Introduction
Mesenteric lymph, which is a major source of lymph,
is vital for intestinal fluid and chyle transport and immune cell
trafficking, thus aiding in homeostatic maintenance (1,2).
Mesenteric lymph is also involved in the pathophysiological changes
associated with hemorrhagic shock, and is considered to be an
important source of factors that link gut ischemia and acute lung
injury following hemorrhagic shock (3,4). The
ischemic gut is a source of tissue-toxic factors, which lead to
distant organ dysfunction, including lung dysfunction, via
mesenteric lymphatics. Mesenteric lymph components, including
electrolytes, lipids, proteins and immune cells, have previously
been studied under physiological and pathophysiological conditions
(1,2). MicroRNAs (miRNAs) are small
non-coding RNAs, which are involved in the post-transcriptional
regulation of their target genes at the mRNA and/or protein level
(5). miRNAs are not only localized
intracellularly, but are also secreted into extracellular fluids,
including plasma, serum, saliva and urine, via exosomes (30–100 nm)
of multivesicular body origin (6).
However, there have been few reports on the profiling and
circulatory dynamics of miRNAs in mesenteric lymph.
The aim of the present study was to provide a
comprehensive analysis of miRNAs in normal rodent mesenteric lymph
using a reverse transcription-quantitative polymerase chain
reaction (RT-qPCR)-based array. The expression profiles of miRNAs
between lymph and blood plasma were examined for differences, and
lymph miRNAs were characterized in terms of their stability and
fractional distribution. Finally, the in vivo delivery of
lymph miRNAs via mesenteric lymphatics into the systemic
circulation was evaluated.
Materials and methods
Animals
The present study was approved by the animal
research committee of Nippon Medical School (Tokyo, Japan). Male
Sprague-Dawley rats (n=38; weight, 286–417 g; age, 8–10 weeks;
Oriental Yeast Co., Ltd., Tokyo, Japan) were maintained under a
12/12 h light/dark cycle at 23°C and fed standard laboratory rat
chow with access to tap water (Oriental MF; Oriental Yeast) ad
libitum.
Collection of lymph and blood samples
using a mesenteric lymph duct cannulated rat model
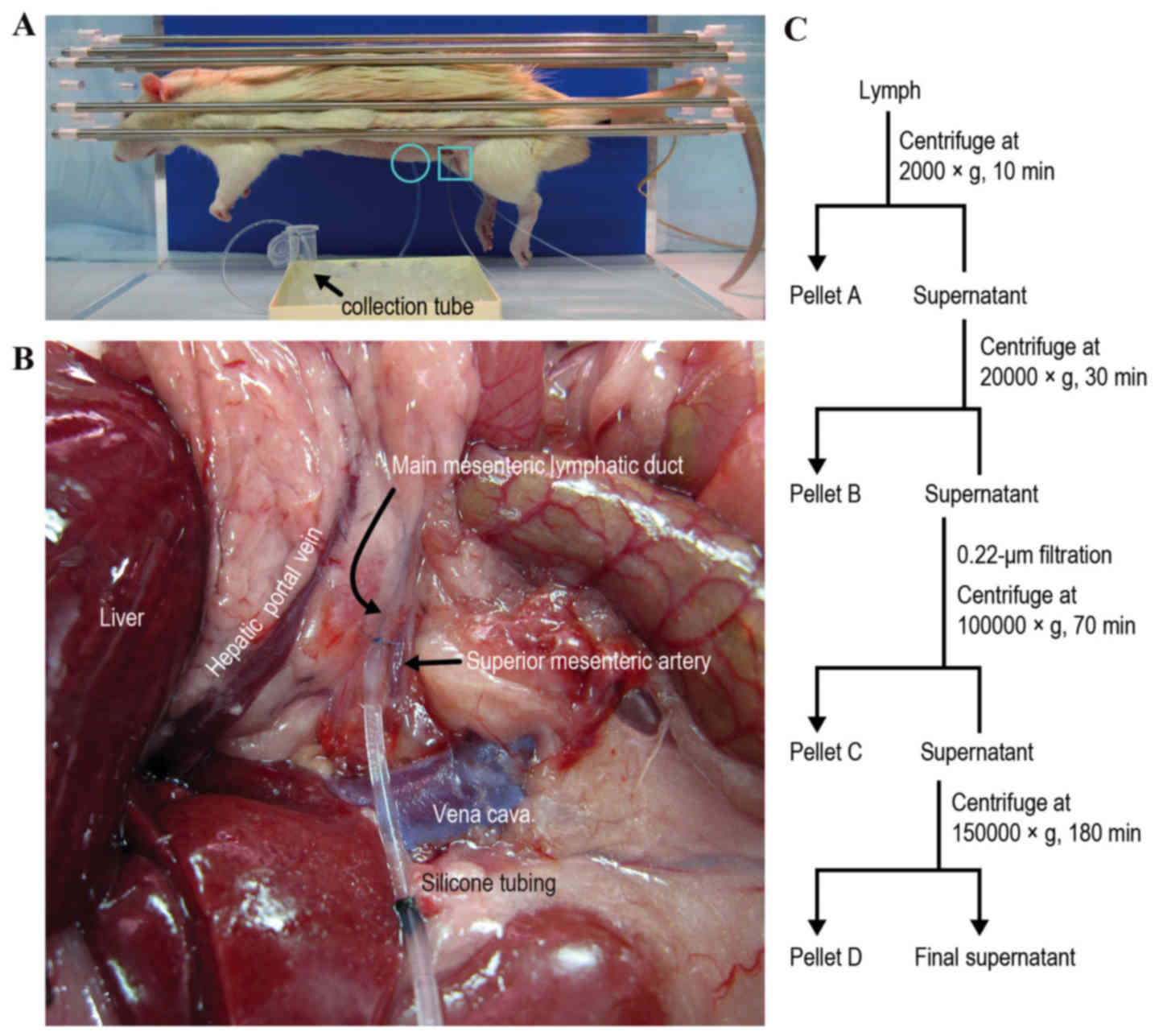
Sample collection was performed using the procedure
employed in our previous rat model study (7). Each rat was anesthetized by
intraperitoneal injection of 65 mg/kg sodium pentobarbital
(Kyoritsu Seiyaku Corporation, Tokyo, Japan). Body temperature was
maintained at 37°C using a heat lamp and a heating blanket
(ATB-1100; Nihon Kohden Corporation, Tokyo, Japan). A midline
incision was made, and a right medial visceral rotation was
performed to expose the main mesenteric lymphatic duct. The
efferent lymphatic duct was cannulated with silicone tubing
(internal diameter, 0.5 mm; external diameter, 1.0 mm; Renover
Science Co., Ltd., Tokyo, Japan) and secured with 8–0 nylon sutures
(Fig. 1A and B). The tubing was
exteriorized via the incision. Mesenteric lymph was continuously
collected into a 1.5 ml nuclease-free centrifuge tube containing
EDTA-2Na (1 mg/tube; Dojindo Molecular Technologies, Inc.,
Mashiki-machi, Kumamoto, Japan) by gravity drip on ice for 1.5 h.
The collected lymph was centrifuged once at 2,000 × g for 10 min at
4°C, and twice at 10,000 × g for 30 min at 4°C to remove cells,
cellular debris and the uppermost white fat layer of the
supernatant. The remaining supernatant was designated as cell-free
lymph and stored at −80°C prior to RNA purification. For lymph
fraction analysis, some of the collected lymph was fractionated by
differential centrifugation as described below.
The right femoral artery and vein were cannulated
with polyethylene catheters (SP-45; Natsume Seisakusho Co., Ltd.,
Osaka, Japan). The arterial catheter was used for continuous blood
pressure monitoring with a carrier amplifier (AP-601G; Nihon Koden
Corporation) and a data acquisition system (PowerLab/8/30; AD
Instruments Japan, Inc., Nagoya-shi, Japan) for blood withdrawal.
Following lymph collection, blood (5 ml) was also obtained from the
femoral artery using an EDTA-containing, vacuum blood-drawing tube
(Venogect II; Terumo Corporation, Shinjuku-ku, Tokyo, Japan) and
centrifuged at 1,700 × g for 15 min at 4°C. Plasma samples
were transferred to a 1.5 ml centrifuge tube and stored at −80°C
prior to RNA purification. Rats were sacrificed by intravenous
administration of 1 ml sodium pentobarbital.
RNA extraction
The results were normalized to allow for
sample-to-sample variation in the RNA isolation procedure, using
the results of the spiked exogenous control cel-miRNAs as described
previously (8,9). Two synthetic RNA oligonucleotides
corresponding to cel-miR-39-3p and cel-miR-238-3p
(Qiagen, Inc., Valencia, CA, USA) were used. The spike-in
oligonucleotides were introduced (as a mixture containing 250 fmol
of each oligonucleotide in 10 µl water) following the addition of
ISOGEN-LS (Nippon Gene Co., Ltd., Tokyo, Japan) to lymph and plasma
samples. Total RNA from lymph and plasma samples was extracted
using ISOGEN-LS, according to the manufacturer's protocol; total
RNA from tissue samples was extracted with RNAiso Plus (Takara Bio,
Inc., Shiga, Japan).
Comprehensive quantitative analysis of
miRNAs using a RT-qPCR-based array
For quantitative analysis, a fixed volume of RNA
eluate from a given volume of starting sample (cell-free lymph and
blood plasma) was used as input for the RT reaction. A total RNA
eluate volume of 20 µl was prepared from a sample in which the
starting volume was 125 µl; an input of 3 µl eluted RNA from lymph
and plasma (n=6 each) was reverse-transcribed using Megaplex RT
primers (Rodent Pool Set v3.0 containing Pool A and B; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) according to the
manufacturer's protocol. cDNA was pre-amplified using Megaplex
PreAmp primers (Rodent Pool Set v3.0; Thermo Fisher Scientific,
Inc.). The pre-amplified products were subjected to RT-qPCR using
TaqMan MicroRNA assays (A and B; version 3.0; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. miRNA
sequences were annotated using the Sanger database (miRBase;
release 15; http://www.mirbase.org/). Data
obtained from this assay were analyzed using RQ Manager 1.2 (Thermo
Fisher Scientific, Inc.). Full array datasets are available upon
request.
RT-qPCR analysis
RT-qPCR was performed using TaqMan MicroRNA assays
(A and B; version 3.0; Thermo Fisher Scientific, Inc.) in a 7300
Real-Time PCR system (Thermo Fisher Scientific, Inc.) or a 7900HT
Fast Real-Time PCR system (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. cel-miRNAs were used as
exogenous controls for lymph and plasma samples to normalize miRNA
expression levels, and U6 small nuclear RNA (Rnu6) was used
as an endogenous internal control for tissue samples. Primers for
miR-145 (assay ID 002278), miR-150 (assay ID 000473),
cel-miR-39-3p (assay ID 000200), cel-miR-238-3p
(assay ID 000248) and Rnu6 (assay ID 001973) were obtained
from Thermo Fisher Scientific, Inc.
Relative quantification of the expression levels of
each miRNA in mesenteric lymph compared with blood plasma was
determined using the comparative cycle quantification (Cq) method
(ΔΔCq method) as described previously (8). Briefly, the Cq values obtained for
the two spike-in cel-miRNAs were averaged to generate a spike-in
control Cq value. The difference (ΔCq) for each sample miRNA was
determined based on the following formula: ΔCq=(miRNA Cq value of a
given sample)-(spike-in control Cq value of the sample).
Subsequently, the ΔΔCq value for each sample was determined using
the following formula: ΔΔCq=ΔCq (lymph sample)-ΔCq (plasma
sample).
miRNA stability assay
miRNA levels in cell-free lymph that had been
incubated at room temperature for 0 to 24 h were measured to
evaluate the stability of miRNAs in mesenteric lymph. The
expression levels of two endogenous miRNAs, miR-145 and
miR-150, were measured, since the RT-qPCR-based miRNA array
analysis revealed that these two miRNAs are highly expressed in
lymph. Following incubation, total RNA from 50 µl of each sample
(n=6) was extracted at the scheduled time points (0, 0.5, 1, 6 and
24 h). Spike-in exogenous cel-miRNAs (250 fmol) were introduced
following the addition of ISOGEN-LS to each sample to normalize the
data. RT-qPCR was performed as aforementioned. Relative
quantification of miRNA expression levels in each incubation sample
vs. the 0 h incubation sample was determined on the basis of the
ΔΔCq method. The stability of the miRNAs in plasma was also
examined, in order to compare with that in lymph.
Lymph fraction analysis
To assess whether lymph miRNAs are present in
exosomes, the lymph samples were fractionated by differential
centrifugation and the expression levels of miR-150 in each
fraction were compared (n=4; Fig.
1C). Pellets collected at low speed (2,000 × g for 10 min at
4°C) were rich in cells and nuclei (defined as Pellet A), and
pellets collected at medium speed (20,000 × g for 30 min at 4°C)
were rich in mitochondria, lysosomes and peroxisomes (Pellet B).
Following removal of the uppermost white fat layer of the
supernatant that formed during the medium speed centrifugation, the
remaining supernatant was diluted five times with
phosphate-buffered saline (PBS) and filtered through a 0.22 µm
filter. Pellets collected at high speed (100,000 × g for 70 min at
4°C) were rich in microsomes and exosomes (Pellet C), and pellets
collected at very high speed (150,000 × g for 180 min at 4°C) were
rich in ribosomes and large macromolecules (Pellet D). To normalize
the data, exogenous cel-miRNAs (250 fmol) were spiked-in following
the addition of ISOGEN-LS to each sample. RT-qPCR was performed as
aforementioned. The relative quantification of miRNA expression
levels in each fraction vs. the Pellet A fraction was determined on
the basis of the ΔΔCq method. The fold difference in the miRNA
level of the final supernatant fraction (soluble protein fraction;
Fig. 1C) relative to the Pellet A
fraction was corrected by the dilution factor used in
ultracentrifugation; therefore, the fold difference was determined,
as follows: 2-ΔΔCq × dilution factor. The expression
level of Rnu6 in each fraction was also examined.
Isolation of exosomes from the IEC-6
rat small intestine epithelial cell line
IEC-6-derived exosomes were prepared from culture
supernatants of IEC-6 cells transfected with cel-miR-238-3p
to trace exogenously administered exosomes in vivo. IEC-6
cells were purchased from the American Type Culture Collection
(Manassas, VA, USA). Cells were cultured in exosome-free Dulbecco's
modified Eagle's medium (Thermo Fisher Scientific, Inc.)
supplemented with 5% fetal bovine serum (Japan Bio Serum Co., Ltd.,
Fukuyama-shi, Japan) and 4 µg/ml insulin (37°C, 5% CO2).
Exosome-free medium was prepared by ultracentrifugation at 100,000
× g for 12 h at 4°C according to the method of Thery et al
(10). IEC-6 cells were
transfected with cel-miR-238-3p (30 nM) using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) for 1 h at 37°C. A total of 48 h
post-transfection, culture supernatants were collected for exosome
isolation. Exosomes were isolated from the culture supernatant as
described previously (10,11). Cell culture supernatants were
sequentially centrifuged at 320 × g for 10 min at 4°C, 2,070 × g
for 10 min at 4°C, and 10,000 × g for 30 min at 4°C to eliminate
cells, dead cells, and cell debris, respectively. Exosomes were
pelleted by ultracentrifugation at 100,000 × g for 100 min at 4°C
and washed twice with PBS. The solution was filtered through a 0.22
µm filter, and exosomes were pelleted by ultracentrifugation
(100,000 × g, 120 min, 4°C) and resolved in 300 µl PBS. The protein
content of the purified exosomes was determined using a
bicinchoninic acid protein assay kit (Thermo Fisher Scientific,
Inc.). RT-qPCR confirmed that cel-miR-238-3p was detected in
IEC-6-derived exosomes (data not shown). Exosomes from
cel-miR-238-3p-transfected IEC-6 cells were designated IEC-6
cel-miR-238-3p-exosomes.
In vivo analysis of exosomal miRNA
delivery from mesenteric lymphatics into the systemic
circulation
The in vivo delivery of lymph miRNAs via
mesenteric lymphatics into the systemic circulation was examined by
injection of rat-derived exosomes (IEC-6
cel-miR-238-3p-exosomes). The mesenteric lymph duct
cannulated rat model was prepared as aforementioned, with the
exception that the afferent lymphatic duct was cannulated with
silicone tubing. IEC-6 cel-miR-238-3p-exosomes were injected
through a catheter that was inserted into the main afferent
mesenteric lymphatic duct for 30 min using a syringe pump (TE-331S;
Terumo Corporation). Each rat received 64.1±16.4 µg (mean ±
standard deviation) exosomes in a final injection volume of
233.3±115.5 µl (n=3). Following an additional 30 min, each rat was
thoroughly perfused with 1 ml/g body weight of 0.9% saline via the
left ventricle to remove circulating blood and minimize
contamination of circulating cel-miR-238-3p-exosomes. The
lung, liver, kidney, and spleen were then excised and homogenized.
Subsequently, tissues underwent total RNA extraction. RT-qPCR for
cel-miR-238 was performed as aforementioned.
Statistical analysis
All analyses were conducted using the SPSS software
version 20 (IBM SPSS, Armonk, NY, USA). The significance of
between-group differences was assessed using Wilcoxon signed rank
test or analysis of variance followed by Dunnett's test or Tukey's
test. P<0.05 was considered to indicate a statistically
significant difference. Data are presented as the mean ± standard
deviation and are representative of at least three independent
experiments.
Results
Comprehensive profile analysis of
lymph miRNAs using an RT-qPCR-based array
An RT-qPCR-based array analysis was performed to
quantitatively examine the expression levels of 375 miRNAs in
normal rat mesenteric lymph, and to compare the miRNA expression
levels in lymph with those in blood plasma. Among the 375 rat
miRNAs preloaded in TaqMan Rodent miRNA Array A Card v3.0 and
TaqMan Rodent miRNA Array B Card v3.0, 287 miRNAs (77% of the
preloaded miRNAs) were detected in the lymph (in at least one of
all six rats), and 267 miRNAs were present in the plasma (in at
least one of all six rats) (71% of the preloaded miRNAs). Although
31 of the 287 lymph miRNAs were detected only in lymph, the
remaining 256 miRNAs were common between lymph and plasma.
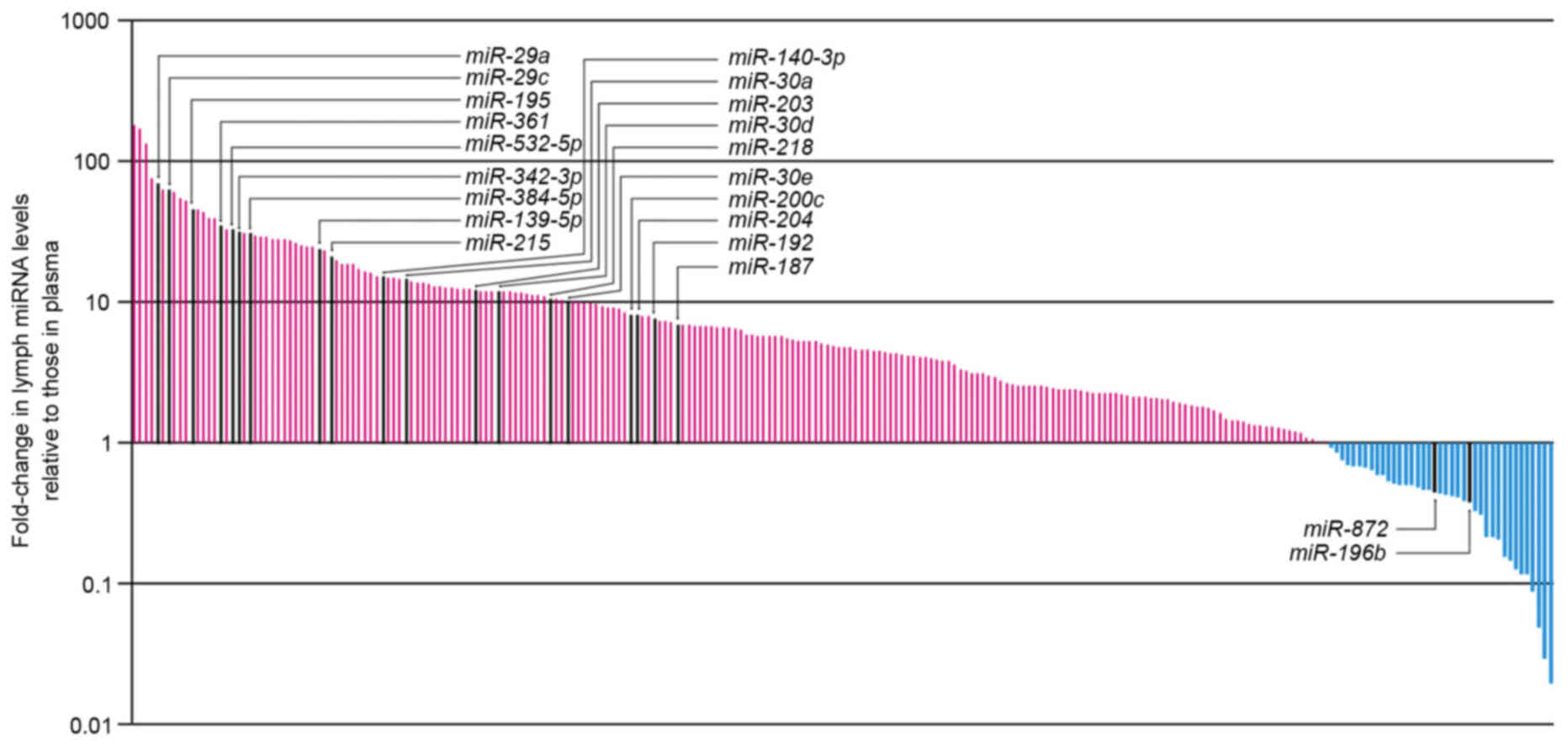
The differential miRNA expression levels in lymph
compared with plasma are presented in Fig. 2; high levels of miRNA expression
were detected in lymph compared with plasma. Among the miRNAs
examined using RT-qPCR in the present study, 218 could be assessed
using the Wilcoxon signed-rank test. A total of 19 of the 218
miRNAs displayed significantly increased expression in the
mesenteric lymph compared with plasma (P<0.05; Table I), whereas two miRNAs displayed
significantly decreased expression in the mesenteric lymph compared
with plasma (P<0.05; Table I).
However, no exclusive miRNAs (i.e., miRNAs detected in all six
samples for one set and undetected in all six samples for the other
set) were detected from either lymph or plasma.
 | Table I.miRNA expression differences between
mesenteric lymph and blood plasma, as revealed by polymerase chain
reaction-based array analysis. |
Table I.
miRNA expression differences between
mesenteric lymph and blood plasma, as revealed by polymerase chain
reaction-based array analysis.
| A, Differentially
expressed miRNAs in lymph vs. plasma |
|---|
|
|---|
| Differentially
expressed miRNAsa | Fold-change
(Lymph/Plasma) | P-value |
|---|
| miR-29a | 69.88 | 0.046 |
| miR-29c | 62.17 | 0.046 |
| miR-195 | 45.84 | 0.046 |
| miR-361 | 34.54 | 0.046 |
|
miR-532-5p | 32.75 | 0.046 |
|
miR-342-3p | 31.22 | 0.046 |
|
miR-384-5p | 30.89 | 0.043 |
|
miR-139-5p | 23.77 | 0.028 |
| miR-215 | 21.18 | 0.046 |
|
miR-140-3p | 15.12 | 0.046 |
| miR-30a | 14.50 | 0.043 |
| miR-203 | 12.08 | 0.046 |
| miR-30d | 11.89 | 0.046 |
| miR-218 | 10.63 | 0.046 |
| miR-30e | 10.19 | 0.046 |
|
miR-200c |
8.11 | 0.046 |
| miR-204 |
8.08 | 0.028 |
| miR-192 |
7.66 | 0.046 |
| miR-187 |
6.96 | 0.043 |
| miR-872 |
0.45 | 0.028 |
|
miR-196b |
0.34 | 0.043 |
|
| B, The 10 most
highly expressed miRNAs in lymph |
|
| miRNAs highly
expressed in lymphb | Fold-change
(Lymph/Plasma) | P-value |
|
| miR-16 | 29.41 | 0.080 |
| miR-24 |
7.35 | 0.075 |
| miR-145 | 15.21 | 0.345 |
| miR-150 | 39.45 | 0.080 |
|
miR-190b |
0.21 | 0.109 |
| miR-191 |
9.11 | 0.463 |
| miR-222 |
3.02 | 0.225 |
| miR-223 |
0.87 | 0.753 |
| miR-632 |
0.52 | 0.080 |
| miR-872 |
0.45 | 0.028 |
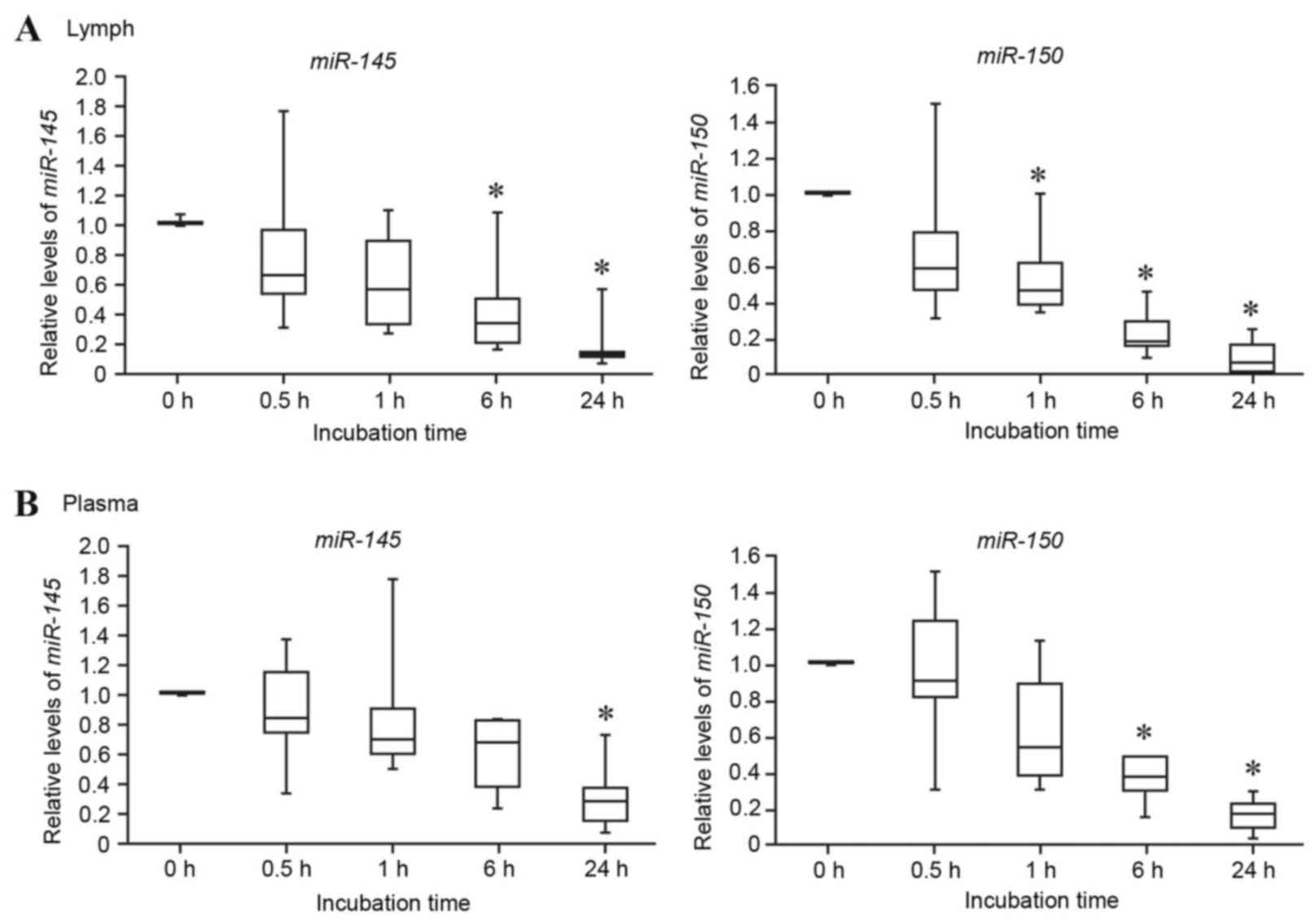
Lymph miRNA stability assay
To the best of our knowledge there is no information
currently available regarding the stability of miRNAs in rat lymph;
therefore, the differences in miRNA stability between lymph and
plasma were compared. As presented in Fig. 3, the lymph miRNAs miR-145
and miR-150 were gradually degraded and decreased to <20%
of the control (0 h) following 24 h incubation (Fig. 3A). The plasma miRNAs miR-145
and miR-150 were also degraded, decreasing to <30% of the
control following 24 h incubation (Fig. 3B). These results suggested that rat
lymph and plasma miRNAs are relatively unstable. However, it should
be noted that the time-dependent decrease in the miRNA levels did
not immediately occur in rat lymph and plasma, it being a gradual
process.
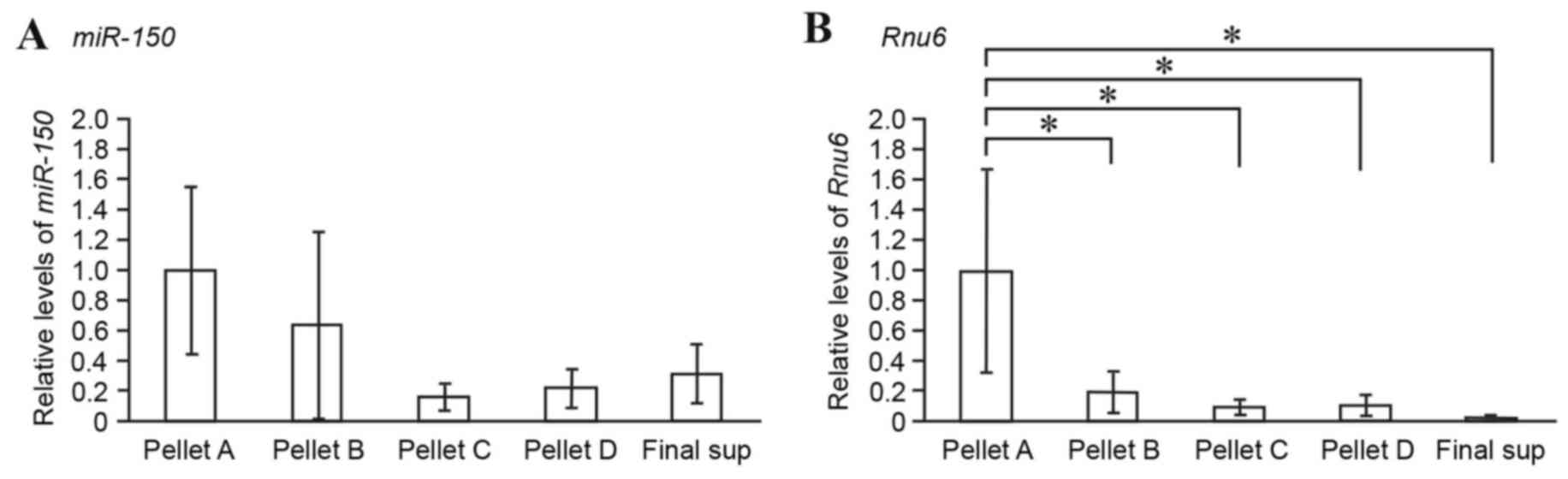
Lymph fraction analysis of miRNAs
Lymph samples were fractionated by differential
centrifugation (Fig. 1C) and the
expression levels of miR-150 in each fraction were compared
by RT-qPCR. miR-150 and Rnu6 were mainly detected in
Pellet A, which was rich in cells and nuclei (Fig. 4A and B). As expected,
miR-150 was detected in Pellet C, which was rich in
microsomes and exosome-containing extracellular vesicles (Fig. 4A). Notably, following
ultracentrifugation at very high speed (150,000 × g for 180 min at
4°C), miR-150 was also detected in Pellet D and the final
supernatant, which included cytosol constituents (Fig. 4A). It is likely that cell-free
lymph miRNAs are present not only as exosome-associated forms but
also as non-vesicle-associated forms.
In vivo analysis of the delivery of
exosomal miRNAs from mesenteric lymphatics into the systemic
circulation
The in vivo delivery of lymph miRNAs via
mesenteric lymphatics into the systemic circulation was evaluated
through injection of rat small intestine epithelial cell
line-derived exosomes, termed IEC-6 cel-miR-238-3p-exosomes.
cel-miR-238-3p can be detected in exosome-capturing organs
since rat cells do not express cel-miRNAs. Following administration
of IEC-6 cel-miR-238-3p-exosomes into the rat via mesenteric
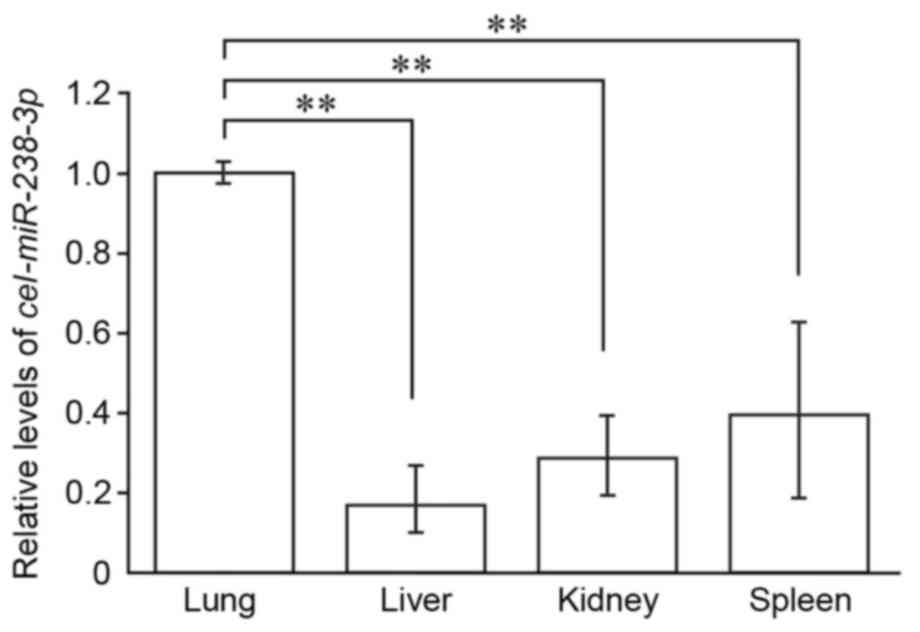
lymphatics, cel-miR-238-3p was detected in all four organs
(lung, liver, kidney and spleen) examined in the present study
(Fig. 5). The levels of captured
cel-miR-238-3p were significantly higher in the lung than
the other organs, (P<0.05; Fig.
5) suggesting that the lung is the major site where exosomal
mesenteric lymph miRNAs are captured in the systemic
circulation.
Discussion
In the present study, the expression levels of 375
miRNAs were quantitatively examined using RT-qPCR-based arrays to
reveal the miRNA expression profiles of normal rodent mesenteric
lymph; in total, 287 miRNAs were detected. Comparison of the
expression profiles of the miRNAs detectable in lymph with those in
plasma revealed that 21 miRNAs were significantly differentially
expressed between lymph and plasma. Furthermore, in vivo
analysis of lymph miRNA delivery by intralymphatic administration
of exosomes demonstrated that exosomal miRNAs were markedly
distributed in the lung, indicating that the lung is the major
organ responsible for clearance of exosomal lymph miRNAs.
To the best of our knowledge, only one previous
report has profiled miRNAs in mesenteric lymph. Blenkiron et
al (12) detected miRNAs in
rat mesenteric lymph during acute pancreatitis using miRNA
microarrays. A total of 85 miRNAs were detected of the 351 miRNAs
probed on the arrays in the lymph samples, and the 10 most abundant
lymph miRNAs were identified (miR-16, miR-23a,
miR-24, miR-26a, miR-145, miR-143,
miR-150, miR-191, miR-320 and let-7b).
In the present study, the 10 most highly expressed lymph miRNAs
(lower Cq values) among 287 lymph miRNAs were revealed to be
miR-16, miR-24, miR-145, miR-150,
miR-190b, miR-191, miR-222, miR-223,
miR-632 and miR-872. Five miRNAs (miR-16,
miR-24, miR-145, miR-150 and miR-191)
are common, and are highly abundant compared with the data
presented in Blenkiron et al (12). There are differences in the types
of detectable miRNAs and abundant miRNAs between the two datasets,
and these differences may be explained by considering the array
methodologies or rat strain differences between each analysis.
The stability of rat lymph miRNAs was investigated
by incubation of lymph at room temperature for up to 24 h. In the
present study, even though rat endogenous lymph miRNA levels
remained high within 1 h of the 24 h incubation, the lymph miRNAs
were moderately unstable (Fig. 3).
Rat plasma miRNAs also exhibited a similar trend with regards to
relatively gradual miRNA degradation (Fig. 3). Mitchell et al (9) demonstrated that human plasma miRNAs
remained stable following incubation at room temperature for 24 h.
In addition, Yamaura et al (13) reported the stability of miRNAs in
rat plasma compared with those in human plasma, and demonstrated
that circulating miRNAs in rat plasma were unstable at room
temperature for 24 h, whereas those in human plasma were stable.
The findings of the present study with regards to the stability of
rat plasma miRNAs are consistent with those of Yamaura et al
(13). To some extent, these
variations in miRNA stability in body fluids may be due to
differences between species.
By differential ultracentrifugation, the extent to
which lymph miRNAs are associated with exosomes was examined. Lymph
miR-150 was detected in the microsomal fraction (~16% of the
miRNA in the cell-containing fraction; Fig. 4), thus suggesting that lymph miRNAs
are present in exosomes. Notably, lymph miR-150 was also
detectable in vesicle-poor cytosol fractions (Fig. 4), thus suggesting that lymph miRNAs
are also present in vesicle-free forms. Therefore, the results from
differential ultracentrifugation suggested that at least two
populations of cell-free miRNAs exist in lymph: Exosomal miRNAs and
vesicle-free miRNAs. Previous studies have indicated that plasma
miRNAs consist of two forms: Extracellular vesicle-associated
miRNAs (14,15) and vesicle-free miRNAs (16,17).
Arroyo et al (16) and
Turchinovich et al (17)
demonstrated that vesicle-free miRNAs were associated with EIF2C2
(also known as protein argonaute-2), a key component of the
RNA-induced silencing complex (18), and were resistant to RNase activity
in plasma by EIF2C2 complexes. Vickers et al (19) demonstrated that high-density
lipoprotein (HDL) had the capacity to bind and deliver plasma
miRNAs to recipient cells with functional targeting capabilities.
HDL is included in mesenteric lymph (20,21)
and is involved in the process of reverse cholesterol transport,
whereby cholesterol is transported from the intestine to the blood
and liver via mesenteric lymphatics (22). Unfortunately, it was not possible
to prepare and investigate the highly purified fraction of HDL from
rat lymph in the present study. Although further biochemical
characterization studies of lymph miRNAs are required, it is
possible that miRNA-binding proteins, such as EIF2C2 and HDL,
prevent lymph miRNA degradation by RNase and transfer lymph miRNAs
to distant organs.
Exosomes (30–100 nm), which are endosomal
membrane-originating extracellular vesicles, are able to transfer
from the cell of origin to adjacent or distant cells, and influence
biological functions of the recipient cells (23). As aforementioned, lymph
fractionation analysis revealed that the cell-free lymph
miR-150 was present in the exosome-containing microsomal
fraction (Fig. 4). Therefore, the
in vivo delivery of lymph miRNAs via mesenteric lymphatics
into the systemic circulation was evaluated. To the best of our
knowledge, the present study is the first to demonstrate the in
vivo delivery of intralymphatically administered exosomes. The
sites where IEC-6 cel-miR-238-3p-exosomes were captured in
the systemic circulation via mesenteric lymphatics were the lung,
liver, kidney and spleen (Fig. 5).
This is consistent with findings demonstrating the in vivo
tissue distribution of intravenously administered exosomes in mice
(24,25). Morishita et al (25) quantitatively evaluated the tissue
distribution of intravenously administered B16-BL6 (murine melanoma
cell line) exosomes, and demonstrated that B16-BL6 exosomes had
rapid clearance in vivo (a half-life of 1.5 min) and were
distributed mainly in the liver, followed by the lung and spleen.
Lai et al (24)
demonstrated that human embryonic kidney 293T cell-derived
extracellular vesicles, including exosomes, were predominantly
distributed in the spleen followed by the liver, then the lungs and
kidney following intravenous injection of exosomes in mice. It is
likely that, in addition to the liver and spleen, lungs contribute
to the accumulation of intravenously administered exosomes in mice.
However, there was a difference in the levels of captured exosomes
in organs between intralymphatic and intravenous administration of
exogenous exosomes. In the present study, the levels of captured
IEC-6 exosomes were significantly higher in the lung than the liver
and spleen following intralymphatic injection of IEC-6 exosomes
(Fig. 5). Mesenteric lymph enters
the blood circulation via the subclavian vein; therefore, the lungs
may serve as the first and most effective filter to trap lymph
exosomes prior to re-entering the systemic circulation. However,
this is unlikely, since the lung also serves as the first filter
for intravenous administration via the tail vein leading to the
inferior vena cava.
Exosomes differ in their cellular origins. It is
possible that exosomes interact with lymph components and form
aggregates prior to entering the blood circulation, which would
cause the marked accumulation of IEC-6 exosomes in the lung.
Another explanation may be that the differences in tissue
distribution of exosomes are due to differences in cell
type-specific cell surface properties of exosomes and recipient
cells (26,27). Notably, Imai et al (28) demonstrated that intravenously
administered B16-BL6 exosomes were taken up by endothelial cells
but not macrophages in the lung. Peinado et al (29) investigated the function of
melanoma-derived exosomes in the formation of primary tumors and
metastases in mice and humans. They analyzed the tissue
distribution of exosomes from a highly metastatic mouse melanoma
cell line (B16-F10) following intravenous injection in mice and
detected B16-F10 exosomes in the lung 24 h after injection.
Furthermore, they performed gene expression profiling of the lung
tissue 24 and 48 h following intravenous injection and discovered
~130 differentially expressed genes in mice injected with B16-F10
exosomes as compared with control particle-injected mice, thus
suggesting that circulating exosomes transfer into the lung and
modulate genes within recipient cells in the lung. Further studies
are required to identify the types of cells that receive IEC-6
exosomes in the lung and to determine whether exosomal lymph miRNAs
modulate target genes within the recipient cells.
In the context of molecular pathogenesis of
post-shock acute lung injury, as mentioned in the introduction,
mesenteric lymph has been considered an important source of factors
that are delivered into the lung and lead to lung dysfunction
(3,4). Candidates for toxic mediators (e.g.,
fatty acids and proteins) in mesenteric lymph have been reported to
participate in the pathogenesis of post-shock acute lung injury
(30–33). In addition to these candidates,
lymph miRNAs may also be toxic mediators that result in post-shock
acute lung injury. At present, a miRNA study of post-shock
mesenteric lymph is in progress to address this possibility.
Acknowledgments
The authors would like to thank Mr. Takuji Kosuge
(Department of Molecular Medicine and Anatomy, Nippon Medical
School, Tokyo, Japan) and Mr. Takayuki Asakura (Department of
Emergency and Critical Care Medicine, Nippon Medical School) for
their technical assistance. The present study was supported by
Grants-in-Aid for Scientific Research (grant no. 24390383 and
26670610 to T.T.) from the Ministry of Education, Culture, Sports,
Science and Technology (MEXT)/Japan Society for the Promotion of
Science, Japan, and the MEXT-Support Program for the Strategic
Research Foundation at Private Universities, 2013–2017 (grant no.
S1311022 to T.T.).
References
|
1
|
Fanous MY, Phillips AJ and Windsor JA:
Mesenteric lymph: The bridge to future management of critical
illness. JOP. 8:374–399. 2007.PubMed/NCBI
|
|
2
|
Barrowman JA and Tso P: Gastrointestinal
lymphaticsHandbook of physiology. Wood JD: American Physiological
Society; Bethesda MD: pp. 1733–1777. 1989
|
|
3
|
Magnotti LJ, Upperman JS, Xu DZ, Lu Q and
Deitch EA: Gut-derived mesenteric lymph but not portal blood
increases endothelial cell permeability and promotes lung injury
after hemorrhagic shock. Ann Surg. 228:518–527. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Deitch EA: Gut lymph and lymphatics: A
source of factors leading to organ injury and dysfunction. Ann N Y
Acad Sci. 1207 Suppl 1:E103–E111. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Weber JA, Baxter DH, Zhang S, Huang DY,
Huang KH, Lee MJ, Galas DJ and Wang K: The microRNA spectrum in 12
body fluids. Clin Chem. 56:1733–1741. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Masuno T, Moore EE, Cheng AM, Sarin EL and
Banerjee A: Bioactivity of postshock mesenteric lymph depends on
the depth and duration of hemorrhagic shock. Shock. 26:285–289.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Takeuchi J, Sakamoto A and Takizawa T:
Sevoflurane anesthesia persistently downregulates muscle-specific
microRNAs in rat plasma. Int J Mol Med. 34:291–298. 2014.PubMed/NCBI
|
|
9
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Thery C, Amigorena S, Raposo G and Clayton
A: Isolation and characterization of exosomes from cell culture
supernatants and biological fluids. Curr Protoc Cell Biol Chapter.
3:Unit 3.222006.
|
|
11
|
Kambe S, Yoshitake H, Yuge K, Ishida Y,
Ali MM, Takizawa T, Kuwata T, Ohkuchi A, Matsubara S, Suzuki M, et
al: Human exosomal placenta-associated miR-517a-3p modulates the
expression of PRKG1 mRNA in Jurkat cells. Biol Reprod. 91:1292014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Blenkiron C, Askelund KJ, Shanbhag ST,
Chakraborty M, Petrov MS, Delahunt B, Windsor JA and Phillips AR:
MicroRNAs in mesenteric lymph and plasma during acute pancreatitis.
Ann Surg. 260:341–347. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yamaura Y, Nakajima M, Takagi S, Fukami T,
Tsuneyama K and Yokoi T: Plasma microRNA profiles in rat models of
hepatocellular injury, cholestasis, and steatosis. PLoS One.
7:e302502012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kosaka N, Iguchi H, Yoshioka Y, Takeshita
F, Matsuki Y and Ochiya T: Secretory mechanisms and intercellular
transfer of microRNAs in living cells. J Biol Chem.
285:17442–17452. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Arroyo JD, Chevillet JR, Kroh EM, Ruf IK,
Pritchard CC, Gibson DF, Mitchell PS, Bennett CF,
Pogosova-Agadjanyan EL and Stirewalt DL: Argonaute2 complexes carry
a population of circulating microRNAs independent of vesicles in
human plasma. Proc Natl Acad Sci USA. 108:5003–5008. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Turchinovich A, Weiz L, Langheinz A and
Burwinkel B: Characterization of extracellular circulating
microRNA. Nucleic Acids Res. 39:7223–7233. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu J, Carmell MA, Rivas FV, Marsden CG,
Thomson JM, Song JJ, Hammond SM, Joshua-Tor L and Hannon GJ:
Argonaute2 is the catalytic engine of mammalian RNAi. Science.
305:1437–1441. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Vickers KC, Palmisano BT, Shoucri BM,
Shamburek RD and Remaley AT: MicroRNAs are transported in plasma
and delivered to recipient cells by high-density lipoproteins. Nat
Cell Biol. 13:423–433. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Green PH, Tall AR and Glickman RM: Rat
intestine secretes discoid high density lipoprotein. J Clin Invest.
61:528–534. 1978. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Forester GP, Tall AR, Bisgaier CL and
Glickman RM: Rat intestine secretes spherical high density
lipoproteins. J Biol Chem. 258:5938–5943. 1983.PubMed/NCBI
|
|
22
|
Randolph GJ and Miller NE: Lymphatic
transport of high-density lipoproteins and chylomicrons. J Clin
Invest. 124:929–935. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Camussi G, Deregibus MC, Bruno S,
Cantaluppi V and Biancone L: Exosomes/microvesicles as a mechanism
of cell-to-cell communication. Kidney Int. 78:838–848. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lai CP, Mardini O, Ericsson M, Prabhakar
S, Maguire CA, Chen JW, Tannous BA and Breakefield XO: Dynamic
biodistribution of extracellular vesicles in vivo using a
multimodal imaging reporter. ACS Nano. 8:483–494. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Morishita M, Takahashi Y, Nishikawa M,
Sano K, Kato K, Yamashita T, Imai T, Saji H and Takakura Y:
Quantitative analysis of tissue distribution of the B16BL6-derived
exosomes using a streptavidin-lactadherin fusion protein and
iodine-125-labeled biotin derivative after intravenous injection in
mice. J Pharm Sci. 104:705–713. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Simpson RJ, Jensen SS and Lim JW:
Proteomic profiling of exosomes: Current perspectives. Proteomics.
8:4083–4099. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Katsuda T, Kosaka N and Ochiya T: The
roles of extracellular vesicles in cancer biology: Toward the
development of novel cancer biomarkers. Proteomics. 14:412–425.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Imai T, Takahashi Y, Nishikawa M, Kato K,
Morishita M, Yamashita T, Matsumoto A, Charoenviriyakul C and
Takakura Y: Macrophage-dependent clearance of systemically
administered B16BL6-derived exosomes from the blood circulation in
mice. J Extracell Vesicles. 4:262382015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Peinado H, Alečković M, Lavotshkin S,
Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M,
Williams C, García-Santos G, Ghajar C, et al: Melanoma exosomes
educate bone marrow progenitor cells toward a pro-metastatic
phenotype through MET. Nat Med. 18:883–891. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gonzalez RJ, Moore EE, Biffl WL, Ciesla DJ
and Silliman CC: The lipid fraction of post-hemorrhagic shock
mesenteric lymph (PHSML) inhibits neutrophil apoptosis and enhances
cytotoxic potential. Shock. 14:404–408. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kaiser VL, Sifri ZC, Dikdan GS, Berezina
T, Zaets S, Lu Q, Xu DZ and Deitch EA: Trauma-hemorrhagic shock
mesenteric lymph from rat contains a modified form of albumin that
is implicated in endothelial cell toxicity. Shock. 23:417–425.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Penn AH and Schmid-Schönbein GW: The
intestine as source of cytotoxic mediators in shock: Free fatty
acids and degradation of lipid-binding proteins. Am J Physiol Heart
Circ Physiol. 294:H1779–H1792. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jordan JR, Moore EE, Sarin EL, Damle SS,
Kashuk SB, Silliman CC and Banerjee A: Arachidonic acid in
postshock mesenteric lymph induces pulmonary synthesis of
leukotriene B4. J Appl Physiol (1985). 104:1161–1166. 2008.
View Article : Google Scholar : PubMed/NCBI
|