Introduction
Ximenynic acid (also termed santalbic acid) is a
conjugated enyne fatty acid that predominantly exists in the seed
oil of the Santalaceae, Olacaceae, and Opiliaceae families
(1). Previous studies demonstrated
that ximenynic acid has antibacterial (2,3),
antifungal and anti-inflammatory activities (3,4).
Among these properties, the anti-inflammatory activity of ximenynic
acid has been widely studied.
The anti-inflammatory activity of ximenynic acid has
been reported since the 1980s. Nugteren and Christ-Hazelhof
(5) demonstrated that ximenynic
acid inhibits the activity of COXs in the sheep vesicular gland
microsomes (5). In rat peritoneal
leukocytes, ximenynic acid inhibited the phospholipase activity and
the production of inflammatory factors, including thromboxane B2,
6-ketoprostaglandin F1α and leukotriene B4 (4). It was observed that the levels of
leukotriene B4, thromboxane B2, prostaglandin F2α and prostaglandin
E2 (PGE2) in the sandalwood seed oil feeding group, which contains
a high percentage of ximenynic acid, were significantly lower in
rat liver and plasma compared with soybean oil and safflower oil
feeding groups after feeding for 8 weeks (6). These reports suggested that ximenynic
acid exerts anti-inflammatory activity by affecting the metabolism
of arachidonic acid.
Arachidonic acid metabolism pathways, induced by
lipoxygenase, cyclooxygenase (COX) and cytochrome P450, are
important for regulating the inflammatory response (7), and they generate several eicosanoid
products, including leukotrienes and prostanoids, which are closely
associated with the inflammatory response. It is established that
lipoxygenase, COX and cytochrome P450 are key targets in various
pathologies, including pain, cardiovascular disease, inflammation
and cancer (7). The eicosanoids
metabolism induced by COX has been previously reported to be
involved in various types of cancer (8). Most nonsteroidal anti-inflammatory
drugs (NSAIDs) are inhibitors of COXs, and they can reduce the risk
of cancer, which was supported by preclinical and clinical studies
(9). Furthermore, many natural and
synthetic acetylenic acids exert inhibitory effects on cancer cells
by suppressing key enzymes of the arachidonic acid metabolism
pathways (10).
There are two common types of COX: COX-1 and COX-2.
Although COX-2 inhibitors for treating cancer have been a research
focus for many years, suppression of COX-1 was also demonstrated to
exhibit anti-cancer properties (11,12),
and alter the distribution of cell cycle (13) and effect angiogenesis (14). Furthermore, inhibition of
angiogenesis and arrest of the cell cycle in oncocytes may induce
cell apoptosis (15,16).
There are few reports that directly demonstrate the
anti-cancer activity of ximenynic acid, thus, it was hypothesized
that ximenynic acid may have antitumor properties through
inhibition of COX activation. The current study investigated the
anti-cancer activity of ximenynic acid in HepG2, and analyzed the
underlying mechanism through determining its effects on cell cycle,
angiogenesis and COXs pathways.
Materials and methods
Fatty acid preparation
Ximenynic acid was obtained (99.5% purity) from the
seed oil of Santalum spicatum by low temperature
recrystallization, as described previously (17). Oleic acid (Sigma-Aldrich; Merck
Millipore, Darmstadt, Germany) was selected as the
non-alkynyl-fatty acid control for its similar structure to
ximenynic acid. All other chemicals were purchased commercially as
analytical grade reagents.
Cell culture and cell treatment
HepG2 human hepatoma cell line was obtained from the
Type Culture Collection of the Chinese Academy of Sciences
(Shanghai, China). Cells were grown in high glucose Dulbecco's
modified Eagle's medium (DMEM; Hyclone; GE Healthcare Life
Sciences, Logan, UT, USA) supplemented with 110 mg/l sodium
pyruvate, 4 mM L-glutamine, 10% fetal bovine serum (Biowest,
Nuaillé, France), 10 mM HEPES (Corning Incorporated, Corning, NY,
USA), 100 U/ml penicillin and 0.1 mg/ml streptomycin at 37°C in a
humidified incubator with 5% CO2. The medium was changed every 2
days. Cells were passaged when they reached 70–80% confluence, and
the culture medium was changed 1 day before.
Cells were seeded overnight in standard medium and
then change to serum-free medium supplemented with 1%
insulin-transferrin-selenium (ITS; Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), 0.1 mg/ml fatty acid-free
bovine serum albumin (BSA; MP Biomedicals, LLC, Santa Ana, CA, USA)
for synchronizing cell cycle division. After 24 h serum starvation,
cells were incubated in experimental media for indicated times. The
experimental media was DMEM supplemented with 1% ITS, 0.1 mg/ml BSA
and various concentrations of ximenynic acid or oleic acid with a
molar ratio of 4:1 to BSA. Cells treated with medium not containing
fatty acids were termed the vehicle control group.
Cell viability assay
Cells were cultured in 96-well plates overnight with
~1500 cells/well. After serum-starving for 24 h, serum-free medium
was removed and ximenynic acid or oleic acid at 25, 50, 100 and 150
µM were added to the cells. After 72 h incubation, the media were
replaced with serum-free medium diluted methyl thiazolyl
tetrazolium (MTT; Amresco, LLC, Solon, OH, USA) solution (5 mg/ml)
and incubated at 37°C for 4 h. Subsequently, MTT medium was removed
and the cells were dissolved with dimethyl sulfoxide followed by
detection of absorption at 595 nm with the Bio-Rad iMark Microplate
Absorbance Reader (Bio-Rad Laboratories, Inc., Hercules, CA,
USA).
Flow cytometry analysis
Cell cycle distribution was analyzed by propidium
iodide (PI) staining. Cells were seeded in the 6-well plates
overnight, and then incubated with serum-free media containing 50
or 150 µM ximenynic acid or oleic acid for 24 h and 36 h following
rinsing with phosphate-buffered saline (PBS). The harvested cells
were slowly fixed in pre-chilled 70% ethanol following washing with
PBS. The fixed cells were incubated at −20°C overnight.
Subsequently to rinsing with PBS, cells were re-suspended with PI
solution from the Cell Cycle Staining kit [CCS012; Multi Sciences
(Lianke) Biotech Co., Ltd., Hangzhou, China] and incubated in a
dark at room temperature for 30 min. Cell cycle distribution was
then analyzed using BD FACSCalibur (Becton Dickinson, USA).
Cell apoptosis was analyzed with Annexin V-FITC
(fluorescein isothiocyanate) and PI kit (Multisciences, China).
Cells were cultured in 6-well plates overnight, and then starved
for 24 h with serum-free medium followed by culture with different
concentrations of ximenynic acid or oleic acid for 72 h. The
harvested cells were gently rinsed with PBS. Cells were stained
with Annexin V-FITC and PI in dark for 15 min. The level of
apoptosis was then analyzed by BD FACSCalibur (BD Biosciences,
Franklin Lakes, NJ, USA).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Cells were seeded in 6-well plates overnight. After
starving for 24 h with serum-free medium, cells were treated with
50 or 150 µM ximenynic acid or oleic acid for 72 h. Total RNA was
extracted by HP Total RNA kit (Omega Bio-Tek, Inc., Norcross, GA,
USA) according to the manufacturer's protocol, and the RNA
concentration was determined using a NanoDrop 2000 (Thermo Fisher
Scientific, Inc., Wilmington, DE, USA). RT was performed on total
RNA to produce cDNA with using a Transcriptor First Strand cDNA
Synthesis kit (Roche Diagnostics, Basel, Switzerland). The
expression of genes was analyzed by qPCR. The sequences of primers
are presented in Table I. The qPCR
analysis was performed using the SsoFast EvaGreen Supermix (Bio-Rad
Laboratories, Inc.), according to the manufacturer's protocol, with
a CFX96™ Real Time PCR Detection System (Bio-Rad Laboratories,
Inc.). The qPCR program commenced with initial denaturation at 95°C
for 30 sec followed by 40 amplification cycles of 95°C for 5 sec
(denaturation) and 60°C for 10 sec (annealing and extension). The
melting curve program was from 65°C to 95°C for 5 sec with an
increment rate of 0.5°C/sec (18).
The RPLPO gene was used as the reference gene. Data were selected
from a minimum of 3 experiments.
 | Table I.Primers used for reverse
transcription-quantitative polymerase chain reaction analysis. |
Table I.
Primers used for reverse
transcription-quantitative polymerase chain reaction analysis.
| Protein | Gene symbol | Accession no. | Forward primer
(5′-3′) | Reverse primer
(5′-3′) | Product length
(bp) |
|---|
| Cyclin D3 | CCND3 | NM_001287427 |
CATGAACTACCTGGATCGCTAC |
GCCAGGAAATCATGTGCAATC | 243 |
| Cyclin E1 | CCNE1 | NM_001238 |
ACTCAACGTGCAAGCCTCG |
GCTCAAGAAAGTGCTGATCCC | 141 |
| Cyclooxygenase
1 | PTGS1 | NM_000962 |
TTCAATGAGTACCGCAAGAGG |
GAAGCAGTCCAGGGTAGAAC | 139 |
| Cyclooxygenase
2 | PTGS2 | NM_000963 |
CAAGACAGATCATAAGCGAGGG |
GTCTAGCCAGAGTTTCACCG | 92 |
| Prostaglandin E
receptor 2 | EP2 | NM_000956 |
CCTCATTCTCCTGGCTATCATG |
CTTTCGGGAAGAGGTTTCATTC | 94 |
| Prostaglandin E
receptor 4 | EP4 | NM_000958 |
ATCTTACTCATTGCCACCTCC |
TGACTTCTCGCTCCAAACTTG | 106 |
| Vascular
endothelial growth factor B | VEGF-B | NM_003377 |
CTTAGAGCTCAACCCAGACAC |
ACCCTGCTGAGTCTGAAAAG | 76 |
| Vascular
endothelial growth factor C | VEGF-C | NM_005429 |
GGCTGGCAACATAACAGAGA |
GTGGCATGCATTGAGTCTTT | 136 |
| Chemokine (C-C
Motif) ligand 2 | CCL2 | NM_002982 |
TGTCCCAAAGAAGCTGTGATC |
ATTCTTGGGTTGTGGAGTGAG | 150 |
| Hypoxia-inducible
factor-1α | HIF-1α | NM_001243084 |
GAAAACTTGGCAACCTTGGA |
AAATCTCCGTCCCTCAACCT | 196 |
| High Mobility Group
Box 1 | HMGB1 | NM_002128 |
CCATTGGTGATGTTGCGAAG |
TCAGGCTTTCCTTTAGCTCG | 145 |
| Transforming growth
factor β1 | TGF-β1 | NM_000660 |
TTCAACACATCAGAGCTCCG |
TGAGGTATCGCCAGGAATTG | 145 |
| Ribosomal protein,
large, P0 | RPLP0a | NM_001002 |
GAAACTCTGCATTCTCGCTTC |
GGTGTAATCCGTCTCCACAG | 150 |
Western blot analysis
Cells were cultured in 100 mm dishes overnight and
then starved with serum-free medium for 24 h. After treatment with
50 or 150 µM ximenynic acid or oleic acid for 72 h, cells were
harvested and proteins were extracted using cell lysis buffer
supplemented with phenylmethylsulfonyl fluoride (Beyotime Institute
of Biotechnology, Haimen, China). Whole-cell lysates were
centrifuged at 20,000 × g at 4°C, for 10 min to collect the
supernatant. The protein concentration was determined by
bicinchoninic acid protein assay (Beyotime Institute of
Biotechnology). Protein (20 µg) were subjected to sodium dodecyl
sulfate-polyacrylamide gel electrophoresis separation and
transferred to polyvinylidene difluoride membrane (Bio-Rad
Laboratories, Inc.) for probing with antibodies. The antibodies for
caspase-3 (cat. no. 9662s), silent information regulator T1 (SIRT1;
cat. no. 2496p), COX-2 (cat. no. 12282s), general control of amino
acid synthesis yeast homolog like 2 (GCN5L2; cat. no. 3305p), and
GAPDH (cat. no. 2118s) were purchased from Cell Signaling
Technology, Inc. (Danvers, MA, USA), and COX-1 (cat. no. ab109025)
from Abcam (Cambridge, UK). The secondary antibodies were
anti-mouse IgG (cat. no. #7076) and anti-rabbit IgG (cat. no.
#7074) from Cell Signaling Technology, Inc. These antibodies were
diluted in 5% BSA (1:1,000; Beyotime Institute of Biotechnology).
The immunoreactive bands were incubated with the primary antibodies
overnight at 4°C, then were washed three times with PBS with
Tween-20 (PBST). The secondary antibody was incubated for 1 h at
room temperature, and also washed three time with PBST. The protein
bands were visualized using the ECL reagent (Bio-Rad Laboratories,
Inc.). The intensity of immunoreactive bands was analyzed by
Quantity One software (Bio-Rad Laboratories, Inc.). Western
blotting data were selected from a minimum of three independent
experiments.
Statistical analysis
Data are expressed as the mean ± standard deviation
of three independent experiments. The significance of the
difference between groups was analyzed by SPSS software version 18
(SPSS, Inc., Chicago, IL, USA) using the Bonferroni or Dunnett's T3
method of single factor analysis of variance (19). P<0.05 was considered to indicate
a statistically significant difference.
Results
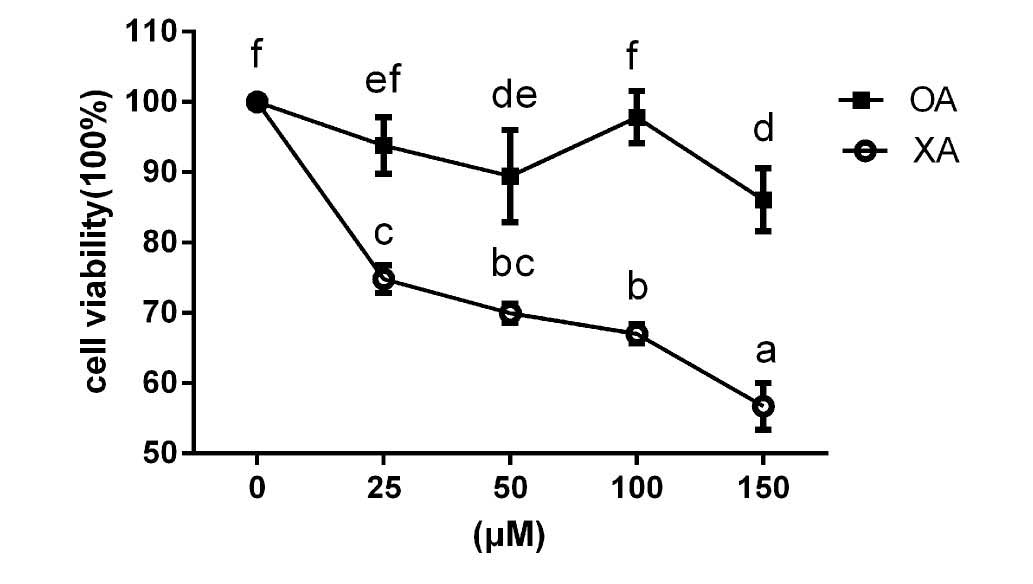
Inhibition of proliferation by
ximenynic acid
HepG2 cells were treated with different
concentrations (0–150 µM) of ximenynic acid or oleic acid for 72 h,
and the cell viability was measured by MTT assay. As shown in
Fig. 1, ximenynic acid
significantly inhibited the growth of HepG2 cells from 0 to 150 µM
in a dose-dependent manner compared with the control group (P=0.12,
25 µM; P=0.04, 50 µM; P=0.03, 100 µM; P=0.12, 150 µM), while oleic
acid could not inhibit HepG2 cell growth at every concentration
compared with the control group (Fig.
1).
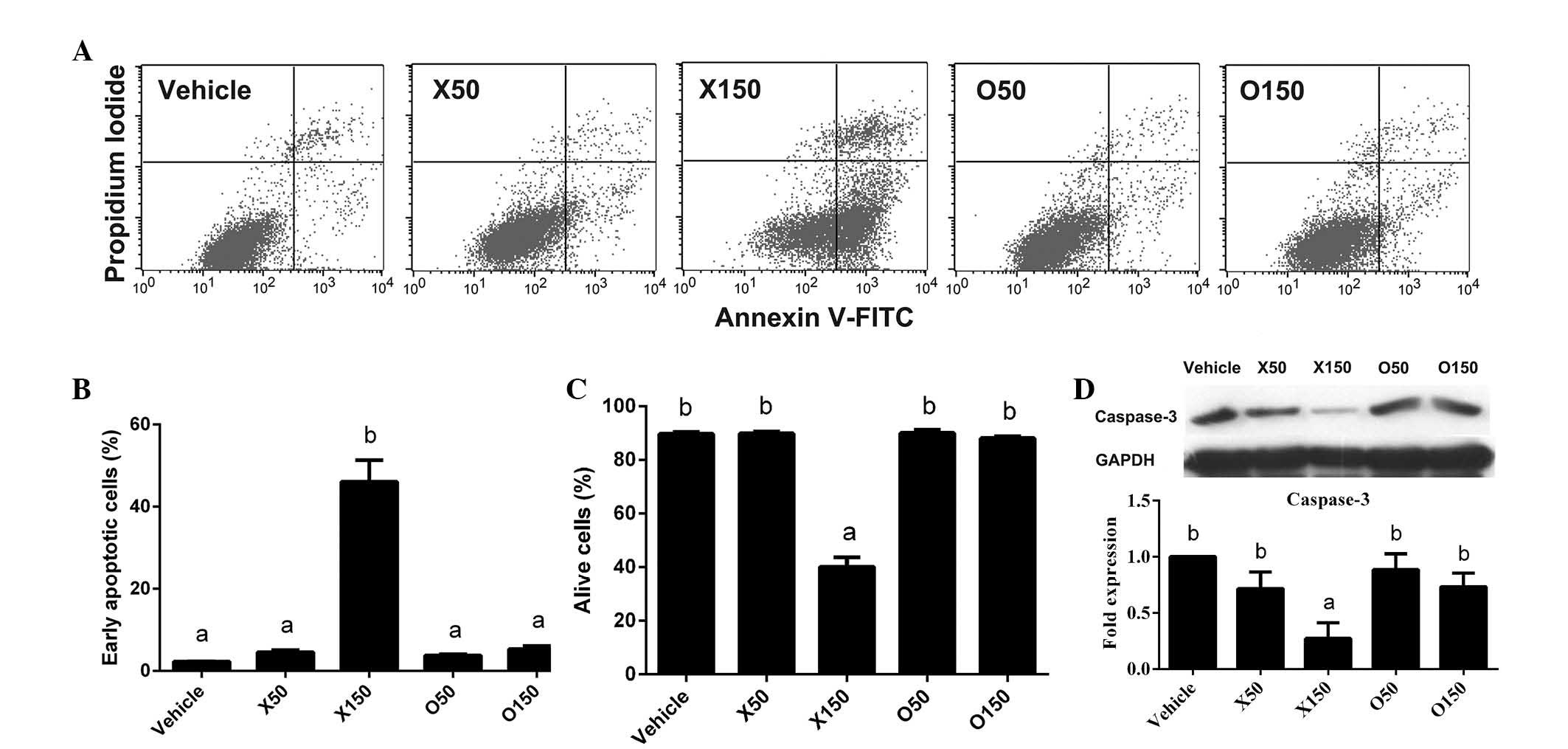
Induction of HepG2 apoptosis and
caspase-3 activation by ximenynic acid
Considering the anti-proliferation activity of
ximenynic acid demonstrated in Fig.
1, it was suspected that ximenynic acid may also lead to
apoptosis of HepG2 cells. Thus, the HepG2 cells were treated with
ximenynic acid or oleic acid (50 and 150 µM) for 72 h followed by
PI staining and flow cytometry analysis (Fig. 2A). It was observed that the
percentage of apoptotic cells was significantly increased by
ximenynic acid at a concentration of 150 µM in a dose-dependent
manner (P=0.018; Fig. 2B), and
accordingly the percentage of live cells was significantly reduced
at the same concentration (P=0.004; Fig. 2C) compared with the vehicle control
group. In the oleic acid group, the percentage of apoptotic cells
and living cells was not significantly different compared with the
control group (Fig. 2B and C). The
anti-cancer activity of 150 µM ximenynic acid was greater than that
of oleic acid.
The degradation of pro-caspase-3 protein occurs
during the process of cell apoptosis, which results in protein
cleavage into several fragments (20) and indirectly induces a decrease in
the pro-protein. The cleaved caspase-3 (17 KDa) was not detected in
the ximenynic acid group, however western blot analysis
demonstrated that the level of pro-caspase-3 (35 KDa) was
significantly decreased by 150 µM ximenynic acid compared with the
vehicle control (P=0.001, P<0.05; Fig. 2D). Together, flow cytometry and
western blot analyses indicated that ximenynic acid promotes
apoptosis of HepG2 cells in a dose-dependent manner, and was more
effective than oleic acid.
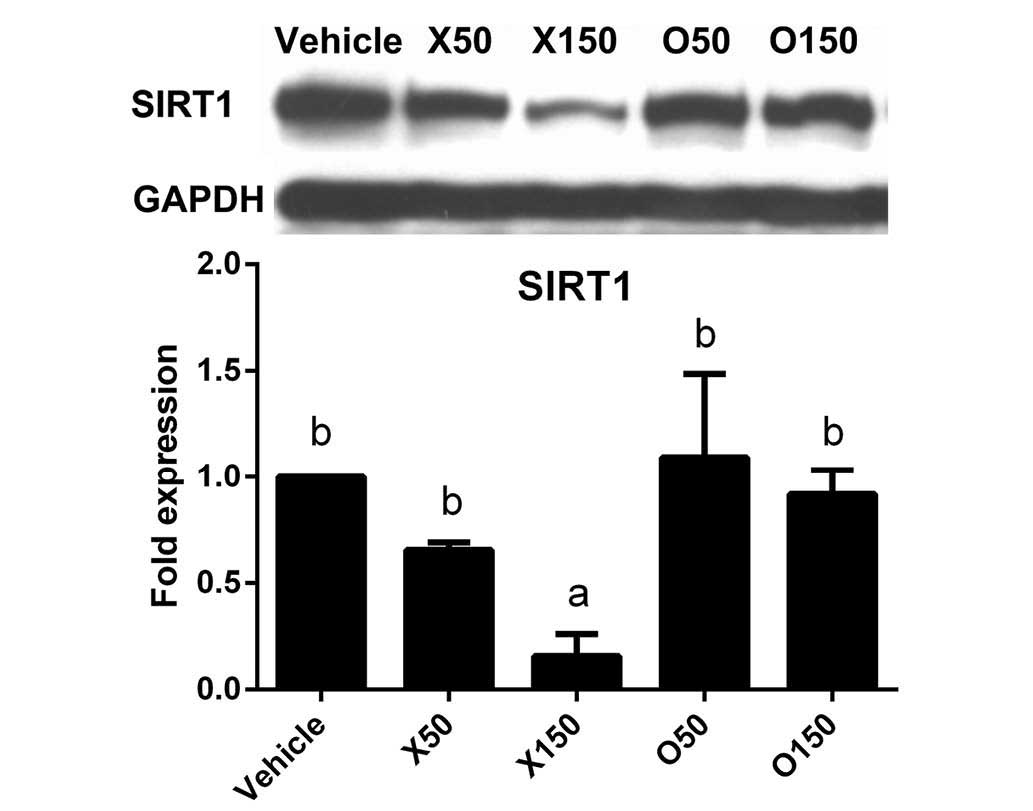
Effect of ximenynic acid on SIRT1
protein expression
The upstream signals that mediate the effect of
ximenynic acid on apoptosis remain unclear. SIRT1 is a
NAD-dependent deacetylase that inhibits apoptosis by downregulating
transcriptional activity of p53 (21). In addition, SIRT1 silencing induces
activation of caspase-3, indicating it is a downstream target of
SIRT1 (22). The current study
demonstrated that SIRT1 protein expression was significantly
reduced by 150 µM ximenynic acid compared with the vehicle control
group (P=0.013, 50 µM; P=0.019, 150 µM), whereas there was no
significant difference in the oleic acid groups compared with
vehicle (Fig. 3). These results
indicate that ximenynic acid-induced apoptosis is associated with
SIRT1 inhibition in HepG2 cells.
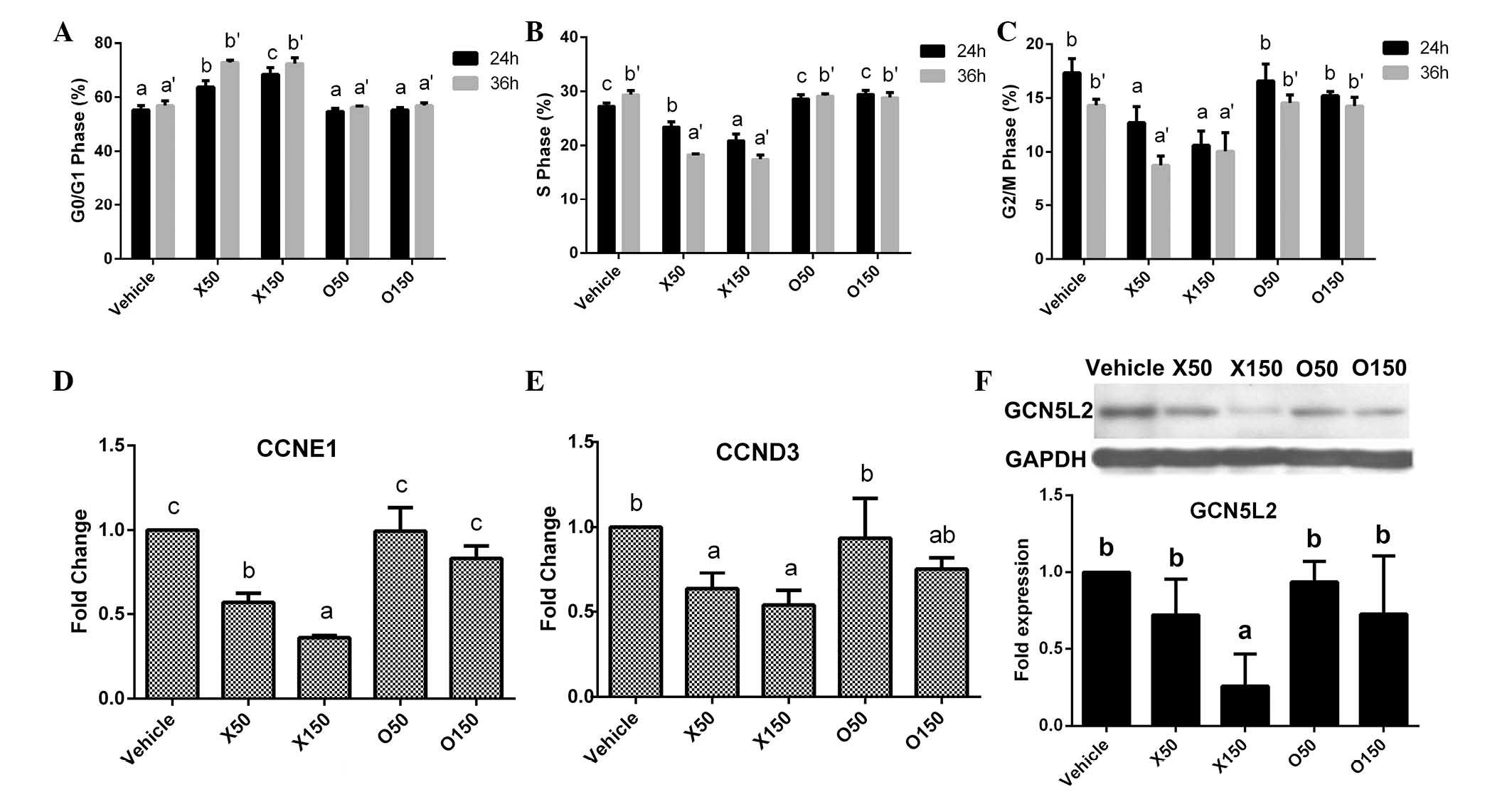
Cell cycle changes and expression
variation of cell cycle-associated genes and proteins in HepG2
cells
Cell cycle arrest in G1/S transition can induce
cancer cell apoptosis (16). The
current study investigated whether ximenynic acid affects the cell
cycle distribution. After treatment with ximenynic acid for 24 or
36 h, the cell cycle distributions of HepG2 cells were observably
changed (Fig. 4A-C). The
percentage of cells in G0/G1 phase was significantly increased in
the ximenynic acid groups (P=0.002, 24 h, 50 µM; P=0.001, 24 h, 150
µM; P=0.001, 36 h, 50 µM; P=0.001, 36 h, 150 µM; Fig. 4A), and significantly decreased in S
phase (P=0.002, 24 h, 50 µM; P=0.001, 24 h, 150 µM; P=0.001, 36 h,
50 µM; P=0.001, 36 h, 150 µM; Fig.
4B) and G2/M phase (P=0.014, 24 h, 50 µM; P=0.001, 24 h, 150
µM; P=0.001, 36 h, 50 µM; P=0.005, 36 h, 150 µM; Fig. 4C) compared with the vehicle control
group in a dose- and time-dependent manner. The structures of
ximenynic acid and oleic acid are similar, however, oleic acid did
not observably alter the cell cycle distributions of HepG2 cells at
50 or 150 µM (Fig. 4A-C). These
results indicate that ximenynic acid change the cell cycle
distribution of HepG2 cells depending on the dose and duration of
ximenynic acid treatment.
To understand whether ximenynic acid affects the
expression of cell cycle-associated genes in HepG2 cells, qPCR
analysis was performed to determine the expression variation of
cyclin genes, including cyclin E1 (CCNE1) and cyclin D3
(CCND3). After treatment for 36 h, the mRNA levels of
CCNE1 and CCND3 were significantly decreased in the50
and 150 µM ximenynic acid group compared with the vehicle control
group (CCNE1, P=0.001; CCND3, P=0.043; Fig. 4D and E), and its inhibitory effect
on CCND3 and CCNE1 was stronger than oleic acid.
GCN5L2 (also termed lysine acetyltransferase 2A) is
a typical histone acetyltransferase (HAT), which promotes cell
cycle progression by acetylation and deacetylation of histones
(23). The expression of GCN5L2
protein was significantly downregulated in the 150 µM ximenynic
acid group compared with the vehicle control group (P=0.026;
Fig. 4F).
Effect of ximenynic acid on
cyclooxygenase pathway
COX-1 and COX-2 mRNA and protein were demonstrated
to be expressed in the HepG2 cells (Fig. 5). After serum starvation for 24 h,
HepG2 cells were incubated with fatty acids for 72 h. Although
COX-2 protein expression was significantly increased in oleic acid
group at 50 µM (P=0.01, P<0.05), it was not changed in the
ximenynic acid group compared with the vehicle control group
(Fig. 5A). These results were
largely in keeping with the COX-2 gene expression of PTGS2
[prostaglandin-endoperoxide synthase 2 (PTGS2)] in HepG2 (Fig. 5C).
Unexpectedly, compared with COX-2, ximenynic acid
exerted a greater effect on COX-1, with PTGS1 mRNA (P=0.009,
50 µM; P=0.001, 150 µM; Fig. 5D)
and COX-1 protein (P=0.005, 50 µM; P=0.001, 150 µM; Fig. 5B) levels significantly decreased by
50 to 150 µM ximenynic acid compared with the vehicle control
group, whereas PTGS1 levels were significantly increased by
oleic acid treatment at the concentrations of 50 and 150 µM
(P=0.001, 50 µM; P=0.001, 150 µM; Fig.
5D). These results indicate that ximenynic acid selectively
inhibits COX-1 expression.
PGE2 is the product of arachidonic acid metabolism
by COXs (24). Prostaglandin E
receptor 2 (EP2) a prostaglandin E receptor 4 (EP4) are PGE2
receptors involved in various physiological and pathophysiological
processes (25). EP2 and
EP4 mRNAs were significantly downregulated in the 150 µM
ximenynic acid group compared with the vehicle control (EP2,
P=0.03; EP4, P=0.001), while they were unchanged by oleic
acid (Fig. 5E and F). It was
indicated that the inhibitory effect of ximenynic acid on
EP2 and EP4 mRNA levels was more effective than that
of oleic acid.
Effect of ximenynic acid on
angiogenesis pathways
Vascular endothelial growth factor (VEGF)-B and
VEGF-C are two members of the VEGF family that induce angiogenesis
and inhibit apoptosis (26). The
VEGF-B and VEGF-C mRNA levels were significantly
decreased in the 50 and 150 µM ximenynic acid groups compared with
the vehicle control group (P<0.05) and oleic acid (P<0.05)
groups (VEGF-B: P=0.001, 50 µM; P=0.001, 150 µM;
VEGF-C: P=0.049, 50 µM; P=0.044, 150 µM), whereas
VEGF-B was significantly upregulated in the oleic acid
(P<0.05) groups (Fig. 6A and
B).
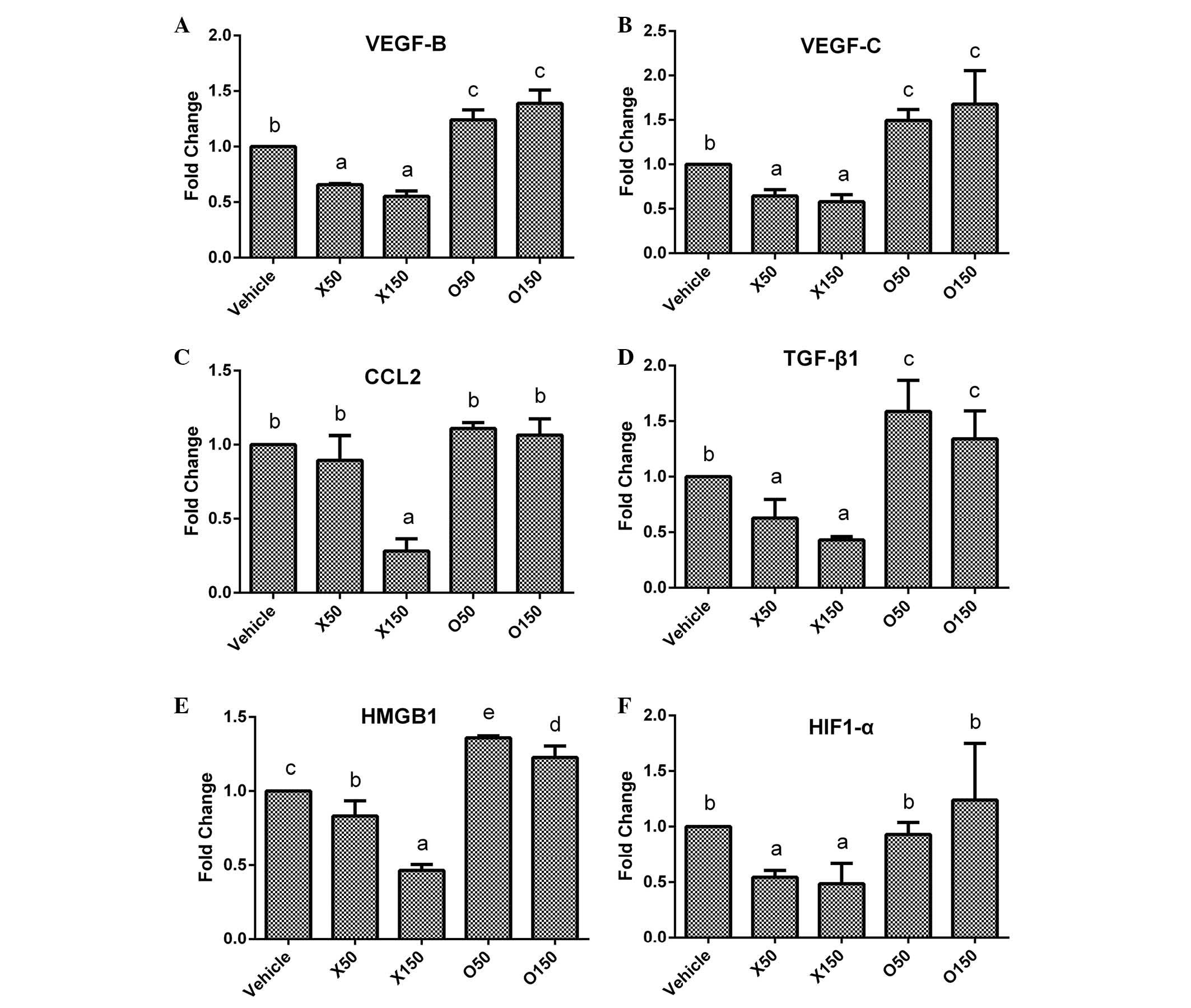 | Figure 6.Expression variations of
angiogenesis-associated genes in HepG2 cells following fatty acids
treatment. Following incubation with ximenynic acid or oleic acid
for 72 h, the expression of angiogenesis-associated genes including
(A) VEGF-B, (B) VEGF-C, (C) CCL2, (D)
TGF-β1, (E) HMGB1 and (F) HIF-1α were
analysed by reverse transcription-polymerase chain reaction. O50
and O150 groups were treated with oleic acid at 50 and 150 µM,
respectively. X50 and X150 groups were treated with ximenynic acid
at 50 and 150 µM, respectively. Data are presented as the mean ±
standard deviation; n=3. *P<0.05 vs. the vehicle group. VEGF,
vascular endothelial growth factor; CCL2, chemokine (C-C motif)
ligand 2; TGF-β1, transforming growth factor-β1;
HMGB1, high mobility group box 1; HIF-1α, hypoxia inducible
factor-1α. |
Numerous factors modulate angiogenesis in cancer.
The current study investigated the expression of several
angiogenesis-associated genes including chemokine (C-C motif)
ligand 2 (CCL2), Hypoxia inducible factor 1, α submit
(HIF-1α), high mobility group box 1 (HMGB1), and
transforming growth factor-β1 (TGF-β1). CCL2 is
recognized for its role in the inflammatory response and modulation
of angiogenesis (27). HMGB1, a
proinflammatory cytokine, is crucial for inflammation-associated
diseases and involved in cell apoptosis and angiogenesis (28). HIF-1α, an oxygen balance regulator,
regulates VEGF transcription and is involved in numerous cell
functions, including angiogenesis (29). TGF-β1, a multifunction factor for
cells, has also been previously demonstrated to induce angiogenesis
progression (30). In the present
study, these four mRNA levels were significantly inhibited by 150
µM ximenynic acid compared with the vehicle control group
(CCL2, P=0.016; TGF-β1, P=0.004; HMGB1,
P=0.007; HIF-1α, P=0.044; Fig.
6C-F). By contrast, oleic acid acid significantly promoted the
expression of HMGB1 genes at 50 µM (P=0.002), however did
not show significant effects on TGF-β1, CCL2 and
HIF-1α genes compared with the control group (Fig. 6C-F).
Discussion
Ximenynic acid is one of main components of
sandalwood seed fatty acids, and remains to be fully investigated.
Sandalwood is widely used in perfume, artwork, furniture and for
religious reasons, however, the sandalwood tree is a slow-growing
plant and requires decades or even centuries for full growth. For
the sustainable development of the sandalwood industry, it the
value of the sandalwood seed should be investigated.
Sandalwood seeds are rich in oil, and the main fatty
acids of the oil are oleic acid and ximenynic acid. The structure
of oleic acid is very similar to ximenynic acid, excluding a triple
bond. The anti-inflammatory and anti-cancer properties of oleic
acid are inferior compared with ximenynic acid, which suggests that
the conjugated enyne structure of ximenynic acid is crucial for the
medicinal properties. Thus, ximenynic acid may be the key
functional factor of sandalwood seed oil. In addition, numerous
acetylenic fatty acids exert important pharmacological functions,
including antibacterial (2),
anti-inflammatory (5) and
anti-cancer (10) activities.
Ximenynic acid has been demonstrated to be anti-bacterial (3) and anti-inflammatory (5), whereas the anti-cancer properties
remain unclear. In the current study MTT assay and flow cytometry
results demonstrated the anti-proliferative and apoptosis-promoting
activities of ximenynic acid on HepG2 cells. Western blotting and
qPCR analysis revealed the suppressive effect of ximenynic acid on
the expression of apoptosis- and angiogenesis-associated factors.
These suggest that the anti-cancer activity of ximenynic acid is
mediated by regulating cell cycle, apoptosis and angiogenesis
pathways.
Ximenynic acid induced G1/G0 phase arrest in HepG2
cells and inhibited the expression of CCND3 and CCNE1
genes, and GCN5L2 protein. G1/G0 phase is the critical stage for
DNA replication, therefore, arrest of cancer cells in G1/G0 phase
may contribute to inhibition of cancer cell proliferation (31). D type-cyclins (D1, D2, D3) are
important for cell cycle progression, inducing the transition from
G1 to S phase (32). It previously
demonstrated that downregulation of cyclin D3 was correlated with
cell cycle arrest and apoptosis (33). Cyclin E is the key factor of G1/S
transition, and overexpression of cyclin E protein can induce
tumorigenesis (34) and shorten
the cell cycle (35). In addition,
GCN5L2 was previously reported to promote G1/S transition and
promote the expression of cell cycle-associated factors, including
cyclin D3 (23) and cyclin E1
(36). The present study
demonstrated that the protein levels of GCN5L2 were reduced by
ximenynic acid. This suggests that the anti-cancer activity of
ximenynic acid is associated with histone acetylation via
downregulation of GCN5L2 that inhibits cyclin expression to arrest
cancer cells in G0/G1 phase.
Ximenynic acid induced HepG2 cell apoptosis and
inhibited SIRT1 expression. SIRT1 has been previously reported to
inhibit cellular senescence and suppress cell apoptosis (37). Overexpression of SIRT1 affects
histone deacetylation and inhibits deacetylation of certain tumor
repressor proteins, including p53 (38). The results of the present study
demonstrated that ximenynic acid downregulates the expression of
SIRT1 in a dose-dependent manner. The progression of cancer is
promoted by accumulation of epigenetic and genetic changes
(39). Acetylation and methylation
are post-translational histone modifications that modulate
transcriptional activity, DNA recombination and repair. The results
of the present study suggest that the anti-cancer activity of
ximenynic acid may be mediated through the deacetylation or
methylation of certain critical factors in tumor progression.
Ximenynic acid selectively inhibited COX-1
expression, however it exhibited no effect on COX-2. Typically,
ximenynic acid has been previously used for anti-inflammatory
treatment and decreases the activity of COXs (4,5). It
was previously reported that NSAIDs inhibit cancer cell growth by
suppressing COX-2 activity (40),
but selective inhibition of COX-1 may achieve the same effect
(11,12). As a typical example, aspirin, a
COX-1 relative inhibitor, was demonstrated to be beneficial for
inhibiting a wide variety of cancers (9). In addition, selective inhibition of
COX-1 expression induces apoptosis in certain types of cancer cells
(41). The present study
demonstrated that ximenynic acid did not alter the COX-2 level,
however had resulted in clear inhibition of COX-1 expression. COX-2
was increased following ximenynic acid treatment, however, the
expression of COX-1 was reduced. This indicates that ximenynic acid
is a selectively inhibits COX-1 expression. Ximenynic acid may
suppress angiogenesis of HepG2 cells through inhibition of COX-1
expression. Angiogenesis is involved in de novo generation
of blood vessels, and is considered crucial for tumor growth and
metastasis (42). Overexpression
of COX-1 has previously been demonstrated to promote the expression
of angiogenic growth factors (11). In the current study, ximenynic acid
suppressed COX-1 protein expression, and inhibited the gene
expression of TGF-β1, and HMGB1. As a
multifunctional cytokine, TGF-β1 has previously been revealed to
induce angiogenesis (30) and be
involved in the progression of angiogenesis induced by COX-1
(43) or HMGB1 (44). In addition, overexpression of HMGB1
has been observed to induce proliferation of cancer cells and
angiogenesis (28). Gene and
protein expression of HIF-1α is also affected by HMGB1 (45). This suggests that the anti-cancer
activity of ximenynic acid in the HepG2 cell line may be achieved
through inhibiting angiogenic factors.
The anti-cancer activity of ximenynic acid is
predominantly attributed to the inhibition of COX-1, which may be
associated with histone acetylation modification (46,47).
A previous report demonstrated that selective inhibition of COX-1
could suppress angiogenesis (14)
and induce G0/G1 phase arrest of the cell cycle due to suppression
of cyclin E, which is the key protein for G1/S transition (13). Furthermore, inhibition of COX-1 has
previously been observed to induce apoptosis and activation of
capspase-3 (48). Additionally,
ximenynic acid may affect the histone acetylation. The association
between COX-1, GCN5L2 and SIRT1 proteins remains unclear, however,
the latter two are involved in histone acetylation. In addition,
aspirin, a relative COX-1 inhibitor, has an enhanced
anti-carcinogenic activity when it was used in combination with a
histone deacetylase inhibitor (49). Taken together, the results of the
present study indicate that ximenynic acid may regulate the
acetylation of histones to exert anti-cancer and anti-inflammatory
activities.
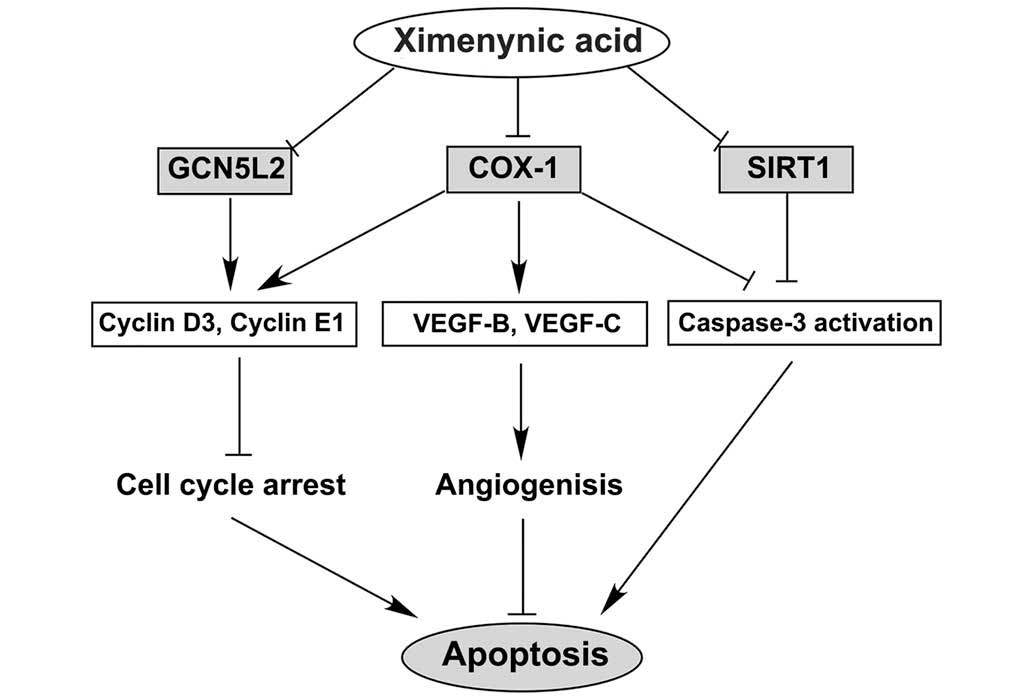
The anti-cancer activity of ximenynic acid may be
associated with the cell cycle, angiogenesis and cell apoptosis
pathways. The putative pathways of ximenynic acid are presented in
Fig. 7, and the anti-cancer
activity of ximenynic acid may be associated with the suppression
of the expression of COX-1, GCN5L2 and SIRT1 proteins, promotion of
cell cycle arrest in G0/G1 phase through suppression cell cycle
regulatory factors, blocking of angiogenesis by inhibiting the VEGF
family, and the acceleration of cell apoptosis via caspase-3
activation. In summary, ximenynic acid inhibited proliferation and
induced apoptosis of HepG2 cells, altered cell cycle distribution
and inhibited the expression of angiogenesis-associated
factors.
Acknowledgements
This work was supported by a grant from the National
Program on Key Basic Research Project of China (973 Program; grant
no. 2015CB553600).
References
|
1
|
Aitzetmuller K: Santalbic acid in the
plant kingdom. Plant Syst Evol. 298:1609–1617. 2012. View Article : Google Scholar
|
|
2
|
Shawar RM, Humble DJ, Van Dalfsen JM,
Stover CK, Hickey MJ, Steele S, Mitscher LA and Baker W: Rapid
screening of natural products for antimycobacterial activity by
using luciferase-expressing strains of Mycobacterium bovis BCG and
Mycobacterium intracellulare. Antimicrob Agents Chemother.
41:570–574. 1997.PubMed/NCBI
|
|
3
|
Jones GP, Rao KS, Tucker DJ, Richardson B,
Barnes A and Rivett DE: Antimicrobial activity of santalbic acid
from the oil of Santalum-Acuminatum (Quandong). Int J Pharmacogn.
33:120–123. 1995. View Article : Google Scholar
|
|
4
|
Croft KD, Beilin LJ and Ford GL:
Differential inhibition of thromboxane B2 and leukotriene B4
biosynthesis by two naturally occurring acetylenic fatty acids.
Biochim Biophys Acta. 921:621–624. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nugteren DH and Christ-Hazelhof E:
Naturally occurring conjugated octadecatrienoic acids are strong
inhibitors of prostaglandin biosynthesis. Prostaglandins.
33:403–417. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li G, Singh A, Liu Y, Sunderland B and Li
D: Comparative effects of sandalwood seed oil on fatty acid
profiles and inflammatory factors in rats. Lipids. 48:105–113.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Imig JD and Hammock BD: Soluble epoxide
hydrolase as a therapeutic target for cardiovascular diseases. Nat
Rev Drug Discov. 8:794–805. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang D and Dubois RN: Eicosanoids and
cancer. Nat Rev Cancer. 10:181–193. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rothwell PM, Price JF, Fowkes FG,
Zanchetti A, Roncaglioni MC, Tognoni G, Lee R, Belch JF, Wilson M,
Mehta Z and Meade TW: Short-term effects of daily aspirin on cancer
incidence, mortality, and non-vascular death: Analysis of the time
course of risks and benefits in 51 randomised controlled trials.
Lancet. 379:1602–1612. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dembitsky V: Anticancer activity of
natural and synthetic acetylenic lipids. Lipids. 41:883–924. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gupta RA, Tejada LV, Tong BJ, Das SK,
Morrow JD, Dey SK and DuBois RN: Cyclooxygenase-1 is overexpressed
and promotes angiogenic growth factor production in ovarian cancer.
Cancer Res. 63:906–911. 2003.PubMed/NCBI
|
|
12
|
Cho M, Kabir SM, Dong Y, Lee E, Rice VM,
Khabele D and Son DS: Aspirin blocks EGF-stimulated cell viability
in a COX-1 dependent manner in ovarian cancer cells. J Cancer.
4:671–678. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wu WK, Sung JJ, Wu YC, Li HT, Yu L, Li ZJ
and Cho CH: Inhibition of cyclooxygenase-1 lowers proliferation and
induces macroautophagy in colon cancer cells. Biochem Biophys Res
Commun. 382:79–84. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sano H, Noguchi T, Miyajima A, Hashimoto Y
and Miyachi H: Anti-angiogenic activity of basic-type, selective
cyclooxygenase (COX)-1 inhibitors. Bioorg Med Chem Lett.
16:3068–3072. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Carmeliet P and Jain RK: Angiogenesis in
cancer and other diseases. Nature. 407:249–257. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schwartz GK and Shah MA: Targeting the
cell cycle: A new approach to cancer therapy. J Clin Oncol.
23:9408–9421. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu YD, Longmore RB and Fox JED:
Separation and identification of ximenynic acid isomers in the seed
oil of Santalum spicatum RBr as their 4,4-dimethyloxazoline
derivatives. J Am Oil Chem Soc. 73:1729–1731. 1996. View Article : Google Scholar
|
|
18
|
Sun G, Zhou Y, Li H, Guo Y, Shan J, Xia M,
Li Y, Li S, Long D and Feng L: Over-expression of microRNA-494
up-regulates hypoxia-inducible factor-1 alpha expression via
PI3K/Akt pathway and protects against hypoxia-induced apoptosis. J
Biomed Sci. 20:1002013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Herrmann E, Call J, Hernandez-Lloreda MV,
Hare B and Tomasello M: Humans have evolved specialized skills of
social cognition: The cultural intelligence hypothesis. Science.
317:1360–1366. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Han Z, Hendrickson EA, Bremner TA and
Wyche JH: A sequential two-step mechanism for the production of the
mature p17:p12 form of caspase-3 in vitro. J Biol Chem.
272:13432–13436. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vaziri H, Dessain SK, Ng Eaton E, Imai SI,
Frye RA, Pandita TK, Guarente L and Weinberg RA: hSIR2 (SIRT1)
functions as an NAD-dependent p53 deacetylase. Cell. 107:149–159.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ford J, Jiang M and Milner J:
Cancer-specific functions of SIRT1 enable human epithelial cancer
cell growth and survival. Cancer Res. 65:10457–10463. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kikuchi H, Takami Y and Nakayama T: GCN5:
A supervisor in all-inclusive control of vertebrate cell cycle
progression through transcription regulation of various cell
cycle-related genes. Gene. 347:83–97. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Williams CS, Mann M and DuBois RN: The
role of cyclooxygenases in inflammation, cancer, and development.
Oncogene. 18:7908–7916. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Regan JW: EP2 and EP4 prostanoid receptor
signaling. Life Sci. 74:143–153. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Neufeld G, Cohen T, Gengrinovitch S and
Poltorak Z: Vascular endothelial growth factor (VEGF) and its
receptors. Faseb J. 13:9–22. 1999.PubMed/NCBI
|
|
27
|
Salcedo R, Ponce ML, Young HA, Wasserman
K, Ward JM, Kleinman HK, Oppenheim JJ and Murphy WJ: Human
endothelial cells express CCR2 and respond to MCP-1: Direct role of
MCP-1 in angiogenesis and tumor progression. Blood. 96:34–40.
2000.PubMed/NCBI
|
|
28
|
Volp K, Brezniceanu ML, Bösser S, Brabletz
T, Kirchner T, Göttel D, Joos S and Zörnig M: Increased expression
of high mobility group box 1 (HMGB1) is associated with an elevated
level of the antiapoptotic c-IAP2 protein in human colon
carcinomas. Gut. 55:234–242. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Carmeliet P, Dor Y, Herbert JM, Fukumura
D, Brusselmans K, Dewerchin M, Neeman M, Bono F, Abramovitch R,
Maxwell P, et al: Role of HIF-1alpha in hypoxia-mediated apoptosis,
cell proliferation and tumour angiogenesis. Nature. 394:485–490.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nakagawa T, Li JH, Garcia G, Mu W, Piek E,
Böttinger EP, Chen Y, Zhu HJ, Kang DH, Schreiner GF, et al:
TGF-beta induces proangiogenic and antiangiogenic factors via
parallel but distinct Smad pathways. Kidney Int. 66:605–613. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Grana X and Reddy EP: Cell cycle control
in mammalian cells: Role of cyclins, cyclin dependent kinases
(CDKs), growth suppressor genes and cyclin-dependent kinase
inhibitors (CKIs). Oncogene. 11:211–219. 1995.PubMed/NCBI
|
|
32
|
Hunter T and Pines J: Cyclins and cancer.
II: Cyclin D and CDK inhibitors come of age. Cell. 79:573–582.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fimognari C, Nüsse M, Berti F, Iori R,
Cantelli-Forti G and Hrelia P: Cyclin D3 and p53 mediate
sulforaphane-induced cell cycle delay and apoptosis in
non-transformed human T lymphocytes. Cell Mol Life Sci.
59:2004–2012. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Keyomarsi K, Tucker SL, Buchholz TA,
Callister M, Ding Y, Hortobagyi GN, Bedrosian I, Knickerbocker C,
Toyofuku W, Lowe M, et al: Cyclin E and survival in patients with
breast cancer. N Engl J Med. 347:1566–1575. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ohtsubo M, Theodoras AM, Schumacher J,
Roberts JM and Pagano M: Human cyclin E, a nuclear protein
essential for the G1-to-S phase transition. Mol Cell Biol.
15:2612–2624. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen L, Wei T, Si X, Wang Q, Li Y, Leng Y,
Deng A, Chen J, Wang G, Zhu S and Kang J: Lysine acetyltransferase
GCN5 potentiates the growth of non-small cell lung cancer via
promotion of E2F1, cyclin D1, and cyclin E1 expression. J Biol
Chem. 288:14510–14521. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Longo VD and Kennedy BK: Sirtuins in aging
and age-related disease. Cell. 126:257–268. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu T, Liu PY and Marshall GM: The
critical role of the class III histone deacetylase SIRT1 in cancer.
Cancer Res. 69:1702–1705. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Baylin SB and Herman JG: DNA
hypermethylation in tumorigenesis: Epigenetics joins genetics.
Trends Genet. 16:168–174. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Fosslien E: Molecular pathology of
cyclooxygenase-2 in neoplasia. Ann Clin Lab Sci. 30:3–21.
2000.PubMed/NCBI
|
|
41
|
Daikoku T, Wang D, Tranguch S, Morrow JD,
Orsulic S, DuBois RN and Dey SK: Cyclooxygenase-1 is a potential
target for prevention and treatment of ovarian epithelial cancer.
Cancer Res. 65:3735–3744. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wu Q, Wang H, Zhao X, Shi Y, Jin M, Wan B,
Xu H, Cheng Y, Ge H and Zhang Y: Identification of
G-protein-coupled receptor 120 as a tumor-promoting receptor that
induces angiogenesis and migration in human colorectal carcinoma.
Oncogene. 32:5541–5550. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yang DH, Hsu CF, Lin CY, Guo JY, Yu WC and
Chang VH: Kruppel-like factor 10 upregulates the expression of
cyclooxygenase 1 and further modulates angiogenesis in endothelial
cell and platelet aggregation in gene-deficient mice. Int J Biochem
Cell Biol. 45:419–428. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Pittet JF, Koh H, Fang X, Iles K,
Christiaans S, Anjun N, Wagener BM, Park DW, Zmijewski JW, Matthay
MA and Roux J: HMGB1 accelerates alveolar epithelial repair via an
IL-1β- and αvβ6 integrin-dependent activation of TGF-β1. PloS One.
8:e639072013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kim HY, Park SY, Lee SW, Lee HR, Lee WS,
Rhim BY, Hong KW and Kim CD: Inhibition of HMGB1-induced
angiogenesis by cilostazol via SIRT1 activation in synovial
fibroblasts from rheumatoid arthritis. PloS One. 9:e1047432014.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Taniura S, Kamitani H, Watanabe T and
Eling TE: Transcriptional regulation of cyclooxygenase-1 by histone
deacetylase inhibitors in normal human astrocyte cells. J Biol
Chem. 277:16823–16830. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Omura N, Griffith M, Vincent A, Li A, Hong
SM, Walter K, Borges M and Goggins M: Cyclooxygenase-deficient
pancreatic cancer cells utilize exogenous sources of
prostaglandins. Mol Cancer Res. 8:821–832. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kolaczkowska E, Koziol A, Plytycz B,
Arnold B and Opdenakker G: Altered apoptosis of inflammatory
neutrophils in MMP-9-deficient mice is due to lower expression and
activity of caspase-3. Immunol Lett. 126:73–82. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Son DS, Wilson AJ, Parl AK and Khabele D:
The effects of the histone deacetylase inhibitor romidepsin (FK228)
are enhanced by aspirin (ASA) in COX-1 positive ovarian cancer
cells through augmentation of p21. Cancer Biol Ther. 9:928–935.
2010. View Article : Google Scholar : PubMed/NCBI
|