Introduction
Cancer is recognized as a major cause for mortality
worldwide. In men, prostate cancer is one of the most prevalent of
these malignancies, and is considered to be the second leading
cause of cancer-associated death in males in many western countries
(1). The morbidity rate of
prostate cancer is currently increasing at an astonishing rate in
developing countries (2). Prostate
cancer presents clinically as a multifocal disease with a slow
progression, and demonstrates a highly aggressive level of
malignant neoplasia (3). The
five-year survival rate of prostate cancer patients was only 29% in
2008, and the disease has now developed into a critical public
health problem (4). A recent study
has revealed that the 5-year biochemical recurrence-free survival
rate is 82.1% in patients with localized prostate cancer who
received radical prostatectomy treatment (5). Despite intensive study of prostate
cancer, the mechanisms by which tumorigenesis and cancer
progression occur have yet to be fully defined. Although there are
substantial efforts to improve therapy for patients, the options
for treatment of advanced prostate cancer are relatively few in
number. Since 1990, the annual morbidity rate of prostate cancer
has increased by 14% (6). This is
largely attributed to limited knowledge concerning the initiation,
growth, invasion and metastasis of prostate cancer, as well as the
poor range of effective novel therapeutics that are available for
those diagnosed at different stages in progression of the cancer.
The conventional diagnosis for prostate cancer involves
prostate-specific antigen (PSA) measurement, digital rectal
examination and needle biopsy (7).
Whilst the detection of PSA has greatly improved the detection rate
of early prostate cancer, a needle biopsy is often recommended to
further examine the histologic evidence (even in cases when an
increased PSA level is detected). Accumulating evidence suggests
that the PSA measurement is insufficient to predict both aggressive
and indolent prostate cancer, with the U.S. Preventive Services
Task Force suggesting removal of this conventional screening method
from clinical practice (8).
Current clinicopathological parameters, including the Gleason
score, pathological grade, lymph node metastasis and tumor volume,
have been considered as prognostic factors for prostate cancer
(9). However, these parameters
remain inadequate to distinguish between different forms of
prostate cancer (10). It is
therefore crucial to identify novel molecular targets in order to
facilitate the early diagnosis of prostate cancer.
MicroRNAs (miRNAs) are small 18–25 nucleotide
non-coding RNA molecules with highly conserved features. They serve
important roles in the initiation, development and metastasis of
many cancers, and are associated with the regulation of cell
growth, migration and invasion (11). It has been demonstrated that
various cancers exhibit aberrant expression of miRNAs (12). Deregulated miRNAs are essential
mediators for cancer pathogenesis by functioning as oncogenes or
tumor suppressor genes (13). An
increasing number of studies have explored the association between
cancer and miRNAs, and several miRNAs have been identified as
biomarkers or therapeutic targets for a number of human cancers
(14). Recent studies have
demonstrated that miR-129 serves an important role in tumor cell
growth and invasion in hepatocellular carcinoma (15) and in lung cancer cells (16). However, the role of miR-129 in
prostate cancer remains largely elusive. In the present study, the
association between miR-129 expression, clinicopathological
features and the prognosis of prostate cancer patients was
investigated.
Materials and methods
Ethics statement
Approval of this study protocol was obtained from
the Ethics Committee of Jinling Hospital (Nanjing, China), and
written informed consent was provided from all subjects. All
experiments were performed in accordance with the relevant
guidelines and regulations of Nanjing University (Nanjing, China).
This study conformed to the principles outlined in the Declaration
of Helsinki adopted by the World Medical Association's General
Assembly (17).
Subjects
A total of 118 prostate cancer patients admitted to
Jinling Hospital (Department of Urology, Nanjing University)
between 2000 and 2007, were included in this study. All patients
had undergone a radical prostatectomy, and had not received
chemotherapy, radiation therapy or androgen-deprivation treatment.
Prostate cancer tissue samples were placed into 10% buffered
formalin, and then subjected to gradient dehydration, wax dipping
and embedding. Sections (5-µm thick) of each sample were placed on
glass slides, dewaxed, dehydrated and stained with
hematoxylin-eosin. All paraffin-embedded tissues for each sample
were subject to histopathological examination by hematoxylin-eosin
staining. The histopathological grading of samples for all cases
was performed by experienced pathologists on hematoxylin-eosin
stained sections to confirm diagnosis and tumor content as >70%
prostate cancer cells in the tissue samples. The
clinicopathological and demographic data pre- and post-operation
were preserved in medical records. The biochemical and
clinicopathological parameters for each patient, including clinical
stage, Gleason score (18), margin
status, angiolymphatic invasion status, seminal vesicle invasion
status and biochemical recurrence were all recorded. The summary of
clinicopathological characteristics for all patients is presented
in Table I. The biochemical
recurrence (BCR) is a surrogate endpoint when PSA levels are ≥0.2
ng/ml in the patient's serum following surgical treatment. The date
of prostatectomy was recognized as the beginning of the follow-up
period. Any patients that did not survive due to unexpected events
or diseases other than prostate cancer were excluded from the
study. The primary prostate cancer tissues and paired noncancerous
prostate tissues from 118 cases were collected and frozen in liquid
nitrogen and stored at −80°C prior to use.
 | Table I.Correlation of miR-129 expression
with the clinicopathological characteristics of 118 prostate cancer
patients. |
Table I.
Correlation of miR-129 expression
with the clinicopathological characteristics of 118 prostate cancer
patients.
|
|
| miR-129 expression
(%) |
|
|
|---|
|
|
|
|
|
|
|---|
| Variable | Cases (%) | Low | High | χ2 | P-value |
|---|
| Age |
|
|
|
|
|
|
≤60 | 71 (60.2) | 41 (57.8) | 30 (42.2) | 0.238 | 0.626 |
|
>60 | 47 (39.8) | 25 (53.2) | 22 (46.8) |
| Histological
grade |
|
|
|
|
|
|
G1+G2 | 65 (55.1) | 25 (38.5) | 40 (61.5) | 17.921 | <0.001 |
| G3 | 53 (44.9) | 41 (77.4) | 12 (22.6) |
|
|
| Preoperative
PSA |
|
|
|
|
|
| <4
ng/ml | 4 (3.4) | 0 (0) | 4 (100) | 33.332 | <0.001 |
|
4–10ng/ml | 30 (25.4) | 5 (16.7) | 25 (83.3) |
|
|
| >10
ng/ml | 84 (71.2) | 61 (72.6) | 23 (27.4) |
|
|
| Pathological
stage |
|
|
|
|
|
| I +
II | 77 (65.3) | 32 (41.6) | 45 (58.4) | 18.576 | <0.001 |
| III +
IV | 41 (34.7) | 34 (82.9) | 7 (17.1) |
|
|
| Gleason score |
|
|
|
|
|
|
<7 | 36 (30.5) | 11 (30.6) | 25 (69.4) | 25.345 | <0.001 |
| 7 | 50 (42.4) | 26 (52.0) | 24 (48.0) |
|
|
|
>7 | 32 (27.1) | 29 (90.6) | 3 (9.4) |
|
|
| Lymph node
metastasis |
|
|
|
|
|
|
Negative | 103 (87.3) | 52 (50.5) | 51 (49.5) | 9.753 | 0.002 |
|
Positive | 15 (12.7) | 14 (93.3) | 1 (6.7) |
|
|
| Surgical margin
status |
|
|
|
|
|
|
Negative | 99 (83.9) | 56 (56.6) | 43 (43.4) | 0.100 | 0.752 |
|
Positive | 19 (16.1) | 10 (52.6) | 9 (47.4) |
|
|
| Angiolymphatic
invasion |
|
|
|
|
|
|
Negative | 82 (69.5) | 40 (48.8) | 42 (51.2) | 5.577 | 0.018 |
|
Positive | 36 (30.5) | 26 (72.2) | 10 (27.8) |
|
|
| Biochemical
recurrence |
|
|
|
|
|
|
Negative | 89 (75.4) | 42 (47.2) | 47 (52.8) | 11.226 | 0.001 |
|
Positive | 29 (24.6) | 24 (82.8) | 5 (17.2) |
|
|
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
The TRIzol reagent (Thermo Fisher Scientific,
Waltham, MA, USA) was used to extract the total RNA from 118 normal
and cancerous prostate tissues, according to the manufacturer's
instructions. The concentration and purity of extracted RNA were
measured at 260 and 280 nm optical densities. Reverse transcription
of the RNA was then performed using the PrimeScript RT-PCR kit
(Takara Bio Inc., Otsu, Japan), according to the manufacturer's
instructions. The cDNA served as a template for qPCR detection
using SYBR Premix Ex Taq™ (Takara Bio Inc.) and the StepOnePlus
Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The U6 gene served as an internal control, as described
previously (19). The PCR cycling
conditions were as follows: Initial denaturation at 95°C for 5 min,
followed by 40 cycles of denaturation at 94°C for 45 sec, annealing
at 50°C for 1 min and extension at 72°C for 1 min. Samples were
analyzed in triplicate and gene expression was quantified by
normalizing target gene expression to that of the internal control
using the 2−ΔΔCq method (20). The primer sequences used were as
follows: miR-129 forward, 5′-GATACTCACTTTTTGCGGTCT-3′ and reverse,
5′-GTGCAGGGTCCGAGGT-3′; U6 forward, 5′-CGCTTCGGCAGCACATATAC-3′ and
reverse, 5′-CAGGGGCCATGCTAATCTT-3′ (21).
Cell culture and transfection
The prostate cell lines, PC-3 and DU145, were
purchased from the Cell Bank of Chinese Academy of Sciences
(Shanghai, China). The cells were cultured in RPMI 1640 medium
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal
bovine serum (Gibco; Thermo Fisher Scientific, Inc.), 1%
penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.), 1%
nonessential amino acids (Gibco; Thermo Fisher Scientific, Inc.)
and 1% (1 mg/ml) sodium pyruvate (Gibco; Thermo Fisher Scientific,
Inc.) at 37°C in a humidified incubator with 5% CO2. The
day before transfection, PC-3 and DU145 cells (2×106)
were seeded into a six-well plate, and the transient transfection
of miR-129 precursor or scramble mimic miRNA (Ambion, Carlsbad, CA,
USA) was conducted using Lipofectamine 2000 Transfection Reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) for 48 h, according to
the manufacturer's instructions. The cells in the control group
were treated with transfection reagent only.
Cell proliferation assay
Cell proliferation was assessed using the Cell
Counting Kit-8 assay (CCK-8; Molecular Technologies, Inc.,
Kumamoto, Japan), in accordance with the manufacturer's
suggestions. The absorbance, measured at a wavelength of 450 nm,
was recorded on days 1, 2 and 4 post-transfection. At 96 h
following transfection, the cells were harvested to determine the
protein expression levels of proliferating cell nuclear antigen
(PCNA) and phosphorylated histone H3 (P-H3).
Western blot analysis
The PC-3 and DU-145 prostate cell lines were
transfected with either the miR-129 precursor or the scramble mimic
negative control for 96 h and were harvested using a lysis buffer
(Beyotime Institute of Biotechnology, Haimen, China) supplemented
with EDTA-free Halt™ Protease Inhibitor Cocktail (Pierce; Thermo
Scientific, Inc.). The total protein concentration of the
supernatant was quantified using a bicinchoninic protein assay kit
(Pierce; Thermo Scientific, Inc.). Equal amounts of protein (40 µg)
for each sample were loaded onto a 8 or 10% SDS gel and transferred
to Immobilon-P polyvinylidene difluoride membranes (EMD Millipore,
Billerica, MA, USA). The membranes were incubated with the
following primary antibodies overnight at 4°C: Rabbit anti-PCNA
(1:200; catalog no. sc-9857-R), mouse anti-P-H3 (1:200; catalog no.
sc-374669) and mouse anti-GAPDH (1:200; catalog no. sc-47724),
obtained from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA); and
rabbit anti-cyclin E (1:1,000; catalog no. 20808), rabbit
anti-cyclin D1 (1:1,000; catalog no. 2978), rabbit anti-p21
(1:1,000; catalog no. 2947) and rabbit anti-p27 (1:1,000; catalog
no. 3686), purchased from Cell Signaling Technology, Inc.,
(Danvers, MA, USA). Membranes were subsequently incubated with an
anti-rabbit (1:200; catalog no. sc-2385; Santa Cruz Biotechnology,
Inc.) or anti-mouse (1:200; catalog no. sc-2380; Santa Cruz
Biotechnology, Inc.) horseradish peroxidase-conjugated secondary
antibody for 1 h at room temperature. Protein bands were visualized
by use of an enhanced chemiluminescence detection kit (Thermo
Fisher Scientific, Inc.). Densitometric analysis of the band
intensities was measured and normalized to the band intensities of
GAPDH using the ImageJ software version 1.48 (National Institutes
of Health, Bethesda, MA, USA).
Statistical analysis
Continuous variables are expressed as the mean ±
standard deviation. SPSS 19.0 software (SPSS Inc., Chicago, IL,
USA) was used for statistical analysis. The Kolmogorov-Smirnov test
was used to determine the normality of the data distribution.
Comparisons between two groups were assessed using the Student's
t-test. One-way or two-way analysis of variance followed by a
post-hoc Bonferroni test was used for multiple comparisons.
The test for categorical variables was determined using the
χ2 test, and the small cell variables were compared
using Fisher's exact test. Survival analysis was conducted with the
Kaplan-Meier method. Multivariate analyses were performed using the
Cox proportional hazards model. P<0.05 was considered to
indicate a statistically significant difference.
Results
Downregulation of miR-129 expression
in prostate cancer tissues
The miR-129 expression levels in 118 paired prostate
cancer and adjacent non-cancerous prostate tissues were measured by
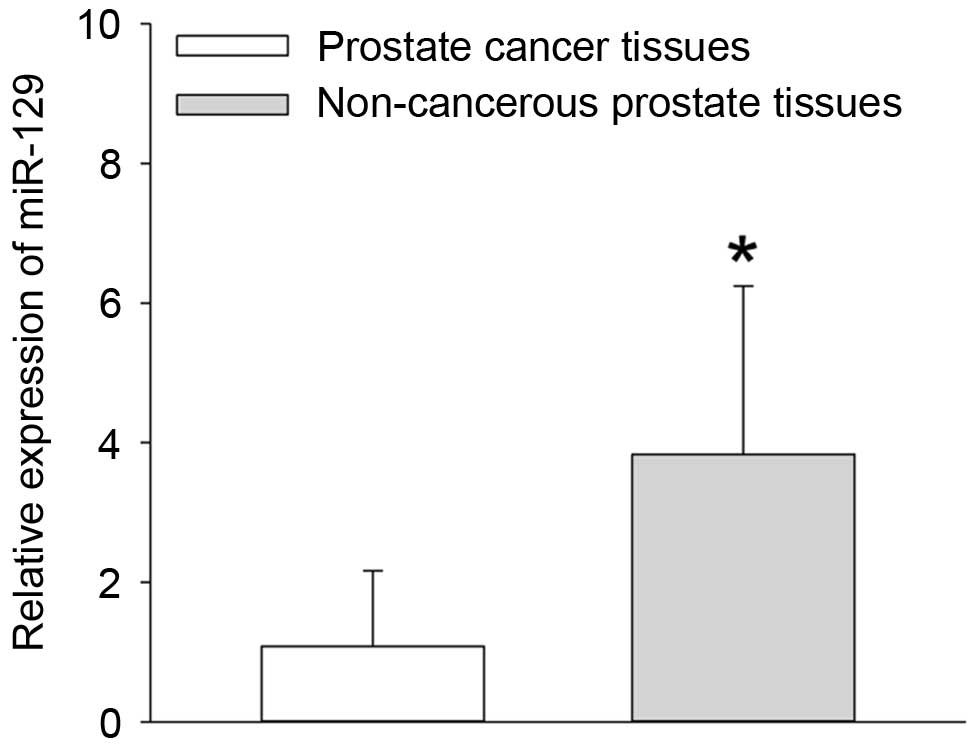
RT-qPCR analysis. The results demonstrated that the expression of
miR-129 at the mRNA level was significantly reduced in the prostate
cancer tissues when compared with non-cancerous prostate tissues
(P=0.013; Fig. 1). In addition,
the median relative quantity of miR-129 expression in the 118
prostate cancer tissues was equal to 1.01 (Fig. 1). Therefore, the number of prostate
cancer patients with low miR-129 expression was 52, and the number
with high miR-129 expression was 66.
Correlation of miR-129 expression with
the clinical parameters of prostate cancer patients
As demonstrated in Table I, low expression of miR-129 in the
prostate cancer tissues was closely correlated with aggressive
clinical pathological parameters including histological grade
(P<0.001), high preoperative PSA level (P<0.001),
pathological stage (P<0.001), a high Gleason score (P<0.001),
lymph node metastases (P=0.002), angiolymphatic invasion (P=0.018)
and BCR (P=0.001). No association between the expression level of
miR-129 and additional clinical factors such as age and surgical
margin status was observed (all P>0.05).
Association between miR-129 expression
and BCR-free survival
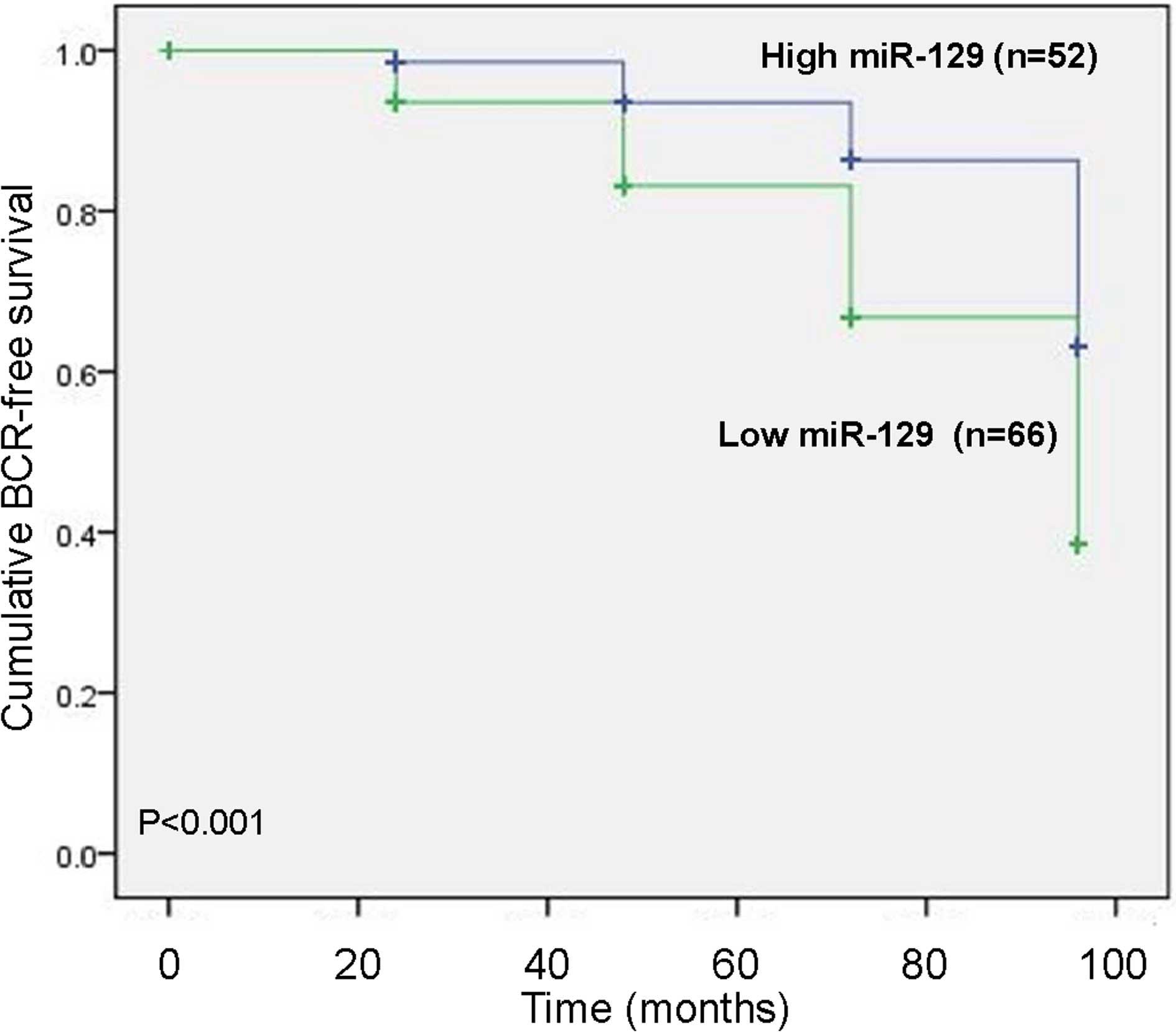
To assess the possible prognostic value of using
miR-129 as a biomarker in prostate cancer tissues, the BCR-free
survival in 118 prostate cancer patients undergoing radical
prostatectomy was performed by calculating the cumulative survival
curves using the Kaplan-Meier method. The Kaplan-Meier curves
plotted with high or low miR-129 expression levels and BCR-free
survival, indicated that prostate cancer patients with low miR-129
expression levels exhibited a significantly shorter BCR-free
survival compared with those with high miR-129 expression levels
(P<0.001; Fig. 2). As
summarized in Table II, the
univariate survival analysis with Cox proportional hazards model
revealed that the impact of well-known clinicopathological
prognostic features, including miR-129 expression (P<0.001),
Gleason score (P<0.001), histological grade (P=0.002),
pathological stage (P<0.001), lymph node metastasis (P=0.006)
and angiolymphatic invasion (P=0.005) were significantly associated
with BCR-free survival in prostate cancer patients. Since variables
that are determined to have a prognostic influence by univariate
analysis may covariate, a multivariate analysis of the association
between miR-129 expression levels and the BCR-free survival of
patients with prostate cancer was performed. The Cox multivariate
analysis confirmed the significance of BCR-free survival with
miR-129 expression (P<0.001), as well as histological grade
(P=0.002), pathological stage (P<0.001), lymph node metastasis
(P<0.001) and angiolymphatic invasion (P<0.001) as
independent prognostic predictors of BCR-free survival of prostate
cancer patients (Table III).
 | Table II.Univariate survival analysis of
BCR-free survival in 118 prostate cancer patients. |
Table II.
Univariate survival analysis of
BCR-free survival in 118 prostate cancer patients.
|
| BCR-free
survival |
|---|
|
|
|
|---|
| Variables | Exp (B) | 95% CI | P-value |
|---|
| Age (≤60 vs.
>60) | 1.067 | 0.802–1.421 | 0.653 |
| Histological grade
(G1+G2 vs. G3) | 2.115 | 0.275–1.224 | 0.002 |
| Preoperative PSA
(<10 ng/ml vs. ≥10 ng/ml) | 1.091 | 0.918–1.294 | 0.317 |
| Pathological stage
(T1-2 vs. T3-4) | 4.524 | 0.883–2.136 | <0.001 |
| Gleason score (4–6
vs. 7–10) | 3.745 | 0.698–1.725 | <0.001 |
| Lymph node
metastasis (negative vs. positive) | 1.941 | 0.189–1.138 | 0.006 |
| Surgical margin
status (negative vs. positive) | 1.015 | 0.714–2.192 | 0.927 |
| Angiolymphatic
invasion (negative vs. positive) | 1.508 | 1.134–2.005 | 0.005 |
| miR-129 expression
(high vs. low) | 5.627 | 1.124–11.392 | <0.001 |
 | Table III.Multivariate survival analysis of
BCR-free survival in 118 prostate cancer patients. |
Table III.
Multivariate survival analysis of
BCR-free survival in 118 prostate cancer patients.
|
| BCR-free
survival |
|---|
|
|
|
|---|
| Variables | Exp (B) | 95% CI | P-value |
|---|
| Histological grade
(G1+G2 vs. G3) | 2.123 | 0.279–1.228 | 0.002 |
| Pathological stage
(T1-2 vs. T3-4) | 3.818 | 0.776–1.913 | <0.001 |
| Lymph node
metastasis (negative vs. positive) | 4.505 | 0.932–2.078 | <0.001 |
| Angiolymphatic
invasion (negative vs. positive) | 3.511 | 0.717–1.795 | <0.001 |
| miR-129 expression
(high vs. low) | 2.692 | 0.441–1.539 | <0.001 |
Effects of miR-129 on prostate cancer
cell growth
The miR-129 precursor or scramble mimic negative
control was transfected to PC-3 and DU-145 prostate cell lines in
order to determine the effect of miR-129 on prostate cancer cell
proliferation in vitro. As shown in Table IV, overexpression of miR-129
effectively attenuated the proliferation rate of PC-3 and DU-145
prostate cell lines at days 2 and 4 post-transfection, as
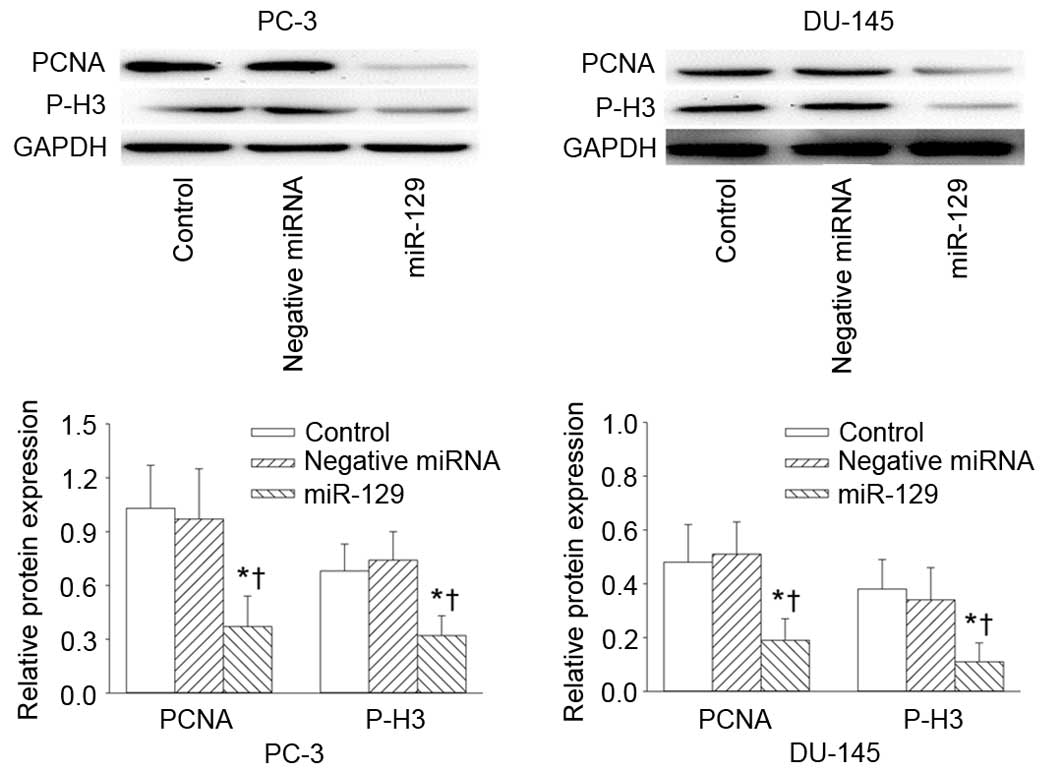
determined using the CCK8 assay. In addition, the protein
expression levels of cell proliferation markers PCNA and P-H3 were
significantly reduced in PC-3 and DU-145 cells at 4 days following
transfection with miR-129 precursor, compared with untreated and
negative controls (Fig. 3).
 | Table IV.CCK-8 assay for the growth of PC-3
and DU-145 prostate cancer cell lines transfected with miR-129
precursor or negative control miRNA and measured at days 1, 2, and
4 post-transfection. |
Table IV.
CCK-8 assay for the growth of PC-3
and DU-145 prostate cancer cell lines transfected with miR-129
precursor or negative control miRNA and measured at days 1, 2, and
4 post-transfection.
| Cell line | Indicated
transfection | OD450 at
day 0 | OD450 at
day 1 | OD450 at
day 2 | OD450 at
day 4 |
|---|
| PC-3 | Negative miRNA | 0.214±0.016 | 0.532±0.045 | 0.834±0.079 | 1.231±0.098 |
|
| miR-129 | 0.225±0.019 | 0.510±0.056 |
0.621±0.066a |
0.813±0.085a |
| DU-145 | Negative miRNA | 0.216±0.021 | 0.652±0.078 | 0.948±0.095 | 1.422±0.113 |
|
| miR-129 | 0.209±0.025 | 0.643±0.069 |
0.724±0.043a |
0.933±0.105a |
Inhibitory effects of miR-129 on the
protein expression levels of cell cycle regulators
Cell cycle progression is associated with cell
proliferation, therefore the present study aimed to clarify whether
inhibition of cell growth by miR-129 involves the regulation of
cell cycle-associated proteins cyclin E, cyclin D1, p27 and p21.
Compared with untreated and negative controls, the protein
expression levels of cyclin E and cyclin D1 were significantly
reduced in PC-3 and DU-145 cells following transfection with
miR-129 (P=0.013; Fig. 4). By
contrast the protein expression levels of p27 and p21 in both cell
lines were significantly increased when compared with untreated and
negative controls (P=0.021; Fig.
4). These results suggest that miR-129 may exert an inhibitory
effect on the proliferation of prostate cancer cells by inducing of
cell cycle arrest.
Discussion
The present study established an association between
miR-129 and aggressive clinicopathological parameters in prostate
cancer patients. miR-129 may be a crucial component in the
pathogenesis and aggressiveness of prostate cancer, and
downregulation of its expression may be closely correlated with
unfavorable prognosis in prostate cancer patients.
Prostate cancer is a prevalent malignancy in
developed and developing countries and is a significant global
clinical burden, accounting for >92,300 mortalities each year
(22). The lack of sufficient
diagnostic and prognostic factors are primary causes for poor
clinical outcome in prostate cancer patients. It has been proposed
that an early and accurate diagnosis of prostate cancer would be
essential for determining an appropriate treatment strategy
(23). Despite the fact that
patients with localized prostate cancer that have undergone a
radical prostatectomy have favorable long-term survival rates,
long-term follow-ups have demonstrated that BCR occurs in ~50% of
patients following surgery (24).
Hence, it is necessary to evaluate the early risk of BCR in order
to improve the prognosis for patients. Conventional prognostic
factors, including the Gleason score or measuring preoperative PSA
levels, are limited for the diagnosis of early prostate cancer
(25). A previous study
demonstrated that Golgi phosphoprotein-3 expression was associated
with androgen-independence, bone metastases, a higher Gleason scale
score and higher baseline PSA levels, and serves as a significant
independent prognostic factor for disease-free survival in prostate
cancer patients (26). In
addition, RNA-binding protein (RBM3) expression was observed to be
increased in the prostate intraepithelium, which was associated
with a prolonged time to disease progression (27). It was therefore suggested that RBM3
may be employed as a useful independent biomarker of favorable
prognosis for prostate cancer (27). In addition, a recent study
discovered that high transformer 2β (Tra2β) expression levels are
significantly associated with a high Gleason score, clinical stage,
lymph node metastases, and BCR in subjects with prostate cancer,
and overexpression of Tra2β may be a possible predictor for poor
BCR-free survival (28). These
studies demonstrate the unremitting efforts that have been made by
many researchers so far in order to attempt to uncover efficient
diagnostic and prognostic biomarkers for prostate cancer.
miRNAs serve an important role in numerous
biological processes in tumors, including cell proliferation,
apoptosis and migration and invasion. Over 1,400 human miRNA
sequences are known to be involved in cancer pathogenesis (29). In recent years, miRNAs have been
demonstrated to function as molecular prognostic biomarkers for a
number of cancers. For example, it has been reported that
miR-188-5p expression levels are significantly downregulated in
metastatic prostate cancer tissues, and miR-188-5p is predicted to
be an independent prognostic marker for BCR-free survival, as well
as overall survival in prostate cancer patients (30). In addition, miR-221 is also
significantly downregulated in prostate cancer patients with lymph
node-metastases, and is employed as a biomarker for the clinical
prognosis of high-risk prostate cancer patients (31). By contrast, miR-96 is overexpressed
in prostate cancer tissues and is closely associated with a poor
median survival of 3 years (32).
The results of these studies suggest that miRNAs are emerging as
potential prognostic biomarkers or useful therapeutic targets in
prostate cancer. miR-129 is a diagnostic and prognostic biomarker
in gastrointestinal cancer, which exhibits suppressive activities
that may lead to inhibition of tumorigenesis, tumor cell
proliferation and invasion, and disease progression (33). In addition, miR-129 was recently
observed to be downregulated in gastric cancer (34,35),
colorectal cancer (34,35), liver cancer (36) and lymphocytic leukemia (37). RT-qPCR results demonstrated that
miR-129-3p gene expression was extensively attenuated in human
renal cell carcinoma, and this low expression was a predictor of
reduced disease-free and overall survival in renal cell carcinoma
patients (37). miR-129 expression
in bladder cancer patients has been demonstrated to be lower when
compared with normal noncancerous controls, thus miR-129 is
suggested to be a predictive marker of the progression of bladder
tumors (38). However, the role of
miR-129 in prostate cancer patients remains largely elusive. The
results of the present study demonstrate a significant reduction in
miR-129 gene expression levels in prostate cancer tissues when
compared with adjacent non-cancerous prostate tissues. Further
analysis indicated that downregulation of miR-129 gene expression
was positively associated with histological grade, high
preoperative PSA levels, pathological stage, a high Gleason score,
lymph node metastases, angiolymphatic invasion and BCR in prostate
cancer patients. In addition, prostate cancer patients with low
miR-129 expression exhibited poor BCR-free survival. The
multivariate analyses clarified that reduced miR-129 expression may
be an independent prognostic indicator of BCR-free survival in
prostate cancer patients. These results suggest that miR-129 may
provide a novel and important prognostic biomarker for prostate
cancer.
Excessive cell proliferation in prostate cancer
tissues is positively associated with tumorigenesis and tumor
progression (39). In bladder
carcinoma cell lines, transfection with a miR-129 precursor
significantly inhibited the growth and induced cell death in T24
and SW780 cells (38). In
hepatocellular carcinomas, transfection with miR-129 mimics
remarkably attenuated the proliferation and invasion of
hepatocellular carcinoma cells (15). Ectopic expression of miR-129-5p
significantly inhibited the growth and migration of medullary
thyroid carcinoma cells by targeting the proto-oncogene pathway
(40). The results of the present
study demonstrated that miR-129 gene overexpression attenuated the
proliferation of PC-3 and DU-145 prostate cancer cell lines. This
was evidenced by the CCK-8 assay and the observed downregulation of
PCNA and P-H3 in response to transfection with an miR-129 mimic.
These results suggest that miR-129 may function as tumor suppressor
gene in prostate cancer, and may be a potential therapeutic target
in prostate cancer.
Abnormal regulation of the cell cycle is an
important factor that is closely associated with the cancer
development (41). Emerging
evidence indicates that deregulated miRNAs are major mediators in
the regulation of tumor-associated cell cycle defects (42). Multiple protein kinases that
contain a regulatory cyclin component and a catalytic
cyclin-dependent kinase (CDK) are necessary for cell cycle
regulation during several major checkpoints of the cell cycle. The
specific CDK inhibitors, p21 and p27, are negative regulators of
cyclins, which inhibit the cell cycle at the G0/G1 phase (43). Overexpression of cyclin D1 is a
critical step in the development of some human cancers, including
prostate cancer, by encoding the regulatory subunit of a holoenzyme
and promoting the progression of the G1/S phase of the cell cycle
(44). Increased cyclin D1
expression is correlated with human tumorigenesis and cellular
metastases, including parathyroid adenoma, breast cancer, colon
cancer, lymphoma, melanoma, and prostate cancer (44). In addition, cyclin E has been
demonstrated to promote the proliferation of prostate cancer cells
(45). Introduction of an
adenovirus vector harboring p27 noticeably reduced the size of
human prostate cancer xenograft tumors (45) and p21 is an established tumor
suppressor gene involved in human prostate cancer development
(46). The present study
demonstrated that miR-129 overexpression in PC-3 and DU-145
prostate cancer cell lines was associated with a reduction in the
expression of cell cycle-associated proteins cyclin E and cyclin
D1, and an increase in the expression of CDK inhibitors p21 and
p27. These results indicate that miR-129 exerts an inhibitory
effect on the growth of prostate cancer cells, potentially via a
direct repressive effect on cell cycle progression. This suggests a
mechanism whereby downregulation of miR-129 may contribute to the
growth of malignant prostate cancer. However, future studies
regarding the role of miR-129 in the migration and invasion of
tumors in nude mice, as well as their underlying molecular
mechanisms, are required.
In conclusion, the results of the present study
demonstrated that low miR-129 expression may serve a integral role
in the progression of prostate cancer, and was significantly
associated with poor prognosis independently of other factors in
prostate cancer. These results raise the possibility that miR-129
may be a useful prognostic parameter for prostate cancer, and may
be considered as a novel molecular target for the diagnosis and
treatment of prostate cancer.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ferlay J, Parkin DM and Steliarova-Foucher
E: Estimates of cancer incidence and mortality in Europe in 2008.
Eur J Cancer. 46:765–781. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Carroll PR: Early stage prostate cancer-do
we have a problem with over-detection, overtreatment or both? J
Urol. 173:1061–1062. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Boehm K, Schiffmann J, Tian Z, Lesmana H,
Larcher A, Mandel P, Karakiewicz PI, Graefen M, Schwarz R, Krüll A
and Tilki D: Five-year biochemical recurrence-free and overall
survival following high-dose-rate brachytherapy with additional
external beam or radical prostatectomy in patients with clinically
localized prostate cancer. Urol Oncol. 34:119.e11–e18. 2016.
View Article : Google Scholar
|
|
6
|
Ishizaki F, Hara N, Koike H, Kawaguchi M,
Tadokoro A, Takizawa I, Nishiyama T, Takahashi K and Hohenfellner
R: Prediction of pathological and oncological outcomes based on
extended prostate biopsy results in patients with prostate cancer
receiving radical prostatectomy: A single institution study. Diagn
Pathol. 7:682012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Velonas VM, Woo HH, dos Remedios CG and
Assinder SJ: Current status of biomarkers for prostate cancer. Int
J Mol Sci. 14:11034–11060. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Moyer VA: U.S. Preventive Services Task
Force: Screening for prostate cancer: U.S. Preventive Services Task
Force recommendation statement. Ann Intern Med. 157:120–134. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dabir PD, Ottosen P, Hoyer S and
Hamilton-Dutoit S: Comparative analysis of three- and two-antibody
cocktails to AMACR and basal cell markers for the
immunohistochemical diagnosis of prostate carcinoma. Diagn Pathol.
7:812012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Humphrey PA and Vollmer RT: Percentage
carcinoma as a measure of prostatic tumor size in radical
prostatectomy tissues. Mod Pathol. 10:326–333. 1997.PubMed/NCBI
|
|
11
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li J, Ju J, Ni B and Wang H: The emerging
role of miR-506 in cancer. Oncotarget. Aug 15–2016.(Epub ahead of
print).
|
|
13
|
Pallante P, Visone R, Ferracin M, Ferraro
A, Berlingieri MT, Troncone G, Chiappetta G, Liu CG, Santoro M,
Negrini M, et al: MicroRNA deregulation in human thyroid papillary
carcinomas. Endocr Relat Cancer. 13:497–508. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ohtsuka M, Ling H, Doki Y, Mori M and
Calin GA: MicroRNA processing and human cancer. J Clin Med.
4:1651–1667. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhai J, Qu S, Li X, Zhong J, Chen X, Qu Z
and Wu D: miR-129 suppresses tumor cell growth and invasion by
targeting PAK5 in hepatocellular carcinoma. Biochem Biophys Res
Commun. 464:161–167. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xiao Y, Li X, Wang H, Wen R, He J and Tang
J: Epigenetic regulation of miR-129-2 and its effects on the
proliferation and invasion in lung cancer cells. J Cell Mol Med.
19:2172–2180. 2015.PubMed/NCBI
|
|
17
|
Malik AY and Foster C: The revised
declaration of Helsinki: Cosmetic or real change? J R Soc Med.
109:184–189. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Miah S, Ahmed HU, Freeman A and Emberton
M: Does true Gleason pattern 3 merit its cancer descriptor? Nat Rev
Urol. 13:541–548. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xu S, Yi XM, Zhou WQ, Cheng W, Ge JP and
Zhang ZY: Downregulation of miR-129 in peripheral blood mononuclear
cells is a diagnostic and prognostic biomarker in prostate cancer.
Int J Clin Exp Pathol. 8:14335–14344. 2015.PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang QY, Tang J, Zhou CX and Zhao Q: The
down-regulation of miR-129 in breast cancer and its effect on
breast cancer migration and motility. Sheng Li Xue Bao. 64:403–411.
2012.(In Chinese). PubMed/NCBI
|
|
22
|
Wilson HC, Shah SI, Abel PD, Price P,
Honeyfield L, Edwards S and Abel RL: Contemporary hormone therapy
with LHRH agonists for prostate cancer: Avoiding osteoporosis and
fracture. Cent European J Urol. 68:165–168. 2015.PubMed/NCBI
|
|
23
|
Mapelli P and Picchio M: Initial prostate
cancer diagnosis and disease staging-the role of choline-PET-CT.
Nat Rev Urol. 12:510–518. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Molitierno J, Evans A, Mohler JL, Wallen
E, Moore D and Pruthi RS: Characterization of biochemical
recurrence after radical prostatectomy. Urol Int. 77:130–134. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Montironi R, Mazzuccheli R, Scarpelli M,
Lopez-Beltran A, Fellegara G and Algaba F: Gleason grading of
prostate cancer in needle biopsies or radical prostatectomy
specimens: Contemporary approach, current clinical significance and
sources of pathology discrepancies. BJU Int. 95:1146–1152. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hua X, Yu L, Pan W, Huang X, Liao Z, Xian
Q, Fang L and Shen H: Increased expression of Golgi
phosphoprotein-3 is associated with tumor aggressiveness and poor
prognosis of prostate cancer. Diagn Pathol. 7:1272012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jonsson L, Gaber A, Ulmert D, Uhlén M,
Bjartell A and Jirström K: High RBM3 expression in prostate cancer
independently predicts a reduced risk of biochemical recurrence and
disease progression. Diagn Pathol. 6:912011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Diao Y, Wu D, Dai Z, Kang H, Wang Z and
Wang X: Prognostic value of transformer 2β expression in prostate
cancer. Int J Clin Exp Pathol. 8:6967–6973. 2015.PubMed/NCBI
|
|
29
|
Sempere LF: Integrating contextual miRNA
and protein signatures for diagnostic and treatment decisions in
cancer. Expert Rev Mol Diagn. 11:813–827. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang H, Qi S, Zhang T, Wang A, Liu R, Guo
J, Wang Y and Xu Y: miR-188-5p inhibits tumour growth and
metastasis in prostate cancer by repressing LAPTM4B expression.
Oncotarget. 6:6092–6104. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Spahn M, Kneitz S, Scholz CJ, Stenger N,
Rüdiger T, Ströbel P, Riedmiller H and Kneitz B: Expression of
microRNA-221 is progressively reduced in aggressive prostate cancer
and metastasis and predicts clinical recurrence. Int J Cancer.
127:394–403. 2010.PubMed/NCBI
|
|
32
|
Haflidadóttir BS, Larne O, Martin M,
Persson M, Edsjö A, Bjartell A and Ceder Y: Upregulation of miR-96
enhances cellular proliferation of prostate cancer cells through
FOXO1. PloS One. 8:e724002013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fesler A, Zhai H and Ju J: miR-129 as a
novel therapeutic target and biomarker in gastrointestinal cancer.
Onco Targets Ther. 7:1481–1485. 2014.PubMed/NCBI
|
|
34
|
Tsai KW, Wu CW, Hu LY, Li SC, Liao YL, Lai
CH, Kao HW, Fang WL, Huang KH, Chan WC and Lin WC: Epigenetic
regulation of miR-34b and miR-129 expression in gastric cancer. Int
J Cancer. 129:2600–2610. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bandres E, Agirre X, Bitarte N, Ramirez N,
Zarate R, Roman-Gomez J, Prosper F and Garcia-Foncillas J:
Epigenetic regulation of microRNA expression in colorectal cancer.
Int J Cancer. 125:2737–2743. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen X, Zhang L, Zhang T, Hao M, Zhang X,
Zhang J, Xie Q, Wang Y, Guo M, Zhuang H and Lu F:
Methylation-mediated repression of microRNA 129-2 enhances
oncogenic SOX4 expression in HCC. Liver Int. 33:476–486. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen X, Ruan A, Wang X, Han W, Wang R, Lou
N, Ruan H, Qiu B, Yang H and Zhang X: miR-129-3p, as a diagnostic
and prognostic biomarker for renal cell carcinoma, attenuates cell
migration and invasion via downregulating multiple
metastasis-related genes. J Cancer Res Clin Oncol. 140:1295–1304.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dyrskjot L, Ostenfeld MS, Bramsen JB,
Silahtaroglu AN, Lamy P, Ramanathan R, Fristrup N, Jensen JL,
Andersen CL, Zieger K, et al: Genomic profiling of microRNAs in
bladder cancer: miR-129 is associated with poor outcome and
promotes cell death in vitro. Cancer Res. 69:4851–4860. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Goździk-Spychalska J, Szyszka-Barth K,
Spychalski L, Ramlau K, Wójtowicz J, Batura-Gabryel H and Ramlau R:
C-MET inhibitors in the treatment of lung cancer. Curr Treat
Options Oncol. 15:670–682. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Duan L, Hao X, Liu Z, Zhang Y and Zhang G:
MiR-129-5p is down-regulated and involved in the growth, apoptosis
and migration of medullary thyroid carcinoma cells through
targeting RET. FEBS Lett. 588:1644–1651. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yang Y, Pan X, Lei W, Wang J, Shi J, Li F
and Song J: Regulation of transforming growth factor-beta 1-induced
apoptosis and epithelial-to-mesenchymal transition by protein
kinase A and signal transducers and activators of transcription 3.
Cancer Res. 66:8617–8624. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bao B, Ahmad A, Kong D, Ali S, Azmi AS, Li
Y, Banerjee S, Padhye S and Sarkar FH: Hypoxia induced
aggressiveness of prostate cancer cells is linked with deregulated
expression of VEGF, IL-6 and miRNAs that are attenuated by CDF.
PloS One. 7:e437262012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Abukhdeir AM and Park BH: P21 and p27:
Roles in carcinogenesis and drug resistance. Expert Rev Mol Med.
10:e192008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Fu M, Wang C, Li Z, Sakamaki T and Pestell
RG: Minireview: Cyclin D1: Normal and abnormal functions.
Endocrinology. 145:5439–5447. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hashimoto Y, Naruyama H, Ando R, Okada S,
Tozawa K and Kohri K: Molecular targeted therapy for prostate
cancer. Hinyokika Kiyo. 54:57–61. 2008.PubMed/NCBI
|
|
46
|
Lee JT, Lehmann BD, Terrian DM, Chappell
WH, Stivala F, Libra M, Martelli AM, Steelman LS and McCubrey JA:
Targeting prostate cancer based on signal transduction and cell
cycle pathways. Cell Cycle. 7:1745–1762. 2008. View Article : Google Scholar : PubMed/NCBI
|