Introduction
Colon cancer is the leading cause of
cancer-associated mortality worldwide, as >1,000,000 individuals
are diagnosed with colorectal cancer annually, resulting in a
mortality rate of ~715,000 in 2010, compared with 490,000 in 1990
(1,2). It is more common in developed
countries, in which >65% of cases are found (3). Colon cancer treatment options include
a combination of surgery, radiation therapy and chemotherapy;
however, options are limited for patients with advanced colon
cancer. Therefore, the identification of novel therapeutic targets
is important to develop a therapeutic strategy, which may improve
survival rates.
Fatty acid synthase (FASN) is a multifunctional
enzyme, which catalyzes fatty acid synthesis from acetyl-CoA,
malonyl-CoA and nicotinamide adenine dinucleotide phosphate (NADPH)
as a cofactor. FASN is the key enzyme for de novo long-chain
fatty acid biosynthesis. In normal human tissues or cells, FASN is
downregulated following the ingestion of a sufficient level of
dietary fatty acids. However, several solid tumor and cell lines
derived from these tumors overexpress FASN (4). The different expression levels of
FASN between cancer and normal tissues suggest that FASN may be a
potential target for cancer therapy (5). FASN relies on enzymatic activity for
survival and proliferation in various types of human cancer
(6). The pharmacological or
genetic inhibition of FASN induces growth arrest and apoptosis in
tumor cells (7–9).
Emodin (1,3,8-trihydroxy-6-methylanthraquinone) is
an anthraquinone found in certain plants and has been evaluated for
its antiproliferative and apoptotic activities in various cancer
cell lines, including breast (10), liver (11), lung (12), prostate (13) and cervical cancer (14), leukemia (15) and colon cancer (16). Emodin has an anticancer effect
based on the suppression of migration, invasion and angiogenesis
(17,18). The underlying mechanism of the
anticancer activities of emodin includes generating reactive oxygen
species (12) and inhibiting the
expression of casein kinase II (19), protein kinase C (20), extracellular-signal regulated
kinase (ERK)1/2 (21), vascular
endothelial growth factor (VEGF) receptor phosphorylation (22) and human epidermal growth factor 2
(HER2)/neu tyrosine kinase (23).
However, the function of emodin in the FASN-induced toxicity of
human colon cancer cells remains to be elucidated.
The present study investigated the effects of FASN
on intracellular fatty acid biosynthesis in emodin-induced
cytotoxicity. The findings suggested that emodin may be a novel
FASN inhibitor and may assist in formulating a therapeutic strategy
for colon cancer.
Materials and methods
Materials
Fetal bovine serum (FBS), Dulbecco's modified
Eagle's medium (DMEM), RPMI-1640, and penicillin/streptomycin were
obtained from HyClone™; GE Healthcare Life Sciences (South Logan,
UT, USA). Trypsin-EDTA was from Gibco; Thermo Fisher Scientific,
Inc. (Waltham, MA, USA). Antibodies targeting FASN (catalog no.
3180), caspase 3 (catalog no. 9662), caspase 7 (catalog no. 9494),
caspase 9 (catalog no. 9502), ubiquitin (catalog no. 3936),
microtubule-associated protein 1A/1B-light chain 3 (LC3) (catalog
no. 4108), Akt (catalog no. 9272), phosphorylated (p) Akt (Ser473)
(catalog no. 9271), phosphatidylinositol 3-kinase (PI3K) (catalog
no. 4292), pPI3K (p85Tyr458/p55Tyr199) (catalog no. 4228), ERK1/2
(catalog no. 4695), pERK1/2 (Thr202/Tyr204) (catalog no. 9101), and
β-actin (catalog no. 4967) were purchased from Cell Signaling
Technology, Inc. (Danvers, MA, USA), and anti-GAPDH (catalog no.
SC-25778) was from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA,
USA). Horseradish peroxidase-conjugated anti-rabbit antibody
(catalog no. 554021) was from Transduction Lab (Lexington, KY,
USA). Super Signal® West Pico Chemiluminescent substrate
was purchased from Pierce; Thermo Fisher Scientific, Inc. The Cell
Counting Kit-8 (CCK8) was from Dojindo Molecular Technologies, Inc.
(Kumamoto, Japan). TRIzol reagent was from Invitrogen™; Thermo
Fisher Scientific, Inc. The Free Fatty Acid Quantification
Colorimetric kit was purchased from BioVision, Inc. (Milpitas, CA,
USA). The EzWay Annexin V-FITC Apoptosis Detection kit was obtained
from KomaBiotech, Inc. (Seoul, Korea). AccuPower®
CycleScript RT PreMix(dT20) was purchased from Bioneer Corporation
(Daejeon, Korea). Emodin, cerulenin, cycloheximide (CHX), NADPH,
palmitic acid, acetyl-CoA, malonyl-CoA, DMSO and other reagents
were purchased from Sigma-Aldrich; Merck Millipore (Darmstadt,
Germany).
Cell culture
The HCT116 and SW480 human colon cancer cell lines
were obtained from American Type Culture Collection (Rockville, MD,
USA), and cultured in RPMI-1640 containing 10% FBS (v/v) and
penicillin (100 U/ml)/streptomycin (100 µg/ml). The SNU-C2A and
SNU-C5 cell lines were purchased from the Korean Cell Line Bank
(Seoul, Korea) and were grown in DMEM supplemented with 10% FBS.
The cells were maintained at 37°C in a humidified atmosphere of 95%
air and 5% CO2.
Cell viability assay
The cells were seeded at a density of 5×103 cells/ml
in 96-well microplates and allowed to attach for 24 h. Following
treatment of emodin (10–50 µM) and/or cerulenin (100 µM) for 6–24
h, cell cytotoxicity and/or proliferation were assessed using CCK8.
Briefly, water-soluble tetrazolium salt,
WST-8[2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium,
monosodium salt], produces an orange water-soluble product,
formazan. The quantity of formazan dye generated by dehydrogenases
in the cells is directly proportional to the number of living
cells. CCK8 (10 µl) was added to each well and incubated for 3 h at
37°C, following which cell proliferation and cytotoxicity were
assessed by measuring the absorbance at 450 nm using a microplate
reader. Three replicate wells were used for each experimental
condition.
Western blot analysis
The cells were harvested using Trypsin-EDTA, washed
twice with cold phosphate-buffered saline (PBS), lysed with lysis
buffer containing 10 mM Tris (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1%
Triton X-100, 0.5% NP-40, 1 mM PI, 1 mM DTT and 1 mM PMSF, and
placed on ice for 1 h with occasional vortexing. Centrifugation
followed at 13,000 × g for 10 min at 4°C to collect the
supernatant. A Pierce BCA Protein Assay kit (Pierce; Thermo Fisher
Scientific Inc.) was used to determine the protein concentration.
The cell lysate (50 µg) was subjected to 6 or 10%
SDS-polyacrylamide gel electrophoresis and transferred onto a
polyvinylidene difluoride membrane in Tris-Glycine buffer (25 mM
Tris, 200 mM Glycine and 20% methanol) for 2 h (60 V). The blots
were blocked with 5% skim milk in PBS containing 0.05% Tween-20 for
1 h at 25°C and were then incubated with primary antibodies
(1:1,000) overnight at 4°C, followed by incubation with anti-rabbit
horseradish peroxidase-conjugated IgG (1:3,000) for 2 h at room
temperature and visualized with enhanced chemiluminescence.
RNA isolation and reverse
transcription-polymerase chain reaction (RT-PCR) analysis
Following drug treatment, the cells were subjected
to total RNA isolation using TRIzol, according to the
manufacturer's protocol. The RNA concentrations were determined by
measuring the absorption at 260 nm in a spectrophotometer. Aliquots
of 1 µg total RNA from each sample were reverse transcribed into
cDNA in a total volume of 20 ul using AccuPower®
CycleScript RT PreMix (dT20) according to the manufacturer's
protocol. The PCR primers (10 pM) used in the present study were as
follows: FASN, forward 5′-CTTGCAGGAGTTCTGGGACA and reverse
5′-CCGTCCACGATGGCTTCATA; GAPDH, forward 5′-TAGACGGGAAGCTCACTGGC-3′
and reverse 5′-AGGTCCACCACCCTGTTGCT-3′. The PCR reactions were
subjected to the following amplification conditions: denaturation
at 94°C for 30 sec, annealing at 60°C for 30 sec and extension at
68°C for 60 sec. FASN and GAPDH were incubated for 40 cycles. The
PCR products (10 µl) were separated on a 1% agarose gel and
detected using ethidium bromide staining.
FASN activity assay
Following exposure to emodin and/or cerulenin, cells
were harvested and suspended with cold PBS. The cells were
sonicated at 4°C and centrifuged at 15,000 × g for 30 min to obtain
particle-free supernatants. FASN activity was determined
spectrophotometrically by measuring the decrease of absorption at
340 nm due to the oxidation of NADPH, as previously described
(24). Particle-free supernatant
(20 µl), 25 mM
KH2PO4-K2HPO4 buffer,
0.25 mM EDTA, 0.25 mM dithiothreitol, 30 µM acetyl-CoA and 350 µM
NADPH (pH 7.0) in a total volume of 200 µl were monitored at 340 nm
for 5 min to measure background NADPH oxidation. Following the
addition of 100 µM of malonyl-CoA, the reaction was assayed for an
additional 3 min to determine FASN-dependent oxidation of NADPH at
340 nm again.
Free fatty acid quantification
assay
Following treatment of cells with emodin and/or
cerulenin at the corresponding concentration and for the indicated
durations, the cells were harvested and washed twice with cold PBS.
The levels of intracellular free fatty acids were measured in the
cells using a Free Fatty Acid Quantification Colorimetric kit
according to the manufacturer's protocol. The samples were measured
against a standard of varying concentrations of palmitic acid
(provided by the kit) and the optical density was measured at 570
nm in a 96-well microplate reader (VersaMax ELISA microplate
reader; VersaMax; Molecular Devices, Sunnyvale, CA, USA). The
levels of free fatty acid in the samples were calculated using the
slope of the standard curve, with the concentration expressed as
nmol/µl.
Annexin V/propidium iodide
staining
The cells were cultured at a 106 density and treated
with emodin and/or or cerulenin for 24 h. The cells were
centrifuged and washed three times with PBS. The supernatant was
discarded and resuspended in 0.5 ml of cold PBS. The cells were
processed and labeled according to the EzWay Annexin V-FITC
Apoptosis Detection kit used for this assay. The labeled cells were
analyzed in a flow cytometer (BD FACSCanto™ II; BD Biosciences,
Franklin Lakes, NJ, USA).
Flow cytometric analysis
The cells (1×105 cells/ml) were suspended in 300 µl
PBS to which 700 µl ethanol was added. The cells were incubated at
4°C for 1 h, washed with PBS and suspended in 250 µl of 1.12%
sodium citrate buffer (pH 8.4) together with 12.5 µg RNase.
Incubation was continued at 37°C for 30 min. The cellular DNA was
stained by applying 250 µl propidium iodide (50 µg/ml) for 30 min
at room temperature. The stained cells were analyzed by fluorescent
activated cell sorting on the BD FACSCanto™ II flow cytometer using
BD FACSDiva™ Software v. 6. 1. 3 (BD Biosciences) to determine the
percentage of apoptotic cells.
Results
Emodin inhibits cell proliferation in
human colon cancer cell lines
Several colon cancer cell lines (HCT116, SW480,
SNU-C2A and SNU-C5) were treated with emodin for 24 h to determine
its effect on cell proliferation (Fig.
1A). Emodin exerted a significant dose-dependent
antiproliferative effect in the HCT116 cell line (30.4 and 24.2%
reduction at concentrations of 25 and 50 µM, respectively; Fig. 1A). Among the cell lines, HCT116
cells expressed the highest protein level of FASN, whereas SW480,
SNU-C2A and SNU-C5 cells expressed lower protein levels of FASN
(Fig. 1B). The results of the
western blot analysis showed that emodin downregulated the protein
expression of FASN, particularly in the HCT-116 cells, moderately
downregulated expression in the SNU-C5 cells, and had no effect in
the SW480 or SNU-C2A cells (Fig.
1C). Cell proliferation decreased and the expression of FASN
was downregulated in SW480 cells following exposure to emodin for
48 h (data not shown). This result showed that emodin-induced cell
death may be associated with the overexpression of FASN.
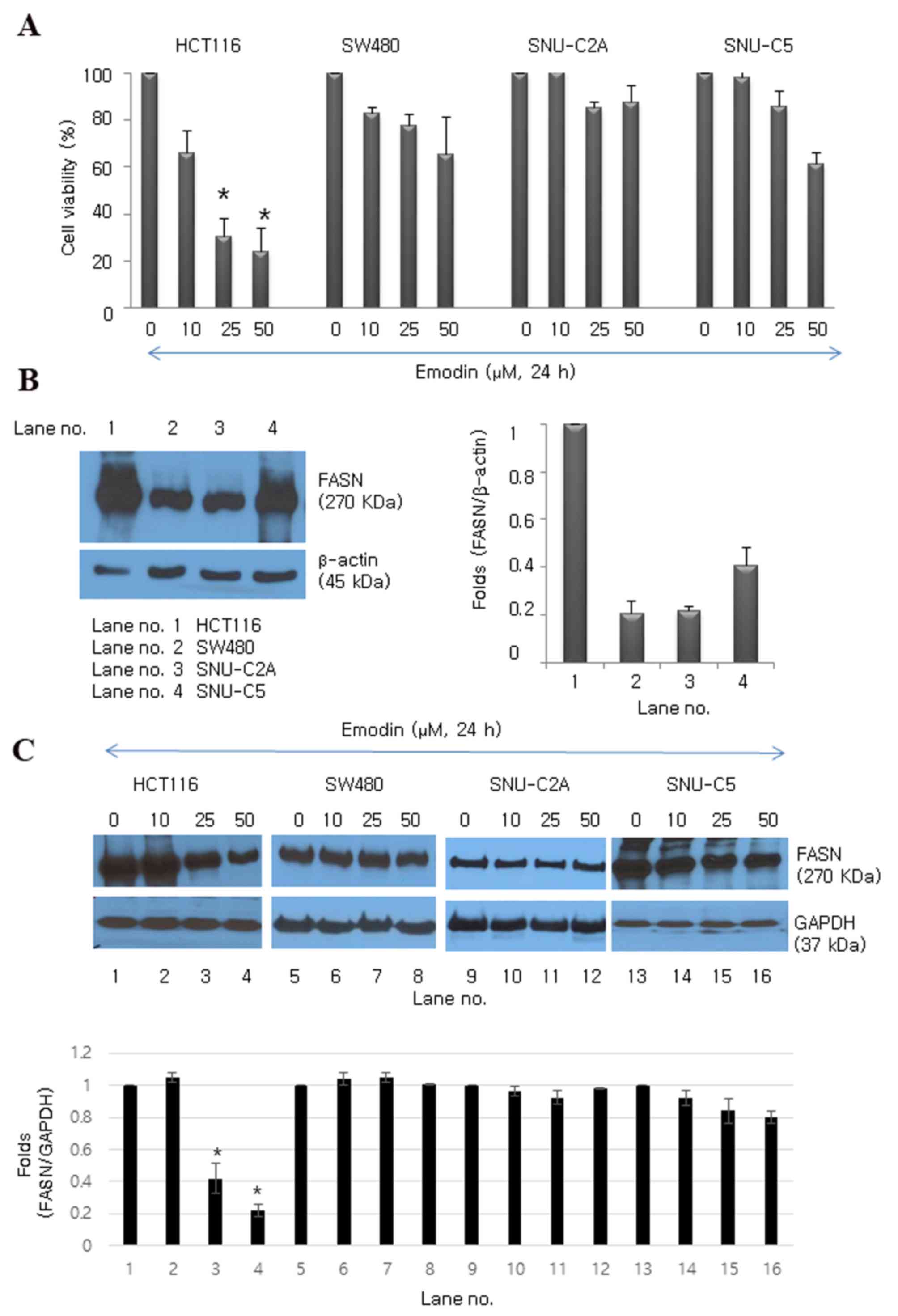 | Figure 1.Emodin inhibits proliferation of
human colon cancer cell lines. (A) Emodin inhibited growth of
HCT116, SW480, SNU-C2A and SNU-C5 human colon cancer cell lines.
Data are presented as the mean ± standard deviation of triplicate
determinations. *P<0.05, vs. control (0 µM). (B) Protein
expression levels of FASN in HCT116, SW480, SNU-C2A and SNU-C5
human colon cancer cells. β-actin (45 kDa) was used as the loading
control. (C) Emodin-induced protein expression levels of FASN at
various concentrations (0, 10, 25 and 50 µM) in HCT116, SW480,
SNU-C2A and SNU-C5 human colon cancer cell lines. GAPDH was used as
the loading control. *P<0.05 vs. control. FASN, fatty acid
synthase; GAPDH, glyceraldehyde 3-phosphate dehydrogenase. |
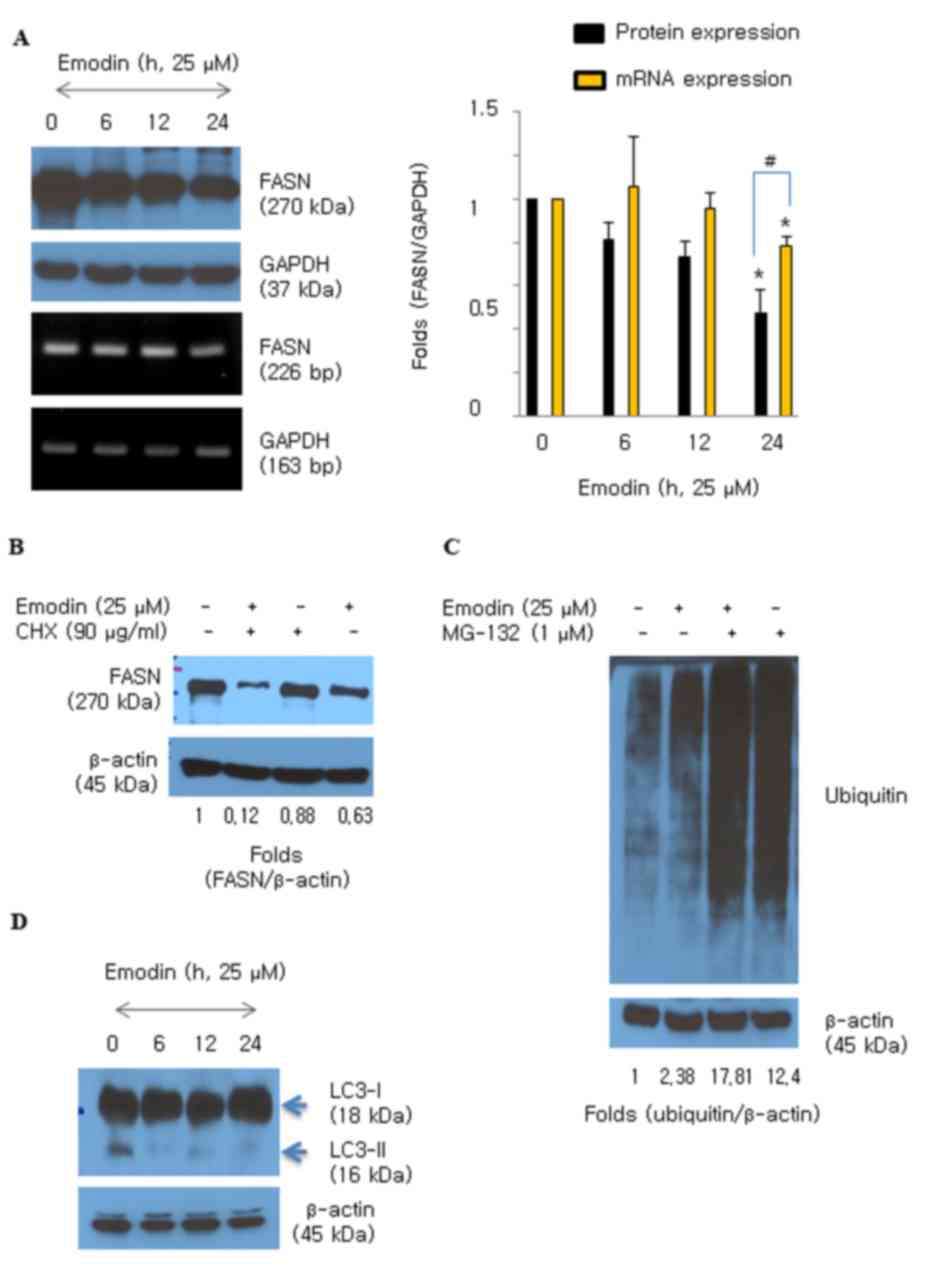
Emodin suppresses the protein level of
FASN in HCT116 cells
Emodin (25 and 50 µM) significantly downregulated
the protein expression of FASN in HCT116 cells (Fig. 1C). The HCT116 cells were then
treated with emodin and the expression of FASN was measured over
time (0–24 h). The protein levels of FASN decreased following
exposure to emodin for 6 h and were significantly decreased at 24 h
(Fig. 2A). The mRNA level of FASN
also decreased following emodin treatment; however, the effect on
protein was more marked, compared with that on mRNA (Fig. 2A). CHX, an inhibitor of de
novo protein synthesis, was added during emodin (25 µM)
exposure to examine whether emodin regulates degradation of the
FASN protein in HCT116 cells. Degradation of the FASN protein
increased with an additive effect in the presence of emodin and CHX
(Fig. 2B). Protein degradation is
usually triggered by ubiquitin moieties attaching to protein.
Therefore, the present study examined whether emodin induced
protein ubiquitination in the HCT116 cells. The results showed that
emodin induced the accumulation of ubiquitinated proteins (Fig. 2C), and MG-132 was used as a potent
proteasome inhibitor. Whether emodin stimulated lysosomal activity
was also examined. Emodin did not convert LC3-I to the smaller
form, LC3-II, which is an autophagy marker (Fig. 2D). This result suggested that
emodin induced the degradation of FASN protein caused by increased
protein ubiquitination activity.
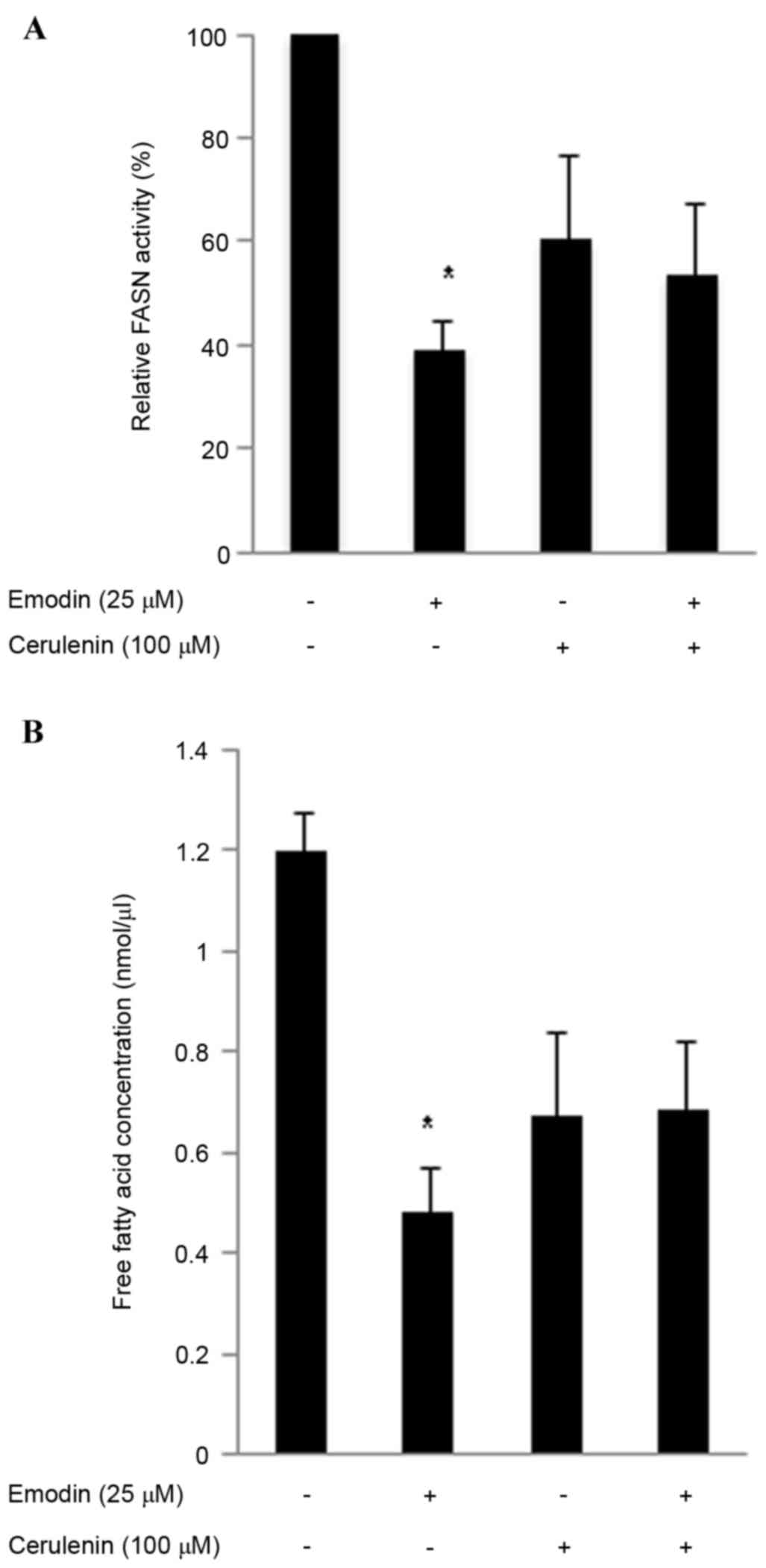
Emodin inhibits intracellular FASN
activity and intracellular free fatty acids
A high expression level of FASN is a molecular
change associated with colon cancer. In the present study, the
cells were incubated with 25 µM emodin and/or the FASN inhibitor
cerulenin (100 µM) for 24 h to evaluate cellular FASN activity.
Emodin reduced cellular FASN activity in the HCT116 cells to 39.2%,
cerulenin reduced FASN activity to 60.4% and the two in combination
reduced FASN activity to 53.8% (Fig.
3A). The overexpression of FASN in several types of cancer
markedly induces de novo lipogenesis, including
phospholipids, which are necessary for de novo synthesis of
the cell membrane (25). Thus,
inhibiting FASN can reduce the levels of phospholipids and free
fatty acids required (26). The
present study measured free fatty acids using a Free Fatty Acid
Quantification lit, as the primary function of FASN is to catalyze
long-chain fatty acid biosynthesis. Emodin treatment reduced the
level of intracellular free fatty acids (Fig. 3B). The emodin-induced (25 µM) FASN
inhibitory activities were more marked, compared with those of
cerulenin (100 µM) in HCT116 cells. These results indicated that
emodin inhibited FASN function.
Combined treatment with emodin and
cerulenin has an additive effect on cell growth and apoptosis
To assess whether the FASN inhibitor, cerulenin,
enhances the emodin-induced expression of FASN in HCT116 cells, the
cells were incubated with emodin (25 µM) and/or cerulenin (100 µM)
for 24 h. The combined treatment of emodin and cerulenin decreased
the protein expression of FASN, compared with either treatment
alone (Fig. 4A). To determine
whether cerulenin enhanced emodin-induced anticancer effects in
HCT116 cells, the cells were incubated with different
concentrations of emodin (10 and 25 µM) for various durations (6,
12 and 24 h), with or without cerulenin (100 µM), and performed a
CCK8 assay. Emodin exhibited cytotoxic effects in a dose-and
time-dependent manner. The combination of emodin and cerulenin
induced higher antiproliferative effects, compared with those of
emodin or cerulenin alone (Fig. 4B and
C). The induction of apoptosis by emodin and/or cerulenin was
concordant with the cell viability results. Apoptosis was
determined using Annexin V/propidium iodide double staining
(Fig. 4D) and caspase cleavage
(Fig. 4E) following exposure of
emodin (25 µM) and/or cerulenin (100 µM) for 24 h. The apoptotic
effect of emodin and cerulenin in combination was higher, compared
with apotosis induced by emodin or cerulenin alone. These results
suggested that the emodin-induced inhibition of FASN enhanced
antiproliferation and apoptosis.
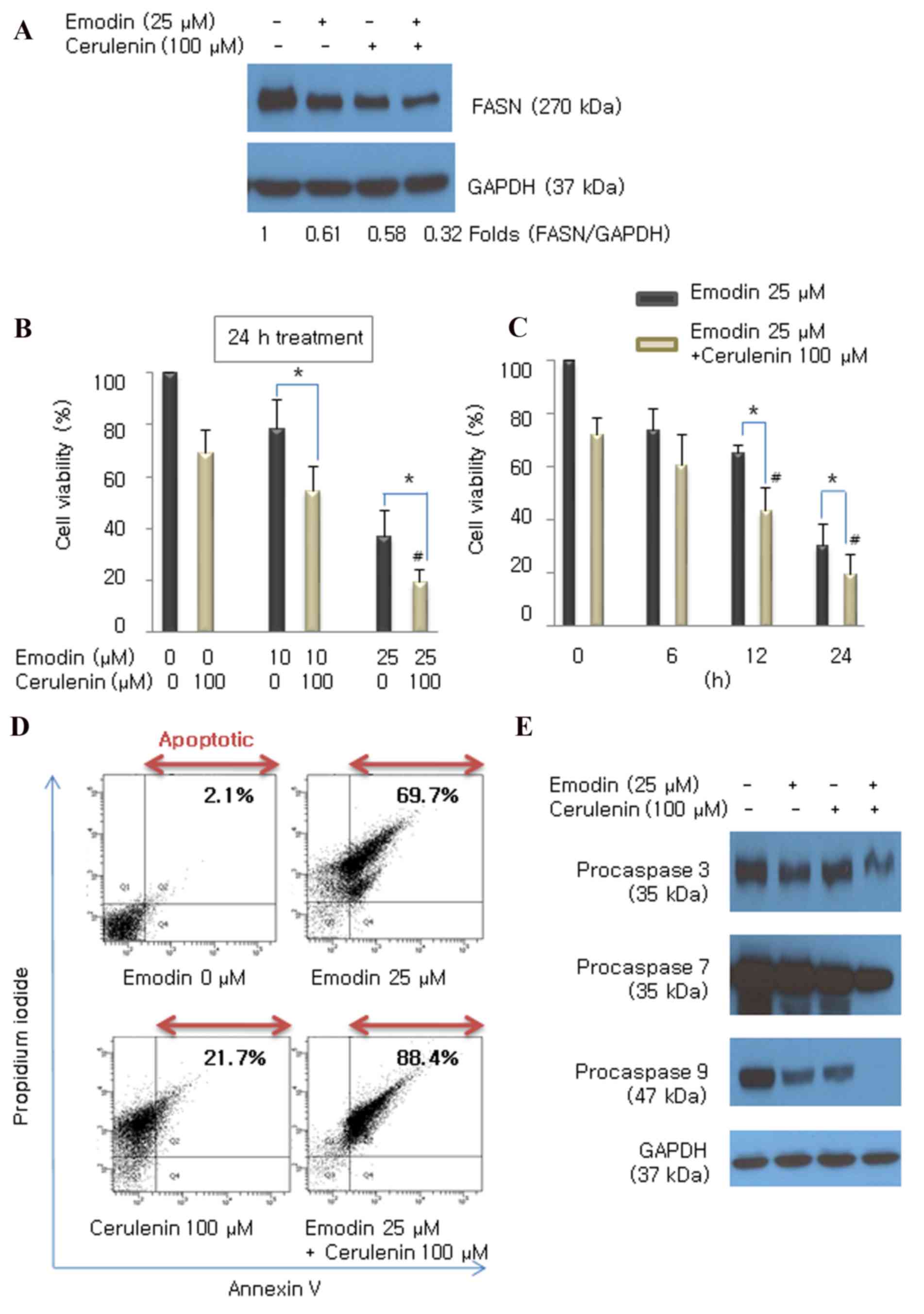 | Figure 4.Emodin combined with the FASN
inhibitor cerulenin enhances antiproliferation and apoptosis,
compared with the effect of either drug alone. (A) Expression of
FASN was measured using western blot analysis following treatment
with emodin (25 µM), cerulenin (100 µM) or the two in combination.
Cells were incubated with (B) 10 and 25 µM emodin for (C) 6, 12 and
24 h, with or without 100 µM cerulenin. Cell viability was
determined using a Cell Counting Kit 8 assay. Values are presented
as the mean ± standard deviation of triplicate determinations.
*P<0.05 vs. emodin single treatment. #P<0.05, vs.
cerulenin single treatment. (D) Following treatment, the cells were
double stained with Annexin V/propidium iodide and analyzed by flow
cytometry. The gate setting distinguished between living (bottom
left), necrotic (top left), early apoptotic (bottom right) and late
apoptotic (top right) cells. (E) Cleavage of caspase 3, 7 and 9 was
determined using western blot analysis following treatment with
emodin (25 µM), cerulenin (100 µM), and the two combined for 24 h.
GAPDH was used as the loading control. FASN, fatty acid synthase;
GAPDH, glyceraldehyde 3-phosphate dehydrogenase. |
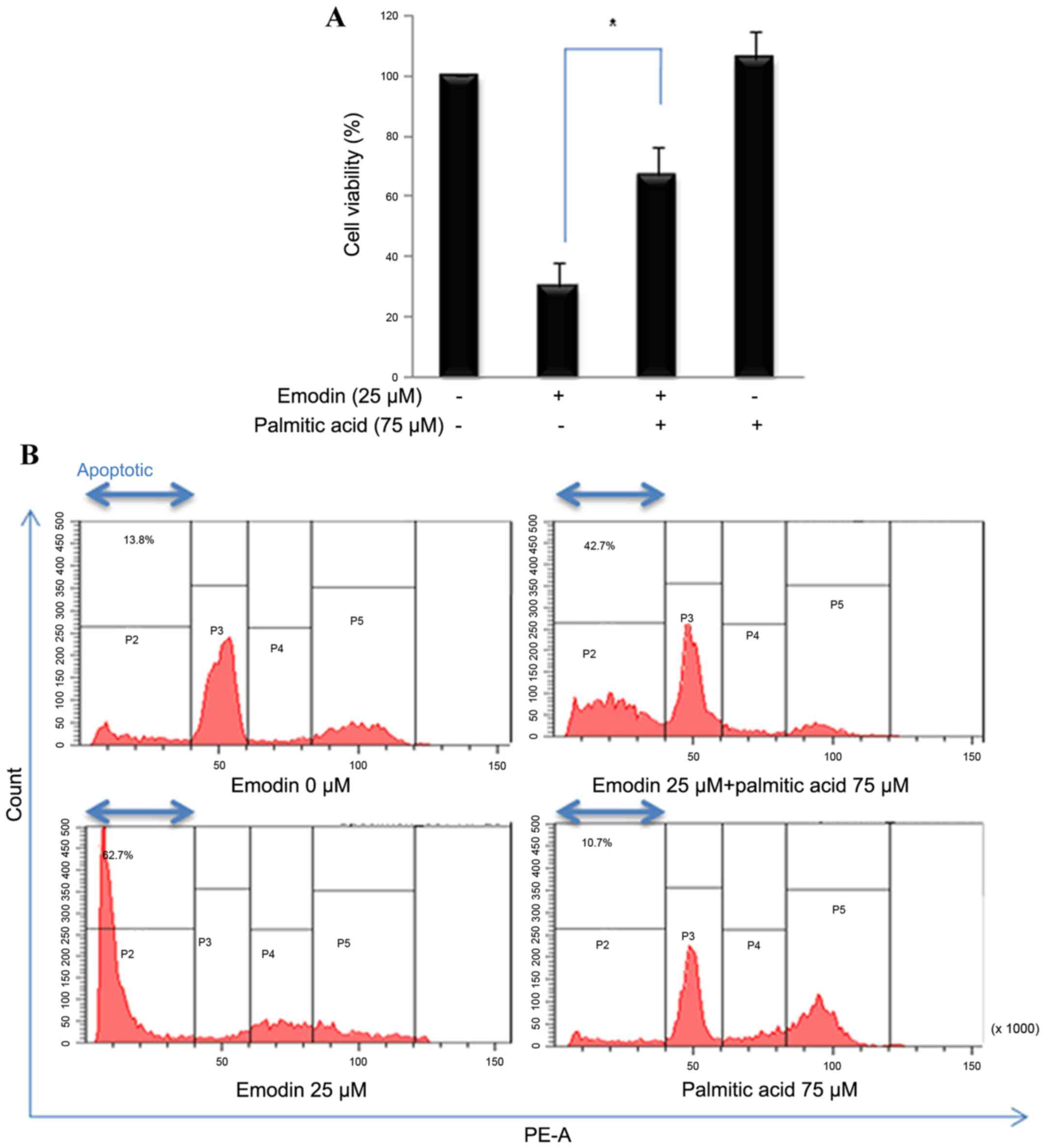
Palmitate rescues emodin-induced
apoptosis and viability
In the present study, cells were also treated with
emodin and palmitate (75 µM), the end product of the FASN reaction,
to determine whether the emodin-induced cell death activities were
associated with inhibiting FASN. In the presence of palmitate,
emodin-induced decreased cell viability was increased (Fig. 5A). Propidium iodide staining was
used to measure the number of apoptotic cells by flow cytometry.
Addition exogenous palmitate reduced the level of emodin-induced
apoptosis (Fig. 5B). These results
suggested that emodin-stimulated cytotoxicity was regulated by
fatty acid synthesis.
Emodin alters PI3K/Akt and ERK1/2
phosphorylation by inhibiting FASN
The PI3K/Akt and MAPK/ERK1/2 cascades are involved
in cell proliferation, survival and apoptosis associated with FASN
regulation in cancer (27–29). The present study examined the
effect of inhibiting FASN by emodin on PI3K/Akt and ERK1/2
phosphorylation. Inhibiting FASN caused a time-dependent decrease
in PI3K/Akt phosphorylation and increase in ERK1/2 phosphorylation
in the HCT116 cells (Fig. 6A). The
combined treatment with cerulenin enhanced these activities
(Fig. 6A). LY294002 (PI3K
inhibitor) or PD98059 (ERK inhibitor) were added with the emodin to
determine whether these two pathways regulated the emodin-induced
inhibition of FASN. The inhibitors altered the phosphorylation of
PI3K/Akt and ERK modulated by emodin (Fig. 6B and C). These data indicated that
the PI3K/Akt and ERK1/2 signaling pathways regulated the reduced
expression of FASN induced by emodin.
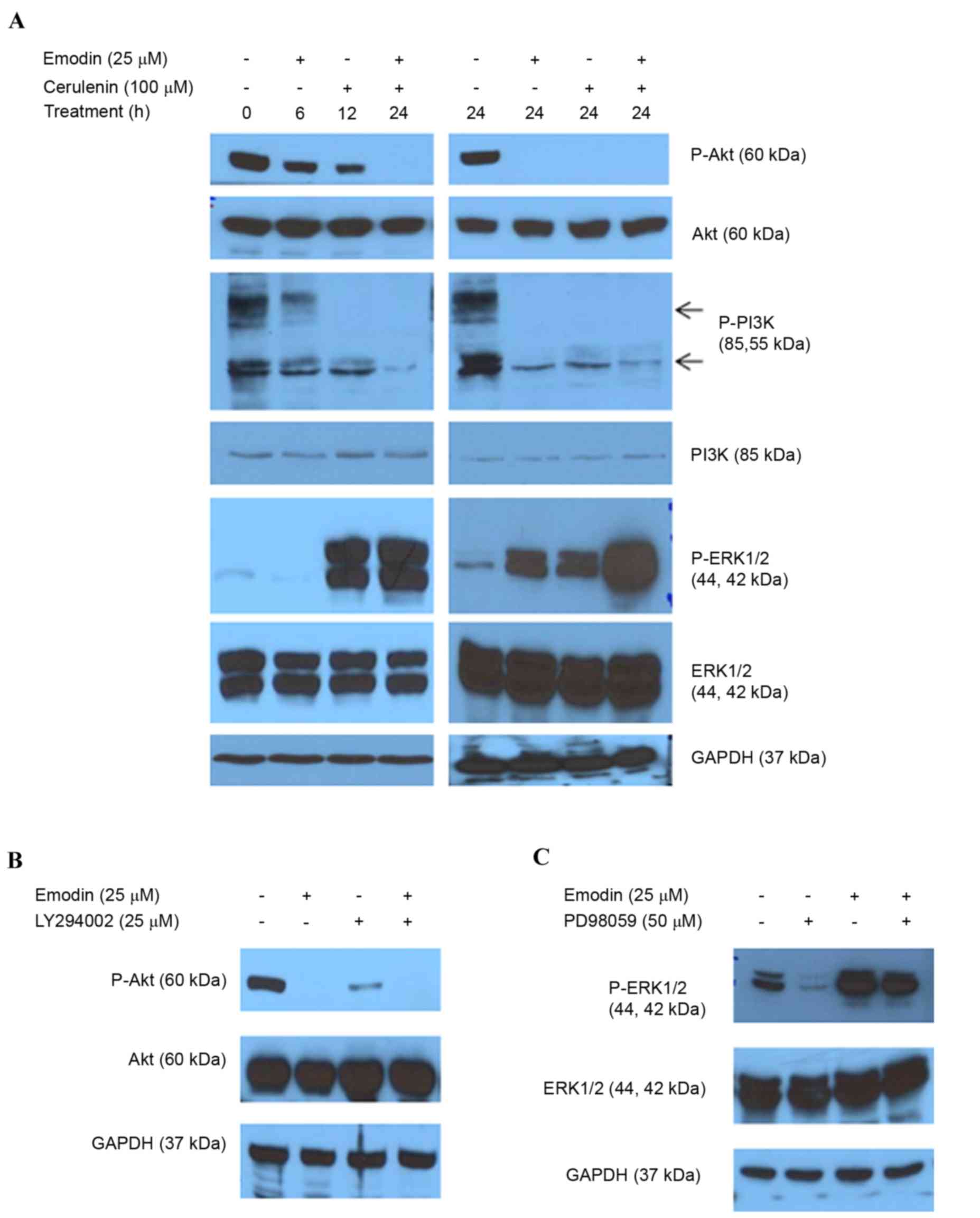 | Figure 6.Emodin alters PI3K/AKT and ERK1/2
phosphorylation. (A) Following treatment with emodin and/or
cerulenin for 6, 12 and 24 h, the levels of AKT, P-AKT, PI3K,
P-PI3K, ERK1/2 and P-ERK1/2 were determined using western blot
analysis. (B) Following treatment with emodin and/or LY294002 (25
µM), AKT and P-AKT (Ser473) were determined using western blot
analysis. (C) Cells were treated with emodin (25 µM) and/or PD98059
(50 µM) as a selective ERK inhibitor for 24 h, and equal quantities
of lysates were immunoblotted with ERK1/2 and P-ERK1/2
(Thr202/Tyr204). GAPDH was used as the loading control. FASN, fatty
acid synthase; GAPDH, glyceraldehyde 3-phosphate dehydrogenase;
PI3K, phosphatidylinositol 3-kinase; ERK, extracellular-signal
regulated kinase; P-, phosphorylated. |
Discussion
Different expression levels of FASN between
cancerous and normal cells have been suggested as a potential
target for anticancer drug development (4). FASN has been implicated in breast
cancer predominantly through its connection with the activity of
HER2 in transcriptional, proteomic and functional analyses
(8,30,31).
The expression of FASN also appears to be important in cell growth
and pathogenesis in colon cancer (32). FASN is expressed in all colorectal
neoplasms and there is a concomitant increase in fatty acid
synthesis (32). The
overexpression of FASN by tumors is associated with improved
survival rates of patients with colon cancer (33), and high serum levels of FASN in
patients with colorectal cancer are associated with tumor events,
lymph node metastasis status, distant metastasis and tumor clinical
stage (5). It has been suggested
that inhibiting FASN pharmacologically reduces cell growth and
survival, and induces the apoptosis of colon cancer cells (34). For example, cerulenin, a natural
FASN inhibitor, enhances antitumor activity when combined with
oxaliplatin in human colon cancer cells (35) and suppresses liver metastasis of
colon cancer (36). C75, a stable
synthetic small molecule developed specifically for inhibiting
FASN, produces a cytotoxic effect modulated by p53 in colon
carcinoma cells (37). Naturally
occurring olive oil polyphenols have the ability to suppress FASN,
providing a well-tolerated novel colon cancer therapy (38). However, no studies have reported
that emodin has FASN inhibitory activity in cancer. It has been
suggested that emodin may be a fat-reducing drug by inhibiting FASN
(39). The findings of the present
study revealed for the first time, to the best of our knowledge,
that emodin suppressed the protein expression of FASN and reduced
its activity in human colorectal cancer cells. As shown in Fig. 1B, the HCT116 cell line had
significantly elevated expression levels of FASN, compared with
levels in the other colon cancer cell lines. This elevated
expression of FASN in HCT116 cells was more effective in reducing
cancer cell death following emodin treatment, compared with cells
with a low expression level of FASN (Fig. 1A). Emodin only downregulated the
expression of FASN in HCT116 cells (Fig. 1C), suggesting that emodin-induced
cell death may be regulated by FASN-involved de novo fatty
acid synthesis. These data suggested that inhibiting FASN may be an
effective strategy for treating colon cancer overexpressing
FASN.
Previous studies have demonstrated that emodin
suppresses tumor growth in LS1034 human colon cancer cells in
vitro and in vivo (40), induces apoptosis triggered by
oxidative stress in colon cancer cells (16) and inhibits colon cancer cell growth
by inhibiting VEGFR signaling (22). However, the molecular mechanisms
underlying emodin-induced cell death in colorectal cancer cells
have not been investigated in detail. Until now, no studies have
reported the effect of emodin on the expression of FASN or its
activity in cancer, including colorectal cancer. The present study
found that emodin suppressed the expression of FASN by degrading
FASN protein, which was caused by elevated protein ubiquitination,
in HCT116 cells (Figs. 1C and
2). Tomek et al (9) reported that inhibiting FASN with C75
results in the accumulation of ubiquitinated proteins, including
PI3K and MAPK signaling proteins, in ovarian cancer (9). Emodin also induces the proteosomal
degradation of EGFR/EGFR variant III in glioma stem cells (41). However, no previous study has
reported that emodin induces the degradation of FASN protein.
In the present study, emodin concomitantly inhibited
FASN activity and downregulated the protein expression of FASN
(Fig. 3A and B). FASN is a key
lipogenic enzyme. The overexpression of FASN in several types of
cancer induces de novo lipogenesis, which is involved in
cell survival, proliferation, migration and invasion (27). Therefore, downregulating the
activity of FASN in cancer cells is necessary for tumor cell death.
The present study determined concentrations of free fatty acids
using a Free Fatty Acid Quantification kit (Fig. 3B). Emodin reduced the concentration
of fatty acids, indicating that emodin inhibited the function of
FASN in HCT116 cells.
FASN is essential for the proliferation and survival
of human colorectal carcinoma cells, as demonstrated by the FASN
inhibitor, which reduces the cell growth and promotes apoptosis
(42). The present study
hypothesized that emodin induces colorectal cancer apoptosis by
inhibiting the expression and function of FASN. It was shown that
emodin produced a dose- and time-dependent decrease in HCT116 cell
viability (Fig. 4B and C) and
induced apoptosis, as shown by Annexin/propidium iodide double
staining and caspase cleavage (Fig. 4D
and E). It was also observed that cell viability and apoptosis
were partially rescued following the addition of palmitate, which
is the final product of FASN activity (Fig. 5A and B). Palmitate-induced toxicity
has been reported in various cell lines; however, the present study
demonstrated that exogenous palmitate and emodin treatment
increased HCT116 cell viability. These results suggested that
inhibiting FASN was a direct contributor to the anticancer effects
of emodin on HCT116 colon cancer cells, and that fatty acid
synthesis was closely associated with colon cancer cell death.
The PI3K/Akt and MAPK/ERK1/2 signaling pathways
promote cell proliferation, survival and the anti-apoptotic
response, and have been implicated in regulating the expression of
FASN in cancer, including breast and ovarian cancer (28,43).
The level of FASN and activity of Akt are higher in HER2-positive
cancer, and inhibiting fatty acid synthesis affects the HER2 and
PI3K/Akt pathways (42). FASN is
regulated in malignancies by growth factor-dependent signaling,
which activates the Ras-Raf-MEK-ERK1/2 and PI3K/Akt pathways
(44). The downregulation of
PI3K/Akt phosphorylation can attenuate the expression of FASN
(27). The FASN inhibitor, C75,
inhibits pAkt, but increases pERK1/2 in ovarian cancer cells
(7); the α-mangostin-induced
inhibition of FASN decreases pAkt, but increases active pERK1/2 in
breast cancer cells (27). In the
present study, it was found that emodin downregulated pPI3K and
pAkt in a time-dependent manner (Fig.
6A). Emodin upregulated active levels of pERK1/2 in HCT116
cells (Fig. 6A). These data
demonstrated that the emodin-induced inhibition of FASN may be
associated with the PI3K/Akt and ERK1/2 signaling pathways in
colorectal cancer.
The findings of the present study suggested that
emodin downregulated the expression of FASN, inhibited
intracellular FASN activity and fatty acid biosynthesis, and
induced antiproliferation and apoptosis in HCT116 human colon
cancer cells. Therefore, the results showed that emodin has
therapeutic potential as a colon cancer treatment, and may provide
a novel method in developing target-directed anticancer drugs for
further investigations.
Acknowledgements
This study was supported by a research fund of
Chungnam National University, Daejeon, Korea (grant no.
2013-2017).
References
|
1
|
Cunningham D, Atkin W, Lenz HJ, Lynch HT,
Minsky B, Nordlinger B and Starling N: Colorectal cancer. Lancet.
375:1030–1047. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lozano R, Naghavi M, Foreman K, Lim S,
Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, et
al: Global and regional mortality from 235 causes of death for 20
age groups in 1990 and 2010: A systematic analysis for the global
burden of disease study 2010. Lancet. 380:2095–2128. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Merika E, Saif MW, Katz A, Syrigos K and
Morse M: Colon cancer vaccines: An update. In vivo. 24:607–628.
2010.PubMed/NCBI
|
|
4
|
Kuhajda FP: Fatty acid synthase and
cancer: New application of an old pathway. Cancer Res.
66:5977–5980. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Long QQ, Yi YX, Qiu J, Xu CJ and Huang PL:
Fatty acid synthase (FASN) levels in serum of colorectal cancer
patients: Correlation with clinical outcomes. Tumour Biol.
35:3855–3859. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mounier C, Bouraoui L and Rassart E:
Lipogenesis in cancer progression (review). Int J Oncol.
45:485–492. 2014.PubMed/NCBI
|
|
7
|
Grunt TW, Wagner R, Grusch M, Berger W,
Singer CF, Marian B, Zielinski CC and Lupu R: Interaction between
fatty acid synthase- and ErbB-systems in ovarian cancer cells.
Biochem Biophys Res Commun. 385:454–459. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee JS, Sul JY, Park JB, Lee MS, Cha EY,
Song IS, Kim JR and Chang ES: Fatty acid synthase inhibition by
amentoflavone suppresses HER2/neu (erbB2) oncogene in SKBR3 human
breast cancer cells. Phytother Res. 27:713–720. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tomek K, Wagner R, Varga F, Singer CF,
Karlic H and Grunt TW: Blockade of fatty acid synthase induces
ubiquitination and degradation of phosphoinositide-3-kinase
signaling proteins in ovarian cancer. Mol Cancer Res. 9:1767–1779.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang Z, Chen G and Shi P: Effects of
emodin on the gene expression profiling of human breast carcinoma
cells. Cancer Detect Prev. 32:286–291. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hsu CM, Hsu YA, Tsai Y, Shieh FK, Huang
SH, Wan L and Tsai FJ: Emodin inhibits the growth of hepatoma
cells: Finding the common anti-cancer pathway using Huh7, Hep3B,
and HepG2 cells. Biochem Biophys Res Commun. 392:473–478. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lai JM, Chang JT, Wen CL and Hsu SL:
Emodin induces a reactive oxygen species-dependent and ATM-p53-Bax
mediated cytotoxicity in lung cancer cells. Eur J Pharmacol.
623:1–9. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cha TL, Qiu L, Chen CT, Wen Y and Hung MC:
Emodin down-regulates androgen receptor and inhibits prostate
cancer cell growth. Cancer Res. 65:2287–2295. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Srinivas G, Anto RJ, Srinivas P,
Vidhyalakshmi S, Senan VP and Karunagaran D: Emodin induces
apoptosis of human cervical cancer cells through poly(ADP-ribose)
polymerase cleavage and activation of caspase-9. Eur J Pharmacol.
473:117–125. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chun-Guang W, Jun-Qing Y, Bei-Zhong L,
Dan-Ting J, Chong W, Liang Z, Dan Z and Yan W: Anti-tumor activity
of emodin against human chronic myelocytic leukemia K562 cell lines
in vitro and in vivo. Eur J Pharmacol. 627:33–41. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xie MJ, Ma YH, Miao L, Wang Y, Wang HZ,
Xing YY, Xi T and Lu YY: Emodin-provoked oxidative stress induces
apoptosis in human colon cancer HCT116 Cells through a
p53-mitochondrial apoptotic pathway. Asian Pac J Cancer Prev.
15:5201–5205. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lin SZ, Wei WT, Chen H, Chen KJ, Tong HF,
Wang ZH, Ni ZL, Liu HB, Guo HC and Liu DL: Antitumor activity of
emodin against pancreatic cancer depends on its dual role:
Promotion of apoptosis and suppression of angiogenesis. PLoS One.
7:e421462012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Manu KA, Shanmugam MK, Ong TH, Subramaniam
A, Siveen KS, Perumal E, Samy RP, Bist P, Lim LH, Kumar AP, et al:
Emodin suppresses migration and invasion through the modulation of
CXCR4 expression in an orthotopic model of human hepatocellular
carcinoma. PLoS One. 8:e570152013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yim H, Lee YH, Lee CH and Lee SK: Emodin,
an anthraquinone derivative isolated from the rhizomes of Rheum
palmatum, selectively inhibits the activity of casein kinase II as
a competitive inhibitor. Planta Med. 65:9–13. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee HZ: Protein kinase C involvement in
aloe-emodin- and emodin-induced apoptosis in lung carcinoma cell.
Br J Pharmacol. 134:1093–1103. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Su YJ, Tsai MS, Kuo YH, Chiu YF, Cheng CM,
Lin ST and Lin YW: Role of Rad51 down-regulation and extracellular
signal-regulated kinases 1 and 2 inactivation in emodin and
mitomycin C-induced synergistic cytotoxicity in human
non-small-cell lung cancer cells. Mol Pharmacol. 77:633–643. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lu Y, Zhang J and Qian J: The effect of
emodin on VEGF receptors in human colon cancer cells. Cancer
Biother Radiopharm. 23:222–228. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang L, Chang CJ, Bacus SS and Hung MC:
Suppressed transformation and induced differentiation of
HER-2/neu-overexpressing breast cancer cells by emodin. Cancer Res.
55:3890–3896. 1995.PubMed/NCBI
|
|
24
|
Menendez JA, Vellon L, Colomer R and Lupu
R: Pharmacological and small interference RNA-mediated inhibition
of breast cancer-associated fatty acid synthase (oncogenic
antigen-519) synergistically enhances Taxol (paclitaxel)-induced
cytotoxicity. Int J Cancer. 115:19–35. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lacroix M and Leclercq G: Relevance of
breast cancer cell lines as models for breast tumours: An update.
Breast Cancer Res Treat. 83:249–289. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Little JL and Kridel SJ: Fatty acid
synthase activity in tumor cells. Subcell Biochem. 49:169–194.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li P, Tian W and Ma X: Alpha-mangostin
inhibits intracellular fatty acid synthase and induces apoptosis in
breast cancer cells. Mol Cancer. 13:1382014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang YA, Han WF, Morin PJ, Chrest FJ and
Pizer ES: Activation of fatty acid synthesis during neoplastic
transformation: Role of mitogen-activated protein kinase and
phosphatidylinositol 3-kinase. Exp Cell Res. 279:80–90. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yellen P and Foster DA: Inhibition of
fatty acid synthase induces pro-survival Akt and ERK signaling in
K-Ras-driven cancer cells. Cancer Lett. 353:258–263. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Menendez JA and Lupu R: Oncogenic
properties of the endogenous fatty acid metabolism: Molecular
pathology of fatty acid synthase in cancer cells. Curr Opin Clin
Nutr Metab Care. 9:346–357. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Menendez JA, Vazquez-Martin A,
Oliveras-Ferraros C, Garcia-Villalba R, Carrasco-Pancorbo A,
Fernandez-Gutierrez A and Segura-Carretero A: Analyzing effects of
extra-virgin olive oil polyphenols on breast cancer-associated
fatty acid synthase protein expression using reverse-phase protein
microarrays. Int J Mol Med. 22:433–439. 2008.PubMed/NCBI
|
|
32
|
Rashid A, Pizer ES, Moga M, Milgraum LZ,
Zahurak M, Pasternack GR, Kuhajda FP and Hamilton SR: Elevated
expression of fatty acid synthase and fatty acid synthetic activity
in colorectal neoplasia. Am J Pathol. 150:201–208. 1997.PubMed/NCBI
|
|
33
|
Ogino S, Nosho K, Meyerhardt JA, Kirkner
GJ, Chan AT, Kawasaki T, Giovannucci EL, Loda M and Fuchs CS:
Cohort study of fatty acid synthase expression and patient survival
in colon cancer. J Clin Oncol. 26:5713–5720. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhan Y, Ginanni N, Tota MR, Wu M, Bays NW,
Richon VM, Kohl NE, Bachman ES, Strack PR and Krauss S: Control of
cell growth and survival by enzymes of the fatty acid synthesis
pathway in HCT-116 colon cancer cells. Clin Cancer Res.
14:5735–5742. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shiragami R, Murata S, Kosugi C, Tezuka T,
Yamazaki M, Hirano A, Yoshimura Y, Suzuki M, Shuto K and Koda K:
Enhanced antitumor activity of cerulenin combined with oxaliplatin
in human colon cancer cells. Int J Oncol. 43:431–438.
2013.PubMed/NCBI
|
|
36
|
Murata S, Yanagisawa K, Fukunaga K, Oda T,
Kobayashi A, Sasaki R and Ohkohchi N: Fatty acid synthase inhibitor
cerulenin suppresses liver metastasis of colon cancer in mice.
Cancer Sci. 101:1861–1865. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li JN, Gorospe M, Chrest FJ, Kumaravel TS,
Evans MK, Han WF and Pizer ES: Pharmacological inhibition of fatty
acid synthase activity produces both cytostatic and cytotoxic
effects modulated by p53. Cancer Res. 61:1493–1499. 2001.PubMed/NCBI
|
|
38
|
Notarnicola M, Pisanti S, Tutino V, Bocale
D, Rotelli MT, Gentile A, Memeo V, Bifulco M, Perri E and Caruso
MG: Effects of olive oil polyphenols on fatty acid synthase gene
expression and activity in human colorectal cancer cells. Genes
Nutr. 6:63–69. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang C, Teng L, Shi Y, Jin J, Xue Y,
Shang K and Gu J: Effect of emodin on proliferation and
differentiation of 3T3-L1 preadipocyte and FAS activity. Chin Med J
(Engl). 115:1035–1038. 2002.PubMed/NCBI
|
|
40
|
Ma YS, Weng SW, Lin MW, Lu CC, Chiang JH,
Yang JS, Lai KC, Lin JP, Tang NY, Lin JG and Chung JG: Antitumor
effects of emodin on LS1034 human colon cancer cells in vitro and
in vivo: Roles of apoptotic cell death and LS1034 tumor xenografts
model. Food Chem Toxicol. 50:1271–1278. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kim J, Lee JS, Jung J, Lim I, Lee JY and
Park MJ: Emodin suppresses maintenance of stemness by augmenting
proteosomal degradation of epidermal growth factor
receptor/epidermal growth factor receptor variant III in glioma
stem cells. Stem Cells Dev. 24:284–295. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chuang HY, Chang YF and Hwang JJ:
Antitumor effect of orlistat, a fatty acid synthase inhibitor, is
via activation of caspase-3 on human colorectal carcinoma-bearing
animal. Biomed Pharmacother. 65:286–292. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang HQ, Altomare DA, Skele KL, Poulikakos
PI, Kuhajda FP, Di Cristofano A and Testa JR: Positive feedback
regulation between AKT activation and fatty acid synthase
expression in ovarian carcinoma cells. Oncogene. 24:3574–3582.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Liu X, Shi Y, Giranda VL and Luo Y:
Inhibition of the phosphatidylinositol 3-kinase/Akt pathway
sensitizes MDA-MB468 human breast cancer cells to cerulenin-induced
apoptosis. Mol Cancer Ther. 5:494–501. 2006. View Article : Google Scholar : PubMed/NCBI
|