Introduction
Osteoporosis and obesity are two of the most common
chronic conditions and pose major health threats worldwide, with
both showing increasing prevalence rates, however, the association
between osteoporosis and obesity is complex. The bone marrow is the
only place in mammalian tissues where bone and fat lie adjacent to
each other, in osteoporosis, adipogenesis is increased at the
expense of osteogenesis from common osteoporotic bone marrow cells
(1,2). Bone marrow-derived mesenchymal stem
cells (BM-MSCs) have the capacity to differentiate into osteoblasts
and adipocytes, and osteoporosis is partially attributable to the
alteration of the balance of BM-MSC differentiation into
osteoblasts and adipocytes. BM-MSC differentiation is regulated by
hormones, cytokines, and genes. The differentiation of BM-MSCs into
adipocytes is accompanied by a marked increase in the expression of
adipocyte markers, including peroxisome proliferator-activated
receptor γ (PPARγ) and CCAAT-enhancer-binding protein α (C/EBPα).
Similarly, the differentiation of BM-MSCs into osteoblasts is
regulated by bone morphogenetic proteins (BMPs) and runt-related
transcription factor-2 (RUNX2) (3–6). A
deeper understanding of the differentiation of BM-MSCs into
osteoblasts or adipocytes will provide insight into the
pathophysiology and treatment of osteoporosis.
S100 calcium-binding protein B (S100B), an important
member of the S100 family, is ubiquitously expressed in human
tissue, including fat tissues, and is associated with a variety of
human diseases such as neurodegenerative disorders (7), malignant melanoma (7), trauma with or without brain injury
(8–10) and obesity (11). Serum S100B levels are positively
correlated with body mass index (11), and S100B expression is increased by
diet-induced obesity (12).
Adipocytes express and secrete S100B protein, which may act as an
adipokine by modulating the immune response and metabolism.
However, the direct effect of S100B on osteoporosis and obesity
remains to be investigated (13).
Therefore, the current study aimed to determine the effect of S100B
on MSC differentiation into adipocytes and osteoblasts.
In the present study, an in vitro model of
osteogenesis and adipogenesis was established using the mouse
embryo cell line C3H/10T1/2 (ATCC number, CCL-226) in a monolayer
and high-density cultures. The current study presents novel
evidence concerning the effect of S100B on cell differentiation,
including adipogenic and osteogenic differentiation.
Materials and methods
Construction of expression plasmids
C57B/6 mice were purchased from Nanjing Qingzilan
Technologies Co., Ltd. (Nanjing, China). Animals were housed at
23±1°C in a 12/12-h light/dark cycle and a humidity of 45±5%, and
allowed free access to a normal chow diet and water. The study was
approved by the ethics committee of the Animal Care Facility of
Nanjing Medical University, Nanjing, China.
Total mRNA was isolated using TRIzol®
reagent (cat. no. 15596-026; Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA). This mRNA was reverse-transcribed into
complementary (c)DNA via reverse transcription-polymerase chain
reaction (RT-PCR) using the ThermoScript™ RT-PCR System for
First-Strand cDNA Synthesis kit; cat. no. 11146024, Thermo Fisher
Scientific, Inc.). Subsequently, this cDNA was used as the template
DNA. PCR (cat. no. 4464268; the Platinum Multiplex PCR Master Mix,
2X; Thermo Fisher Scientific, Inc.) was performed to clone S100B
cDNA using appropriate primers.
Plasmids overexpressing S100B, termed pcDNA3.1(+)
A-S100B, were constructed. The coding sequences of mouse S100B were
amplified using RT-PCR and mRNAs isolated from the white adipose
tissue of mice using TRIzol® reagent (cat. no.
15596-026; Invitrogen; Thermo Fisher Scientific, Inc.). The primer
sequences (including the sites of restriction enzymes) were as
follows: Forward, 5′-CGTGAATTCATGTCCGAGCTGGGAAG-3′ and reverse,
5′-GCTGTCGACGGGTCACTCATGTTCAAAGAAGT-3′. The PCR products were
subcloned into the pcDNA3.1(+)A expression vector and then
confirmed by sequencing.
Subsequently, miRNA-S100B expression plasmids were
constructed. Three distinct domains within the coding region of the
mouse S100B cDNA were targeted for RNA interference. For this
purpose, four pairs of reverse complementary oligonucleotides were
designed and synthesized (Table
I). The thermocycling conditions used were: 95°C for 15 sec;
58°C for 20 sec; and 72°C for 20 sec. The oligonucleotides were
annealed and inserted into the pcDNA6.2-GW/EmGFP-miR expression
vector (Invitrogen; Thermo Fisher Scientific, Inc.) to create
pcDNA6.2-GW/EmGFP-miR-S100B 1, 2 and 3. A scrambled control
construct was also created.
 | Table IReverse complementary
oligonucleotides. |
Table I
Reverse complementary
oligonucleotides.
| Oligo | 5′-3′ sequence |
|---|
| 13MR0109-01-F |
TGCTGAACAACTGCCTTCTCCAGCTCGTTTTGGCCACTGACTGACGAGCTGGAAGGCAGTTGTT |
| 13MR0109-01-R |
CCTGAACAACTGCCTTCCAGCTCGTCAGTCAGTGGCCAAAACGAGCTGGAGAAGGCAGTTGTTC |
| 13MR0109-02-F |
TGCTGTTCTGGATGAGCTTGTCAGCTGTTTTGGCCACTGACTGACAGCTGACACTCATCCAGAA |
| 13MR0109-02-R |
CCTGTTCTGGATGAGTGTCAGCTGTCAGTCAGTGGCCAAAACAGCTGACAAGCTCATCCAGAAC |
| 13MR0109-03-F |
TGCTGTTCGGAAGCTGGACTTGCTGAGTTTTGGCCACTGACTGACTCAGCAAGCAGCTTCCGAA |
| 13MR0109-03-R |
CCTGTTCGGAAGCTGCTTGCTGAGTCAGTCAGTGGCCAAAACTCAGCAAGTCCAGCTTCCGAAC |
| Negative-F |
TGCTGAAATGTACTGCGCGTGGAGACGTTTTGGCCACTGACTGACGTCTCCACGCAGTACATTT |
| Negative-R |
CCTGAAATGTACTGCGTGGAGACGTCAGTCAGTGGCCAAAACGTCTCCACGCGCAGTACATTTC |
Cell culture and stable clone
selection
The mouse embryo cell line C3H/10T1/2 (ATCC number,
CCL-226; American Type Culture Collection, Manassas, VA, USA) was
cultured in Eagle's basal medium (MEM; cat. no. 11095080; Life
Technologies; Thermo Fisher Scientific, Inc.) supplemented with 10%
fetal bovine serum (FBS; cat. no. 10100147; Gibco; Thermo Fisher
Scientific, Inc.), penicillin (100 U/ml) and streptomycin sulfate
(100 µg/ml; cat. no. 15070063; Life Technologies; Thermo
Fisher Scientific, Inc.), and maintained at 37°C in a humid
incubator containing 5% CO2.
The expression constructs were transfected into
C3H/10T1/2 cells using the X-tremeGENE HP DNA Transfection Reagent
(cat. no.; 06366236001; Roche Diagnostics, Basel, Switzerland).
After 48 h, the cells were cultured in a selective medium
containing 300 µg/ml G418 or 3 mg/ml blasticidin (cat. nos.,
N6386 and 11033102, respectively; Sigma-Aldrich; Merck Millipore,
Darmstadt, Germany) for 1 week, and resistant colonies, which
indicated successful transfectants, were selected.
Osteoblast differentiation
Osteogenic differentiation was induced as described
previously (14). Confluent
C3H/10T1/2 cells (the day on which confluence was reached was
considered day 0) were incubated for 12 days in an osteogenic
induction medium consisting of MEM containing 10% FBS, 0.1 mM
dexamethasone (cat. no. D4902; Sigma-Aldrich; Merck Millipore,
Darmstadt, Germany), 10 mM β-glycerophosphate (cat. no. G9422;
Sigma-Aldrich; Merck Millipore) and 50 mM ascorbic acid. The
induction medium was changed every 2 days. The presence and extent
of bone matrix mineralization was evaluated using alizarin red S
staining.
Adipocyte differentiation
Adipocyte differentiation was induced as described
previously (14). C3H/10T1/2 cells
were seeded on plates, and allowed to grow for 2 days to reach
confluence (considered day 0). Cell differentiation was induced by
culturing the cells in MEM containing 10% FBS, 0.5 mM
3-isobutyl-1-methylxanthine (cat. no. I7018; Sigma-Aldrich; Merck
Millipore), 1 µg/ml porcine insulin (cat. no. I0320000;
Sigma-Aldrich; Merck Millipore) and 1 mM dexamethasone. Following
48 h of incubation, the medium was replaced with MEM containing 10%
FBS and 1 µg/ml insulin. On day 4, the medium was replaced
with fresh medium (MEM containing 10% FBS), and the incubation was
continued for 12 days. Lipid droplets were evaluated using oil red
O staining.
Alkaline phosphatase staining
The differentiation of C3H/10T1/2 cells into
osteoblasts. After 4 days, alkaline phosphatase (ALP) staining was
performed according to the protocol described in the
5-Bromo-4-chloro-3′-indolyphosphate p-Toluidine Salt
(BCIP)/Nitro-Blue Tetrazolium Chloride (NBT) ALP Color Development
kit (cat. no. C3206; Beyotime Institute of Biotechnology, Inc.).
The cells were fixed with 10% formalin for 10 min at room
temperature, washed with phosphate-buffered saline (PBS), and
stained with 300 µg/ml BCIP/NBT solution for 30 min at room
temperature. ALP-positive cells were stained blue. Stained cells
were examined using light microscopy (OLYMPUS IX51) and
photographed.
Alizarin red S staining
We induced the differentiation of C3H/10T1/2 cells
into osteoblasts. Twelve days later, the cells were gently washed
three times with PBS. Then, the Alizarin Red S Staining kit
(GMS80046.3v.A; Genmed Scientifics, Inc., Wilmington, DE, USA) was
used according to the manufacturer's instructions. The cells were
carefully rinsed three times with 1.0 ml double-distilled water and
allowed to dry. Stained cells were examined using light microscopy
(IX51; Olympus Corporation, Tokyo, Japan) and were then
photographed.
Oil red O staining
The differentiation of C3H/10T1/2 cells into
adipocytes was induced, then 12 days later, oil red O staining was
performed according to a previously published protocol (14). The cells were washed three times
with PBS and fixed with 10% formalin for 60 min at room
temperature. Subsequent to fixation, the cells were washed twice
with PBS and stained with filtered oil red O solution (cat. no.
O0625; Sigma-Aldrich; Merck Millipore) for 60 min at room
temperature. The cells were then washed with distilled water to
remove unbound dye, visualized using light microscopy (IX51), and
were then photographed.
Triglyceride glycerol phosphate
oxidase-peroxidase (GPO-POD) assay
Cellular triglyceride content was determined using
the Triglyceride GPO-POD Assay kit (cat. no. TR0100; Sigma-Aldrich;
Merck Millipore). At 12 days subsequent to the induction of
C3H/10T1/2 cell differentiation into adipocytes, the cells were
washed twice with PBS, scraped in 500 µl PBS, sonicated to
homogenize the suspension and then assayed to determine the total
triglyceride content.
Western blot analysis
At 0, 4, 8 and 12 days subsequent to the induction
of C3H/10T1/2 cell differentiation into adipocytes or osteoblasts,
the cells were lysed in radioimmunoprecipitation assay buffer
[composition: 50 mM Tris-HCl (pH 7.4), 1% NP-40, 150 mM NaCl, 1 mM
EDTA and 100 µg/ml phenylmethylsulfonyl fluoride]. Equal
amounts of protein (60 µg) were separated using 10% sodium
dodecyl sulfate polyacrylamide gel electrophoresis and were
electrophoretically transferred to polyvinylidene difluoride
membranes (EMD Millipore, Billerica, MA, USA). The membranes were
incubated overnight at 4°C with rabbit monoclonal anti-S100B (cat.
no. 9550), rabbit monoclonal PPARγ (cat. no. 2430), rabbit
polyclonal anti-C/EBPα (cat. no. 2295) and rabbit monoclonal
anti-RUNX2 (cat. no. 8486; all 1:1,000; Cell Signaling Technology,
Inc., Danvers, MA, USA) antibodies, and anti-BMP2 (cat. no.
ab82511, Abcam, Cambridge, MA, USA) and mouse monoclonal β-tubulin
(cat. no. T5168; 1:5,000, Sigma-Aldrich; Merck Millipore)
antibodies in Tris-buffered saline with Tween-20 containing 1%
(w/v) bovine serum albumin (cat. no. 05470; Sigma-Aldrich; Merck
Millipore). The blots were then incubated for 2 h with anti-rabbit
or anti-mouse secondary antibodies [anti-rabbit immunoglobulin G
(IgG), horseradish peroxidase (HRP)-linked antibody; cat. no. 7074;
and anti-mouse IgG, HRP-linked antibody; cat. no. 7076; Cell
Signaling Technology, Inc.]. Immune complexes were detected using a
Pierce ECL Western Blotting Substrate kit (cat. no. 32106; Thermo
Fisher Scientific, Inc.), and analyzed using a scanning
densitometer with molecular analysis software (Bio-Rad
Laboratories, Inc., Hercules, CA, USA).
Statistical analysis
Statistical analyses were performed using SPSS 19
software (IBM SPSS, Armonk, NY, USA). Data were assessed using
one-way analysis of variance with a correction for multiple
comparisons, as appropriate. P<0.05 was considered to indicate a
statistically significant difference.
Results
Measurement of S100B expression
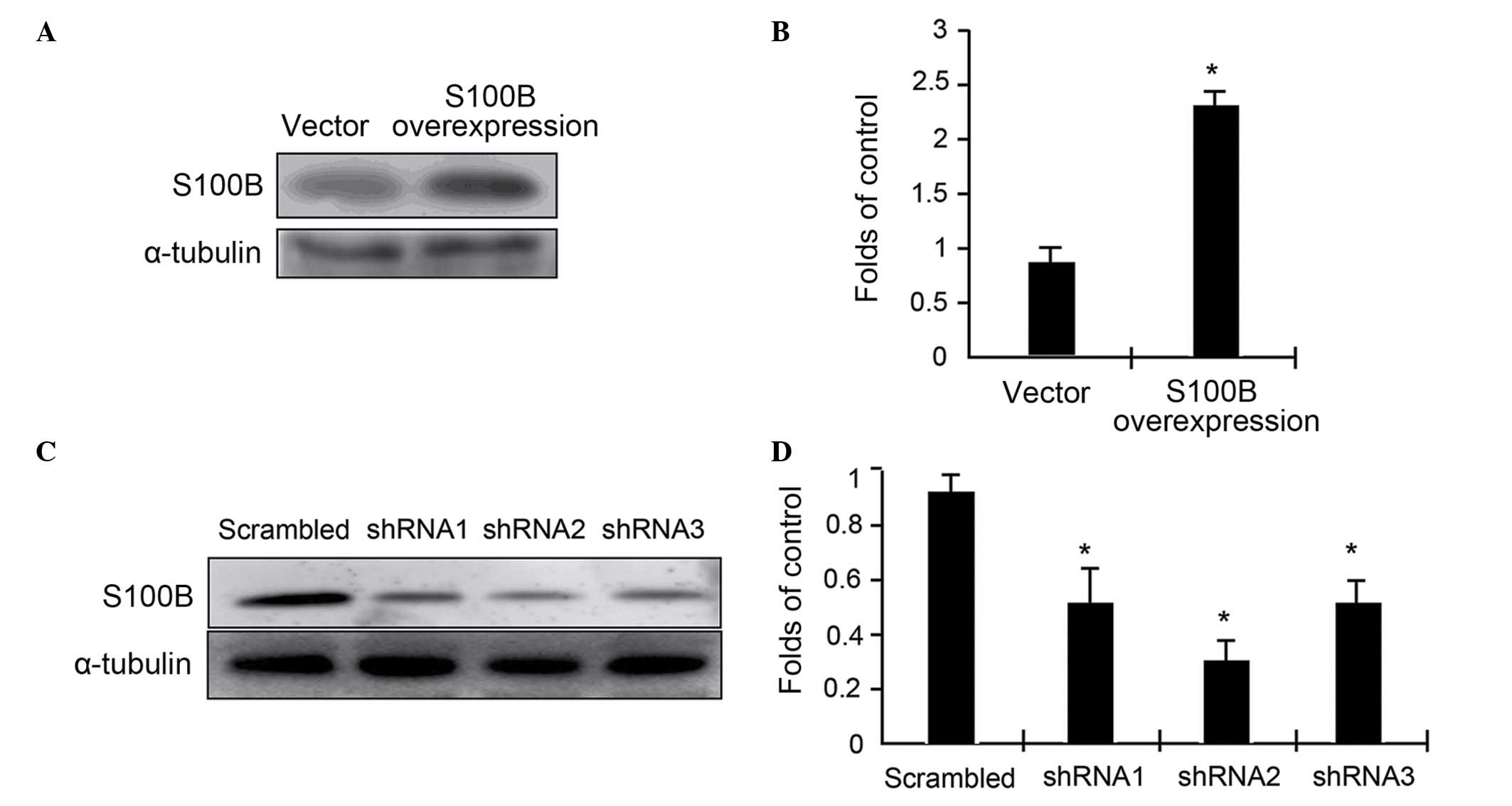
To assess the functional roles of S100B in
C3H/10T1/2 cell differentiation, S100B expression levels were
altered in C3H/10T1/2 cells through either an overexpression system
or RNA interference. In either case, stable transfectants were
selected using G418 and blasticidin, and then expanded for further
studies. The expression levels of S100B were determined using
western blot analysis. The results indicated that overexpression
driven by a cytomegalovirus-promoter resulted in a 2.4-fold
elevation of S100B protein expression (Fig. 1A and B). In contrast, the
expression of three specific miRNAs targeting three different
regions of S100B mRNA resulted in up to 50% reduction in S100B
expression (Fig. 1C and D). Thus,
the cell models were successfully built with varying levels of
S100B protein expression. miRNA2 was used in the following
experiments.
S100B inhibits C3H/10T1/2 cell
differentiation into osteoblasts
To determine the effect of S100B on the
differentiation of C3H/10T1/2 cells into osteoblasts,
differentiation of C3H/10T1/2 cells with different levels of S100B
protein expression into osteoblasts was induced by specific
differentiation protocols (Fig.
2). At 4 days after the induction of differentiation, ALP
activity in the cells was examined using ALP staining. The results
indicated that S100B overexpression suppressed ALP activity, while
S100B underexpression enhanced ALP activity (Fig. 2A and C). At 12 days after the
induction of differentiation, alizarin red S staining was used to
detect calcium nodule formation. Fewer red nodules were observed in
the S100B overexpression group than in the S100B underexpression
group (Fig. 2B and D).
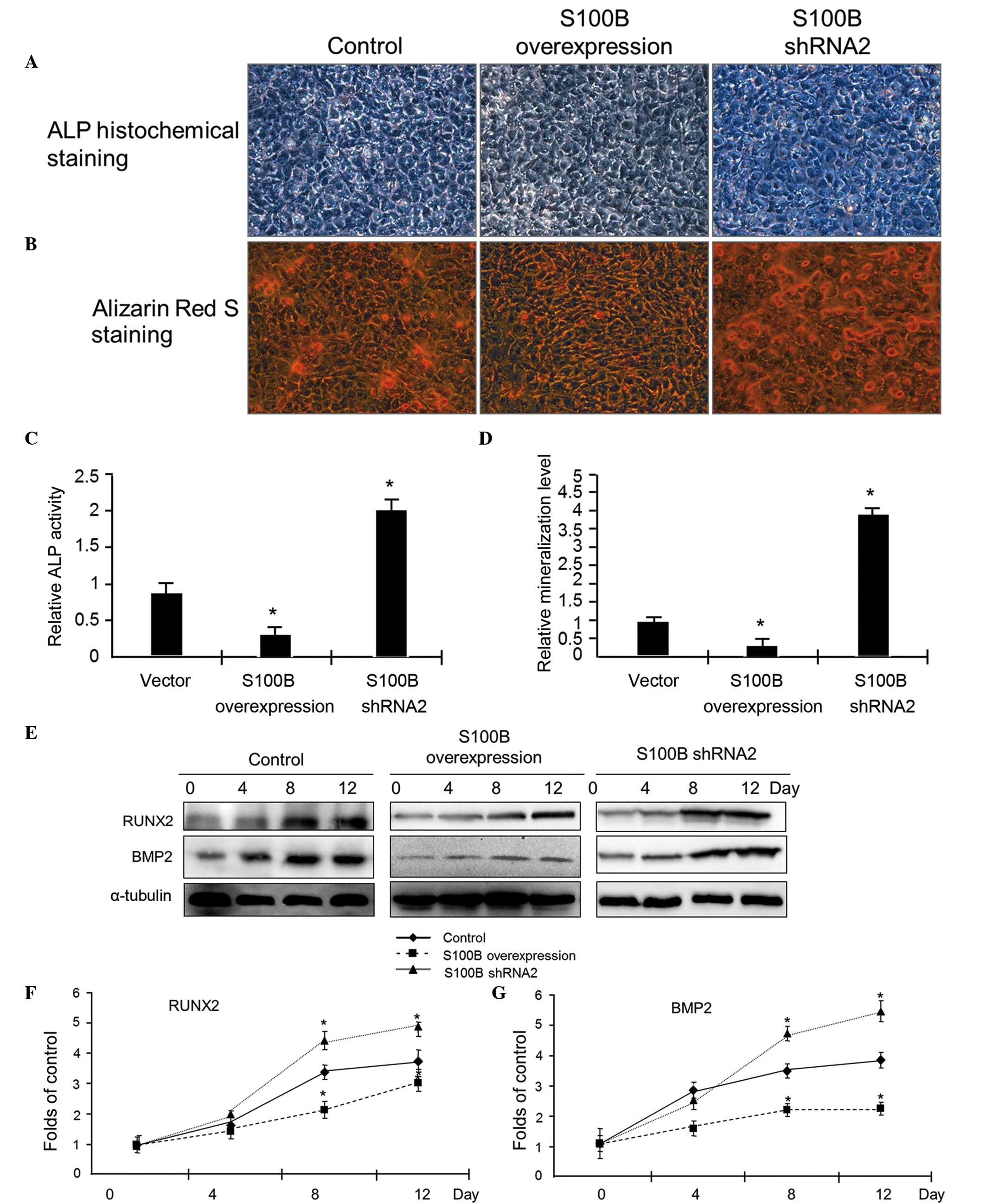 | Figure 2S100B suppresses C3H/10T1/2 cell
differentiation into osteoblasts. (A) Osteogenic differentiation
was induced in C3H/10T1/2 cells with different levels of S100B
protein expression, including control, S100B-overexpressing and
S100B shRNA cells. After 4 days, ALP staining was used to examine
the ALP activity. Photographs were taken using a light microscope
at ×200 magnification. (B) After 12 days, alizarin red S staining
was used to examine the calcium nodules formed. Photographs were
taken using a light microscope at ×200 magnification. (C)
Quantification of the ALP activity presented in (A). (D)
Quantification of the mineralization presented in (B). The results
are presented as the mean ± standard deviation of three independent
experiments. *P≤0.05 vs. the vector control. (E) Western
blot analysis was used to examine the expression of the
osteogenetic markers RUNX2 and BMP2. At different time points (0,
4, 8 and 12 days) after the induction of differentiation, RUNX2 and
BMP2 expression levels were analyzed. Quantification of the (F)
RUNX2 and (G) BMP2 protein expression levels in (D) based on
grayscale analysis (analyzed from three independent experiments).
*P≤0.05 vs. the control. S100B, S100 calcium-binding
protein B; shRNA, small hairpin RNA; ALP, alkaline phosphatase;
RUNX2, runt-related transcription factor 2; BMP2, bone
morphogenetic protein 2. |
To confirm the effect of S100B on osteogenesis, the
expression levels of the markers of osteogenic differentiation were
examined using western blot analysis. At 0, 4, 8 and 12 days
subsequent to the induction of C3H/10T1/2 cell differentiation into
osteoblasts, total protein was extracted to examine the expression
of the osteoblast markers RUNX2 and BMP2. The western blot results
indicated that in the control group, RUNX2 and BMP2 protein
expression increased gradually as the cells differentiated into
osteoblasts. In the S100B overexpression group, there was no
significant increase in RUNX2 and BMP2 expression, however, in the
S100B underexpression group, the magnitude of the increase in RUNX2
and BMP2 expression was greater than in the control group (Fig. 2E–G). These results suggested that
S100B suppressed the osteogenic differentiation of C3H/10T1/2
cells.
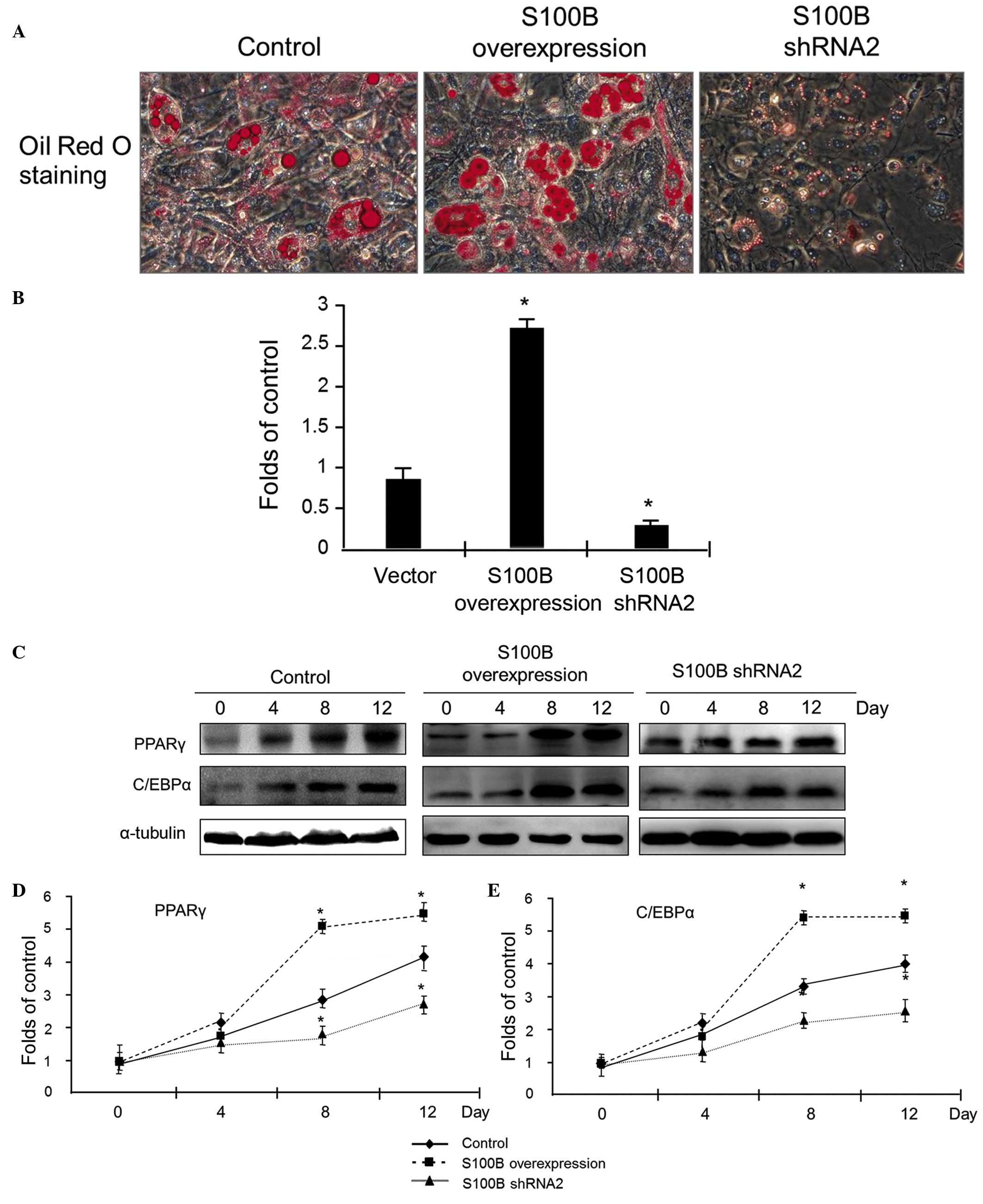
S100B stimulates C3H/10T1/2 cell
differentiation into adipocytes
To investigate the effect of S100B on the
differentiation of C3H/10T1/2 cells into adipocytes, the
differentiation of C3H/10T1/2 cells with different levels of S100B
expression into adipocytes was induced. At 12 days after the
induction of differentiation, oil red O staining was applied to
detect cellular lipid droplets. The results of staining indicated
that S100B overexpression led to a significant increase in oil red
O staining, however, the reduction of S100B expression led to
sparse expression of oil red O staining (Fig. 3A). The quantitative analysis of
cellular triglycerides was used to evaluate the above observations;
the results confirmed that triglyceride accumulation was high in
C3H/10T1/2 cells overexpressing S100B, however was low in cells
with reduced S100B expression (Fig.
3B).
At different time points (0, 4, 8 and 12 days)
subsequent to the induction of C3H/10T1/2 cell differentiation into
adipocytes, proteins were extracted, and western blot analysis was
applied to detect the expression of PPARγ and C/EBPα. The results
indicated that in control cells, PPARγ and C/EBPα protein
expression increased as the cells gradually differentiated into
osteoblasts. Compared with the control group, in the S100B
overexpression group, there was a significant increase in PPARγ and
C/EBPα expression, however in the group with low S100B expression,
the magnitude of the increase in PPARγ and C/EBPα expression was
reduced (Fig. 3C–E). These data
suggest that S100B stimulates the adipogenic differentiation of
C3H/10T1/2 cells.
Extracellular signal-regulated kinase
(ERK) signaling regulates osteogenic C3H/10T1/2 cell
differentiation and JNK signaling regulates adipogenic C3H/10T1/2
cell differentiation
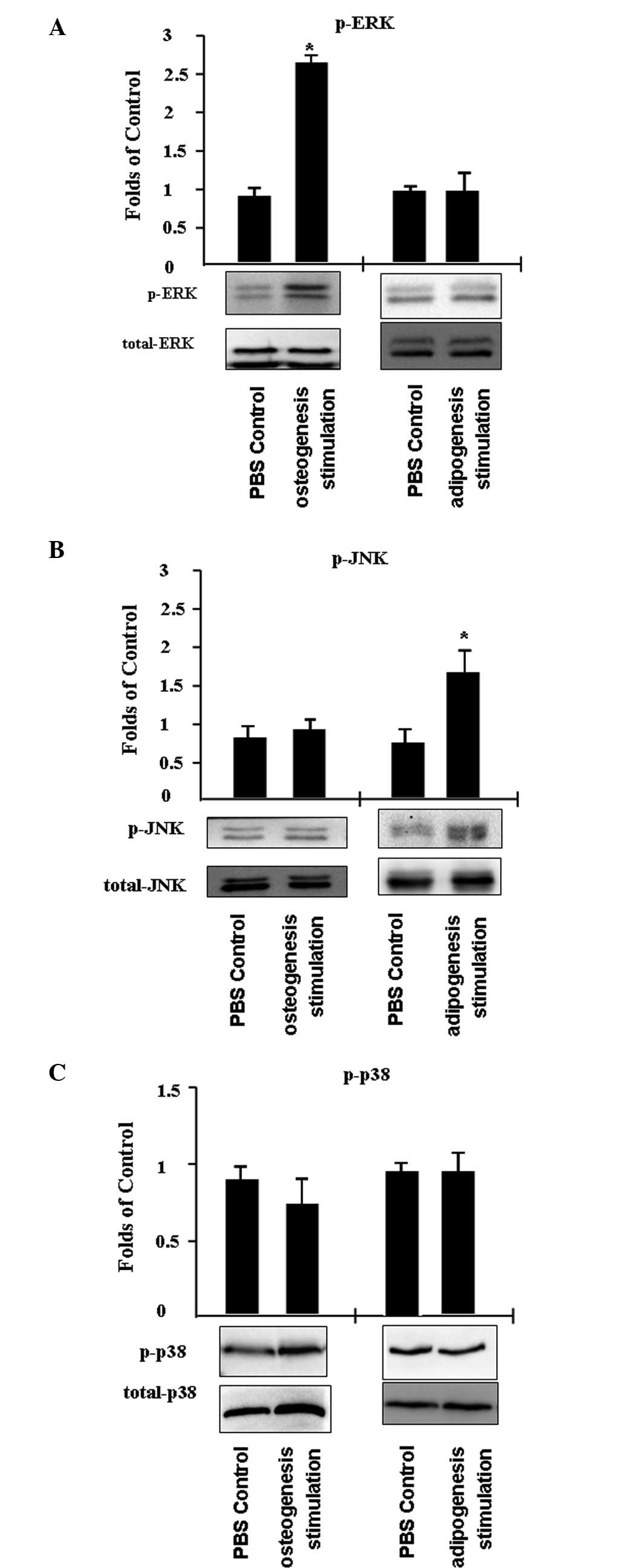
In addition, the activity of mitogen-activated
protein kinases (MAPKs) was investigated, including ERK, JNK and
p38, in C3H/10T1/2 cells that differentiated into adipocytes or
osteoblasts. C3H/10T1/2 cells were treated with PBS (control) or an
inducer of adipogenesis or osteogenesis for 45 min. It was
identified that ERK phosphorylation increased by ~2.7-fold
following the induction of osteogenesis, while JNK and p38 activity
remained unchanged (Fig. 4A and
B). In addition, JNK phosphorylation increased ~1.7-fold
following the stimulation of adipogenesis, while p38 and ERK
activity was unaffected.
Discussion
The bone marrow is the only place in mammalian
tissues where bone and fat lie adjacent to each other. Although
bone marrow adipose tissue was first identified in the 19th
century, the effect and origin of bone marrow-derived adipocytes
remain unclear (2,15,16).
The bone marrow micro-environment includes osteoblasts, adipocytes,
bone lining cells, pre-osteoblasts, pre-adipocytes and BM-MSCs
(17). BM-MSCs differentiate into
osteoblasts and adipocytes, which express osteoblast and adipocyte
markers (18). BM-MSCs can
differentiate into adipocytes in response to injury, aging,
starvation and diabetes, which results in osteoblast reduction and
osteoporosis (19). For example,
aging is associated with a high incidence of obesity and
osteoporosis, which is attributable to the alteration of the
balance between adipocytes and osteoblasts in the bone marrow
(20,21). Thus, the differentiation of BM-MSCs
is crucial for bone metabolism.
S100B is a member of the calcium-regulated protein
S100 family and is characterized by two calcium-binding sites with
EF-hand conformations. The S100 protein family has a minimum of 25
members that are expressed in various tissue types. S100B is
involved in numerous cellular signaling pathways, and previous
studies have indicated that S100B serves an important role in
neurodegenerative disorders, trauma, and obesity (13,22,23).
Serum S100B levels have been suggested to be elevated following
bone fracture (24). Adipose
tissue expresses high levels of S100B, and adipocytes release S100B
protein, however, the role of S100B protein released by adipocytes
remains unclear. S100B has been suggested to act as an adipokine by
modulating local microcirculation, immune response and insulin
resistance (13,25). Plasma S100B levels and S100B gene
expression in white adipose tissue are significantly increased in
obesity, and this increase has been reported to be reversed
following weight loss (12). In
the current study, it was identified that S100B stimulated the
differentiation of C3H/10T1/2 cells, a mouse embryo cell line, into
adipocytes (Fig. 3). The
overexpression of S100B led to a significant increase in oil red O
staining and in the protein expression levels of the adipogenesis
markers PPARγ and C/EBPα. The reduction of S100B expression had the
opposite effects. PPARγ, a marker of adipogenesis, has been
reported to be a promising target for anti-osteoporosis therapy
because of its positive effect on BM-MSC differentiation into
adipocytes (26).
In addition, the effect of S100B on the
differentiation of BM-MSCs into osteoblasts was determined. Using
the C3H/10T1/2 cell model, it was identified that S100B inhibited
the osteogenic differentiation of the cells. S100B overexpression
suppressed and S100B underexpression enhanced ALP activity,
alizarin red S staining, and the expression of the osteogenesis
markers RUNX2 and BMP2 (Fig. 2).
The results indicated that S100B is involved in bone homoeostasis
by regulating BM-MSC differentiation. However, the cell signals
involved in the regulation of BM-MSC differentiation by S100B
remain to be determined.
The cell signals involved in BM-MSC differentiation
are complex. Extracellular Ca2+ has been reported to
induce the differentiation of BM-MSCs into adipocytes by
suppressing ERK activity (27),
and the ERK signaling pathway mediates C/EBPα protein expression in
preadipocyte differentiation (28). S100B induces the nuclear factor κB,
p53, ERK/ribosomal s6 kinase, and signal transducer and activator
of transcription 3 pathways (29–31).
Therefore, in the current study, the status of the MAPK signaling
pathway during C3H/10T1/2 cell differentiation into adipocytes or
osteoblasts was investigated. The results indicated that the
stimulation of osteogenesis increased ERK phosphorylation and the
stimulation of adipogenesis increased JNK phosphorylation (Fig. 4). This suggests that the ERK
pathway is involved in the regulation of osteogenesis, whereas the
JNK pathway is involved in the regulation of adipogenesis.
In summary, the results suggest that S100B inhibits
osteogenesis, however stimulates adipogenesis, and the ERK pathway
is involved in the regulation of osteogenesis, while the JNK
pathway is involved in the regulation of adipogenesis. The results
of the current study indicate that BM-MSC differentiation is
important for bone homeostasis, however further research into the
cell signaling involved in this process is required.
Acknowledgments
The present study was supported by grants from the
Science Development Projects of Nanjing in 2012 (grant no.
ZKX12049) and the Natural Science Foundation of Jiangsu Province
(grant no. BK20141026).
References
|
1
|
Gonnelli S, Caffarelli C and Nuti R:
Obesity and fracture risk. Clin Cases Miner Bone Metab. 11:9–14.
2014.PubMed/NCBI
|
|
2
|
Devlin MJ and Rosen CJ: The bone-fat
interface: Basic and clinical implications of marrow adiposity.
Lancet Diabetes Endocrinol. 3:141–147. 2015. View Article : Google Scholar
|
|
3
|
Chen C, Uludağ H, Wang Z and Jiang H:
Noggin suppression decreases BMP-2-induced osteogenesis of human
bone marrow-derived mesenchymal stem cells in vitro. J Cell
Biochem. 113:3672–3680. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Smith KE, Huang Z, Ma T, Irani A, Lane
Smith R and Goodman SB: Molecular profile of osteoprogenitor cells
seeded on allograft bone. J Tissue Eng Regen Med. 5:704–711. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nathan SS, Pereira BP, Zhou YF, Gupta A,
Dombrowski C, Soong R, Pho RW, Stein GS, Salto-Tellez M, Cool SM
and van Wijnen AJ: Elevated expression of Runx2 as a key parameter
in the etiology of osteosarcoma. Mol Biol Rep. 36:153–158. 2009.
View Article : Google Scholar
|
|
6
|
Gregoire FM, Smas CM and Sul HS:
Understanding adipocyte differentiation. Physiol Rev. 78:783–809.
1998.PubMed/NCBI
|
|
7
|
Faye RS, Paus E, Maelandsmo GM, Berner A,
Høifødt HK, Fodstad Ø and Aamdal S: S100B in bone marrow aspirates
in healthy individuals and malignant melanoma patients. Melanoma
Res. 18:134–140. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Undén J, Bellner J, Eneroth M, Alling C,
Ingebrigtsen T and Romner B: Raised serum S100B levels after acute
bone fractures without cerebral injury. J Trauma. 58:59–61. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Savola O, Pyhtinen J, Leino TK, Siitonen
S, Niemelä O and Hillbom M: Effects of head and extracranial
injuries on serum protein S100B levels in trauma patients. J
Trauma. 56:1229–1234; discussion 1234. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pelinka LE, Szalay L, Jafarmadar M,
Schmidhammer R, Redl H and Bahrami S: Circulating S100B is
increased after bilateral femur fracture without brain injury in
the rat. Br J Anaesth. 91:595–597. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Steiner J, Schiltz K, Walter M, Wunderlich
MT, Keilhoff G, Brisch R, Bielau H, Bernstein HG, Bogerts B,
Schroeter ML and Westphal S: S100B serum levels are closely
correlated with body mass index: An important caveat in
neuropsychiatric research. Psychoneuroendocrinology. 35:321–324.
2010. View Article : Google Scholar
|
|
12
|
Buckman LB, Anderson-Baucum EK, Hasty AH
and Ellacott KLj: Regulation of S100B in white adipose tissue by
obesity in mice. Adipocyte. 3:215–220. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gonçalves CA, Leite MC and Guerra MC:
Adipocytes as an important source of serum S100B and possible roles
of this protein in adipose tissue. Cardiovasc Psychiatry Neurol.
2010:7904312010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li D, Zhang R, Zhu W, Xue Y, Zhang Y,
Huang Q, Liu M and Liu Y: S100A16 inhibits osteogenesis but
stimulates adipogenesis. Mol Biol Rep. 40:3465–3473. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Titorencu I, Pruna V, Jinga VV and
Simionescu M: Osteoblast ontogeny and implications for bone
pathology: An overview. Cell Tissue Res. 355:23–33. 2014.
View Article : Google Scholar
|
|
16
|
Bidwell JP, Alvarez MB, Hood M Jr and
Childress P: Functional impairment of bone formation in the
pathogenesis of osteoporosis: The bone marrow regenerative
competence. Curr Osteoporos Rep. 11:117–125. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Desiderio V, Tirino V, Papaccio G and
Paino F: Bone defects: Molecular and cellular therapeutic targets.
Int J Biochem Cell Biol. 51:75–78. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
James AW: Review of signaling pathways
governing MSC osteogenic and adipogenic differentiation.
Scientifica (Cairo). 2013:6847362013.
|
|
19
|
Chen X, He F, Zhong DY and Luo ZP:
Acoustic-frequency vibratory stimulation regulates the balance
between osteogenesis and adipogenesis of human bone marrow-derived
mesenchymal stem cells. Biomed Res Int. 2015:5407312015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou ZG, Gao M, Liu Q and Tao MD:
Comprehensive transcriptome analysis of mesenchymal stem cells in
elderly patients with osteoporosis. Aging Clin Exp Res. 27:595–601.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Beane OS, Fonseca VC, Cooper LL, Koren G
and Darling EM: Impact of aging on the regenerative properties of
bone marrow-, muscle- and adipose-derived mesenchymal stem/stromal
cells. PLoS One. 9:e1159632014. View Article : Google Scholar
|
|
22
|
Steiner J, Bogerts B, Schroeter ML and
Bernstein HG: S100B protein in neurodegenerative disorders. Clin
Chem Lab Med. 49:409–424. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kartal AG, Yılmaz S, Yaka E, Pekdemir M,
Sarısoy HT, Çekmen MB and Yüksel M: Diagnostic value of S100B
protein in the differential diagnosis of acute vertigo in the
emergency department. Acad Emerg Med. 21:736–741. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhao P, Gao S and Lin B: Elevated levels
of serum S100B is associated with the presence and outcome of
haemorrhagic shock. Clin Lab. 58:1051–1055. 2012.PubMed/NCBI
|
|
25
|
Steiner J, Myint AM, Schiltz K, Westphal
S, Bernstein HG, Walter M, Schroeter ML, Schwarz MJ and Bogerts B:
S100B serum levels in schizophrenia are presumably related to
visceral obesity and insulin resistance. Cardiovasc Psychiatry
Neurol. 2010:4807072010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cao J, Ou G, Yang N, Ding K, Kream BE,
Hamrick MW, Isales CM and Shi XM: Impact of targeted PPARγ
disruption on bone remodeling. Mol Cell Endocrinol. 410:27–34.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hashimoto R, Katoh Y, Miyamoto Y, Itoh S,
Daida H, Nakazato Y and Okada T: Increased extracellular and
intracellular Ca2+ lead to adipocyte accumulation in
bone marrow stromal cells by different mechanisms. Biochem Biophys
Res Commun. 457:647–652. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sayed M, Drummond CA, Evans KL, Haller ST,
Liu J, Xie Z and Tian J: Effects of Na/K-ATPase and its ligands on
bone marrow stromal cell differentiation. Stem Cell Res. 13:12–23.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang L, Liu W, Alizadeh D, Zhao D,
Farrukh O, Lin J, Badie SA and Badie B: S100B attenuates microglia
activation in gliomas: Possible role of STAT3 pathway. Glia.
59:486–498. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hartman KG, Vitolo MI, Pierce AD, Fox JM,
Shapiro P, Martin SS, Wilder PT and Weber DJ: Complex formation
between S100B protein and the p90 ribosomal S6 kinase (RSK) in
malignant melanoma is calcium-dependent and inhibits extracellular
signal-regulated kinase (ERK)-mediated phosphorylation of RSK. J
Biol Chem. 289:12886–12895. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Meghnani V, Vetter SW and Leclerc E: RAGE
overexpression confers a metastatic phenotype to the WM115 human
primary melanoma cell line. Biochim Biophys Acta. 1842:1017–1027.
2014. View Article : Google Scholar : PubMed/NCBI
|