is involved in the miR-588-mediated
suppression of SCC cell metastasis
It has been reported that GRN is critical in
stimulating cell migration and promoting metalloproteinase
activity. In addition, GRN can upregulatet the expression of VEGF
(24). As miR-588 was found to be
downregulated in SCC, and miR-588 downregulated the expression of
GRN, the present study examine whether the GRN was involved in the
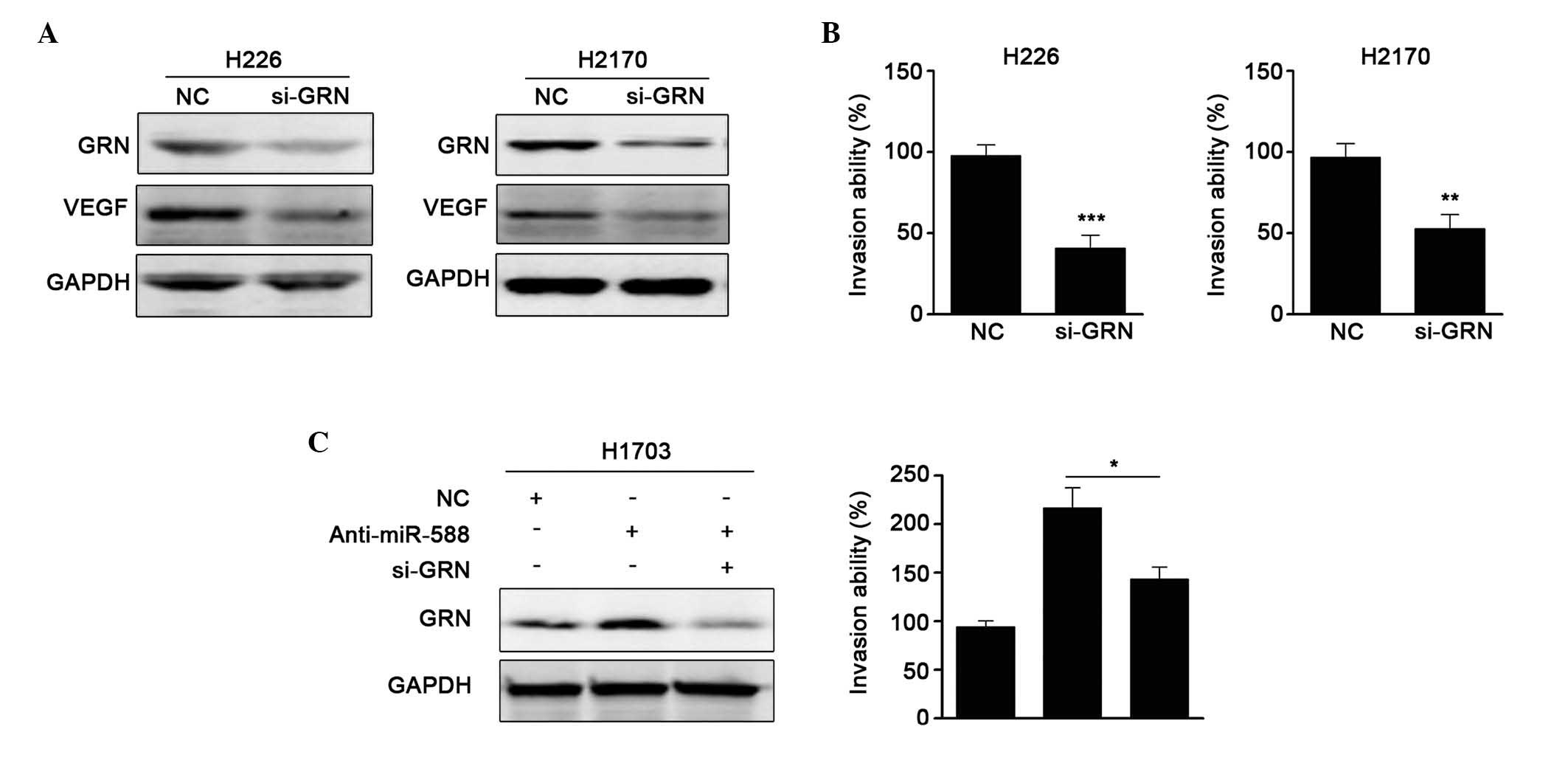
miR-588-mediated suppression of SCC cell metastasis. First, siRNA
against GRN was used, and it was found that si-GRN significantly
reduced the protein expression of GRN and the expression of VEGF.
Invasion assays showed that si-GRN suppressed cell metastasis,
which was similar to the results obtained when miR-588 was
overexpressed (Fig. 5A and B).
Subsequently, anti-miR-588 and si-GRN were transfected into H1703
cells, and the protein levels were confirmed, as shown in Fig. 5C. It was found that the enhanced
metastatic ability mediated by anti-miR-588 was attenuated by
si-GRN. Collectively, these results suggested that GRN was involved
in the miR-588-mediated suppression of SCC cell metastasis.
Discussion
In the present study, it was shown that miR-588 was
significantly decreased in SCC, particularly at advanced stages and
in cases with lymph node invasion. The reinforcement of miR-588
inhibited the migration and invasion abilities of the SCC cells,
and an antagonist of miR-588 promoted cell migration and invasion.
Further investigations revealed that GRN was a direct target of
miR-588. The expression of GRN was significantly higher in the SCC
tissues, compared with the adjacent non-tumorous tissues, and was
negatively correlated with the expression of miR-588. These data
suggested that miR-588 acts as important negative regulator in the
development and progression of SCC.
Metastasis is characterized as an important
life-threatening factor in the majority of types of cancer. Lymph
node invasion and angiogenesis are the major routes for SCC tumor
cell metastasis (25,26). Therefore, it is important to
elucidate the underlying molecular mechanisms. Several studies have
reported that miRNAs are associated with lymph node invasion and
angiogenesis by targeting certain crucial factors. For example,
miR-15a and miR-16 can target VEGF, which subsequently affects the
angiogenesis of multiple myeloma (27). miR-194 downregulates the expression
of matrix metalloproteinase 2/9 through targeting the expression of
bone morphogenetic protein 1 and reducing the activity of
transforming growth factor-β in NSCLC (28). The present study revealed that
miR-588 was complementary to the GRN 3′UTR. The in vitro
restoration of miR-588 significantly suppressed cell migration and
invasion. Thus, miR-588 may have potential as a treatment target
for SCC.
The biological functions on miR-588 remain to be
fully elucidated. Previous studies have shown that miR-588 was
downregulated in cells with high metastatic capacity. Li et
al (29) used an miRNA assay
to scan miRNAs, which were differentially expressed between cells
with high and low metastatic capacities. Their investigation found
that the expression levels of miR-339-5p and miR-588 were decreased
in cells with high metastatic capacity. Almog et al
(30) demonstrated that high
expression levels of miR-588 were shown in dormant tumors, compared
with the fast-growing glioblastoma. In the presents study, miR-588
was inversely correlated with advanced tumor stage and these
results were consistent with previous studies (31), indicating tumor suppression
functions.
GRN has been found to be a target of miR-659 and
contains common genetic variability in its miRNA binding site
(32). miR-659 has been
demonstrated to bind to position 83–89 of the GRN 3′UTR in complex
neurodegenerative disorders, whereas miR-588 binds to position
226–232. GRN has also been reported to be regulated by miR-29b in
frontotemporal dementia (33).
These results suggest that multiple miRNAs may contribute to
modulation of the expression of GRN. GRN has been shown be critical
in pathogenesis, and as an autocrine growth and survival factor in
several types of cancer (15). GRN
is expressed in 70% of lung adenocarcinoma and squamous cell
carcinoma tissues, whereas it is negative in normal lung tissues
and in small cell carcinoma tissues (14). In addition, GRN can stimulate
migration, invasiveness and the expression of VEGF in breast cancer
(34). Therefore, the present
study assessed the expression of VEGF when siRNA was used to
knockdown GRN in SCC cells. The results provided support for the
association between GRN and VEGF. It it also well documented that
the expression of GRN has been associated with drug resistance in
NSCLC (35). Further
investigations are necessary to determine the functions of miR-588
in patients who may benefit from chemical treatment.
In conclusion, the results of the present study
showed that the expression of miR-588 was downregulated in tumor
tissues, compared with normal tissues, and was associated with
lymph node metastasis in SCC. Furthermore, the enforced expression
of miR-588 suppressed SCC cell invasion and migration through the
direct targeting of GRN. These findings indicated that the
downregulation of miR-588 in SCC contributed to SCC metastasis and
progression, suggesting that miR-588 may be useful as a biomarker
and potential therapeutic target in SCC.
Acknowledgments
This study was supported by the Scientific and
Technological Innovation Programs of Higher Education Institutions
in Shanxi (grant no. 20110013).
References
|
1
|
Bueno R, Hughes E, Wagner S, Gutin AS,
Lanchbury JS, Zheng Y, Archer MA, Gustafson C, Jones JT, Rushton K,
et al: Validation of a molecular and pathological model for
five-year mortality risk in patients with early stage lung
adenocarcinoma. J Thorac Oncol. 10:67–73. 2015. View Article : Google Scholar
|
|
2
|
DeSantis CE, Lin CC, Mariotto AB, Siegel
RL, Stein KD, Kramer JL, Alteri R, Robbins AS and Jemal A: Cancer
treatment and survivorship statistics, 2014. CA Cancer J Clin.
64:252–271. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fan XX, Wong MP, Cao ZW, Li N, Wu JL, Zhou
H, Jiang XH, Liu L and Leung ELH: Apoptotic effect of a single
compound derived from natural product in Gefitinib-resistant
non-small cell lung cancer cells. Proceedings of the 105th Annual
Meeting of the American Association for Cancer Research; San Diego,
CA, USA. 74. pp. 31932014
|
|
4
|
Kim HR, Kim DJ, Kang DR, Lee JG, Lim SM,
Lee CY, Rha SY, Bae MK, Lee YJ, Kim SH, et al: Fibroblast growth
factor receptor 1 gene amplification is associated with poor
survival and cigarette smoking dosage in patients with resected
squamous cell lung cancer. J Clin Oncol. 31:731–737. 2013.
View Article : Google Scholar
|
|
5
|
Tsai JH, Donaher JL, Murphy DA, Chau S and
Yang J: Spatiotemporal regulation of epithelial-mesenchymal
transition is essential for squamous cell carcinoma metastasis.
Cancer Cell. 22:725–736. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wood SL, Pernemalm M, Crosbie PA and
Whetton AD: The role of the tumor-microenvironment in lung
cancer-metastasis and its relationship to potential therapeutic
targets. Cancer Treat Rev. 40:558–566. 2014. View Article : Google Scholar
|
|
7
|
Yang L, Li G, Zhao L, Pan F, Qiang J and
Han S: Blocking the PI3K pathway enhances the efficacy of
ALK-targeted therapy in EML4-ALK-positive nonsmall-cell lung
cancer. Tumor Biol. 35:9759–9767. 2014. View Article : Google Scholar
|
|
8
|
Gainor JF, Varghese AM, Ou SH, Kabraji S,
Awad MM, Katayama R, Pawlak A, Mino-Kenudson M, Yeap BY, Riely GJ,
et al: ALK rearrangements are mutually exclusive with mutations in
EGFR or KRAS: An analysis of 1,683 patients with non-small cell
lung cancer. Clin Cancer Res. 19:4273–4281. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Boeri M, Verri C, Conte D, Roz L, Modena
P, Facchinetti F, Calabrò E, Croce CM, Pastorino U and Sozzi G:
MicroRNA signatures in tissues and plasma predict development and
prognosis of computed tomography detected lung cancer. Proc Natl
Acad Sci USA. 108:3713–3718. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Song SJ, Poliseno L, Song MS, Ala U,
Webster K, Ng C, Beringer G, Brikbak NJ, Yuan X, Cantley LC, et al:
MicroRNA-antagonism regulates breast cancer stemness and metastasis
via TET-family-dependent chromatin remodeling. Cell. 154:311–324.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Valeri N, Braconi C, Gasparini P, Murgia
C, Lampis A, Paulus-Hock V, Hart JR, Ueno L, Grivennikov SI, Lovat
F, et al: MicroRNA-135b promotes cancer progression by acting as a
downstream effector of oncogenic pathways in colon cancer. Cancer
Cell. 25:469–483. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kohlhapp FJ, Mitra AK, Lengyel E and Peter
ME: MicroRNAs as mediators and communicators between cancer cells
and the tumor microenvironment. Oncogene. 34:5857–5868. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Edelman MJ, Feliciano J, Yue B, Bejarano
P, Ioffe O, Reisman D, Hawkins D, Gai Q, Hicks D and Serrero G:
GP88 (progranulin): A novel tissue and circulating biomarker for
non-small cell lung carcinoma. Hum Pathol. 45:1893–1899. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang M, Li G, Yin J, Lin T and Zhang J:
Progranulin overexpression predicts overall survival in patients
with glioblastoma. Med Oncol. 29:2423–2431. 2012. View Article : Google Scholar
|
|
16
|
Serrero G, Hawkins DM, Yue B, Ioffe O,
Bejarano P, Phillips JT, Head JF, Elliott RL, Tkaczuk KR, Godwin
AK, et al: Progranulin (GP88) tumor tissue expression is associated
with increased risk of recurrence in breast cancer patients
diagnosed with estrogen receptor positive invasive ductal
carcinoma. Breast Cancer Res. 14:R262012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Abrhale T, Brodie A, Sabnis G, Macedo L,
Tian C, Yue B and Serrero G: GP88 (PC-cell derived growth factor,
progranulin) stimulates proliferation and confers letrozole
resistance to aromatase overexpressing breast cancer cells. BMC
Cancer. 11:2312011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Demorrow S: Progranulin: A novel regulator
of gastrointestinal cancer progression. Transl Gastrointest Cancer.
2:145–151. 2013.PubMed/NCBI
|
|
19
|
Brambilla E, Travis WD, Colby TV, Corrin B
and Shimosato Y: The new World Health Organization classification
of lung tumours. Eur Respir J. 18:1059–1068. 2001. View Article : Google Scholar
|
|
20
|
Goldstraw P, Crowley J, Chansky K, Giroux
DJ, Groome PA, Rami-Porta R, Postmus PE, Rusch V and Sobin L;
International Association for the Study of Lung Cancer
International Staging Committee; Participating Institutions: The
IASLC Lung Cancer Staging Project: Proposals for the revision of
the TNM stage groupings in the forthcoming (seventh) edition of the
TNM Classification of malignant tumours. J Thorac Oncol. 2:706–714.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sartorius CA, Hanna CT, Gril B, Cruz H,
Serkova NJ, Huber KM, Kabos P, Schedin TB, Borges VF, Steeg PS and
Cittelly DM: Estrogen promotes the brain metastatic colonization of
triple negative breast cancer cells via an astrocyte-mediated
paracrine mechanism. Oncogene. 35:2881–2892. 2016. View Article : Google Scholar :
|
|
22
|
Chen Q, Sun W, Liao Y, Zeng H, Shan L, Yin
F, Wang Z, Zhou Z, Hua Y and Cai Z: Monocyte chemotactic protein-1
promotes the proliferation and invasion of osteosarcoma cells and
upregulates the expression of AKT. Mol Med Rep. 12:219–225.
2015.PubMed/NCBI
|
|
23
|
Jiang J, Yu C, Chen M, Tian S and Sun C:
Over-expression of TRIM37 promotes cell migration and metastasis in
hepatocellular carcinoma by activating Wnt/beta-catenin signaling.
Biochem Biophys Res Commun. 464:1120–1127. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tangkeangsirisin W and Serrero G: GP88
(progranulin) confers fulvestrant (Faslodex, ICI 182,780)
resistance to human breast cancer cells. Adv Breast Cancer Res.
3:68–78. 2014. View Article : Google Scholar
|
|
25
|
Liu XG, Zhu WY, Huang YY, Ma LN, Zhou SQ,
Wang YK, Zeng F, Zhou JH and Zhang YK: High expression of serum
miR-21 and tumor miR-200c associated with poor prognosis in
patients with lung cancer. Med Oncol. 29:618–626. 2012. View Article : Google Scholar
|
|
26
|
Zhang BC, Gao J, Wang J, Rao ZG, Wang BC
and Gao JF: Tumor-associated macrophages infiltration is associated
with peritumoral lymphangiogenesis and poor prognosis in lung
adenocarcinoma. Med Oncol. 28:1447–1452. 2011. View Article : Google Scholar
|
|
27
|
Yang T, Thakur A, Chen T, Yang L, Lei G,
Liang Y, Zhang S, Ren H and Chen M: MicroRNA-15a induces cell
apoptosis and inhibits metastasis by targeting BCL2L2 in non-small
cell lung cancer. Tumor Biol. 36:4357–4365. 2015. View Article : Google Scholar
|
|
28
|
Wu X, Liu T, Fang O, Leach LJ, Hu X and
Luo Z: miR-194 suppresses metastasis of non-small cell lung cancer
through regulating expression of BMP1 and p27kip1. Oncogene.
33:1506–1514. 2014. View Article : Google Scholar
|
|
29
|
Li Y, Zhao W, Bao P, Li C, Ma XQ, Li Y and
Chen LA: miR-339-5p inhibits cell migration and invasion in vitro
and may be associated with the tumor-node-metastasis staging and
lymph node metastasis of non-small cell lung cancer. Oncol Lett.
8:719–725. 2014.PubMed/NCBI
|
|
30
|
Almog N, Ma L, Schwager C, Brinkmann BG,
Beheshti A, Vajkoczy P, Folkman J, Hlatky L and Abdollahi A:
Consensus micro RNAs governing the switch of dormant tumors to the
fast-growing angiogenic phenotype. PLoS One. 7:e440012012.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li Y, Zhao W, Bao P, Li C, Ma XQ, Li Y and
Chen LA: miR-339-5p inhibits cell migration and invasion in vitro
and may be associated with the tumor-node-metastasis staging and
lymph node metastasis of non-small cell lung cancer. Oncology Lett.
8:719–725. 2014.
|
|
32
|
Rademakers R, Eriksen JL, Baker M,
Robinson T, Ahmed Z, Lincoln SJ, Finch N, Rutherford NJ, Crook RJ,
Josephs KA, et al: Common variation in the miR-659 binding-site of
GRN is a major risk factor for TDP43-positive frontotemporal
dementia. Hum Mol Genet. 17:3631–3642. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jiao J, Herl LD, Farese RV and Gao FB:
MicroRNA-29b regulates the expression level of human progranulin, a
secreted glycoprotein implicated in frontotemporal dementia. PLoS
One. 5:e105512010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tangkeangsirisin W and Serrero G: PC
cell-derived growth factor (PCDGF/GP88, progranulin) stimulates
migration, invasiveness and VEGF expression in breast cancer cells.
Carcinogenesis. 25:1587–1592. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Edelman MJ, Feliciano J, Yue B, Bejarano
P, Ioffe O, Reisman D, Hawkins D, Gai Q, Hicks D and Serrero G:
GP88 (progranulin): a novel tissue and circulating biomarker for
non-small cell lung carcinoma. Hum Pathol. 45:1893–1899. 2014.
View Article : Google Scholar : PubMed/NCBI
|