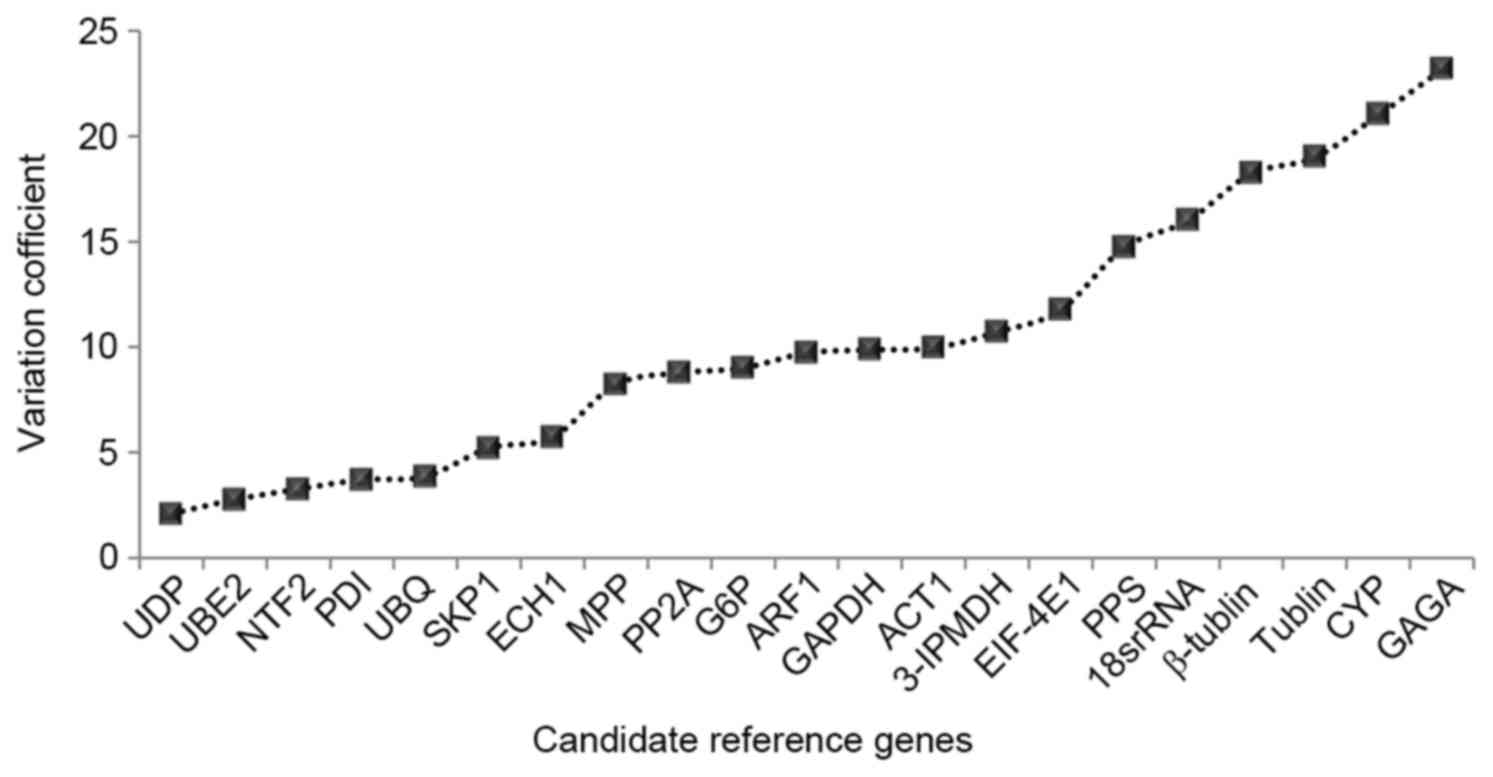
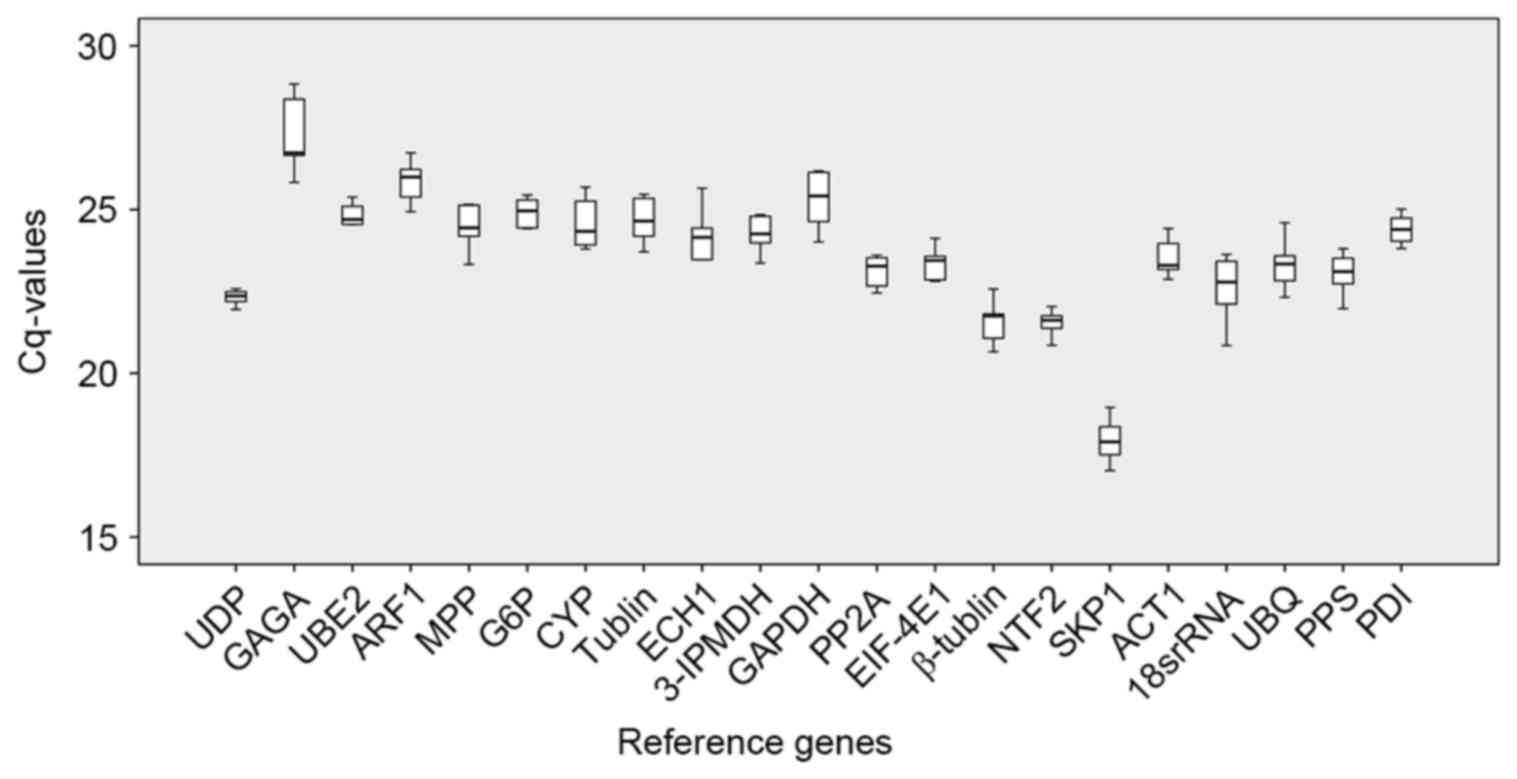
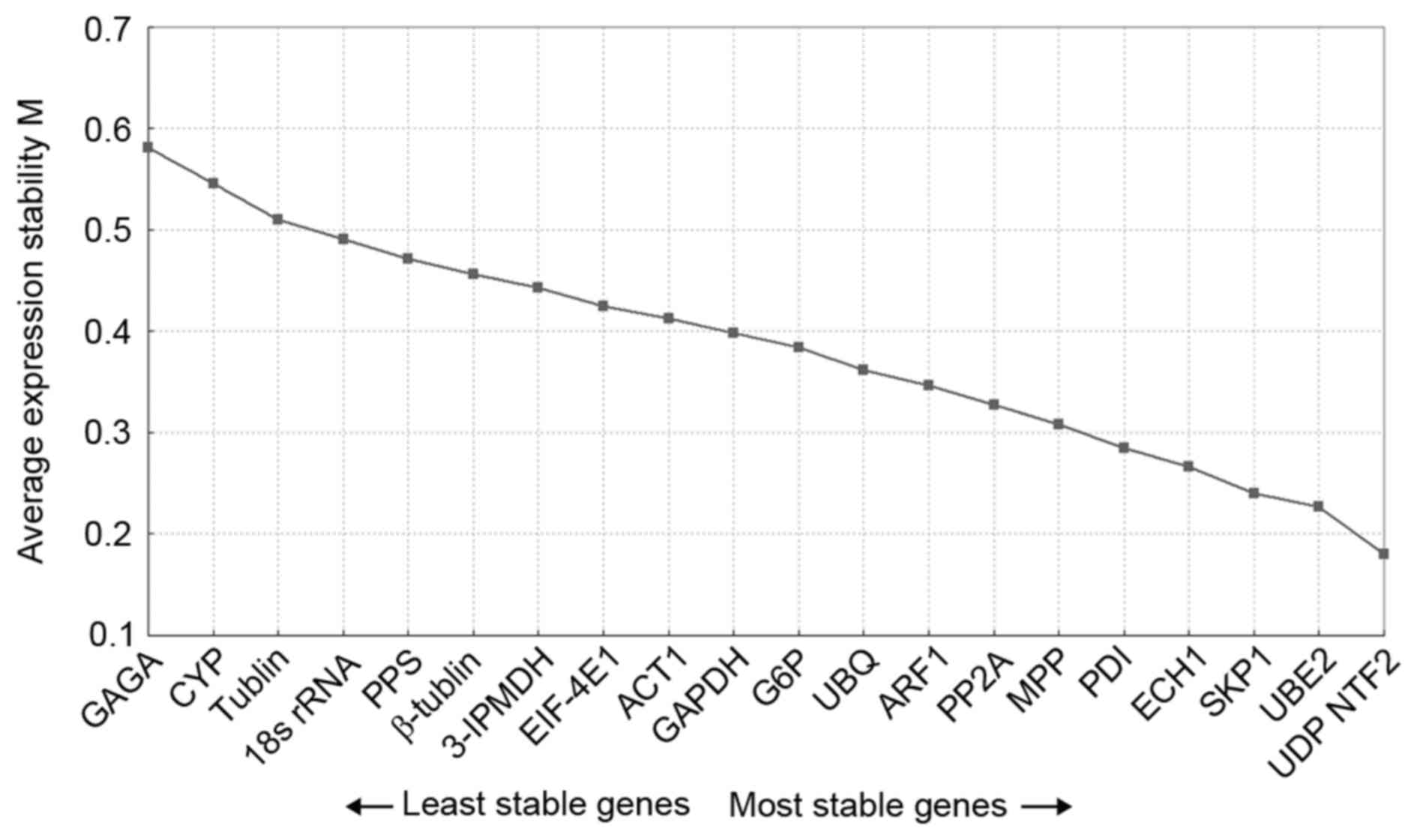
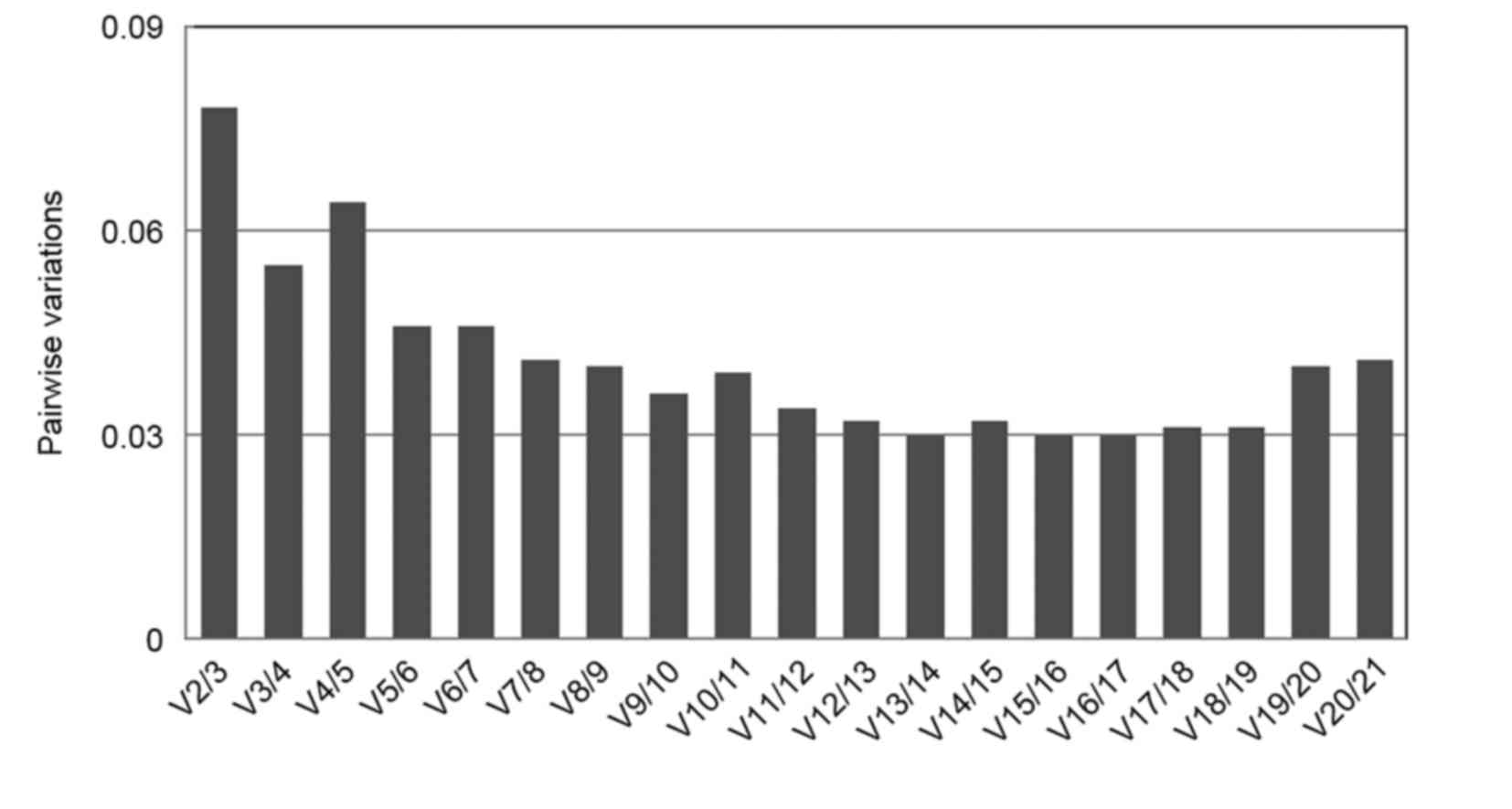
|
1
|
Xiang YZ, Shang HC, Gao XM and Zhang BL: A
comparison of the ancient use of ginseng in traditional chinese
medicine with modern pharmacological experiments and clinical
trials. Phytother Res. 22:851–858. 2008. View Article : Google Scholar
|
|
2
|
James AD: The Green Pharmacy Herbal
Handbook: Your comprehensive reference to the best herbs for
healing. Rodale: Emmaus, USA. 115–116. 2000.
|
|
3
|
Dan M, Xie G, Gao X, Long X, Su M, Zhao A,
Zhao T, Zhou M, Qiu Y and Jia W: A rapid ultra-performance liquid
chromatography-electrospray Ionisation mass spectrometric method
for the analysis of saponins in the adventitious roots of Panax
notoginseng. Phytochem Anal. 20:68–76. 2009. View Article : Google Scholar
|
|
4
|
Shan SM, Luo JG, Huang F and Kong LY:
Chemical characteristics combined with bioactivity for
comprehensive evaluation of Panax ginseng C.A. Meyer in different
ages and seasons based on HPLC-DAD and chemometric methods. J Pharm
Biomed Anal. 89:76–82. 2014. View Article : Google Scholar
|
|
5
|
Wan JY, Fan Y, Yu QT, Ge YZ, Yan CP,
Alolga RN, Li P, Ma ZH and Qi LW: Integrated evaluation of malonyl
ginsenosides, amino acids and polysaccharides in fresn and
processed ginseng. J Pharm Biomed Anal. 107:89–97. 2015. View Article : Google Scholar
|
|
6
|
He JM, Zhang YZ, Luo JP, Zhang WJ and Mu
Q: Variation of ginsenoside in ginseng of different ages. Nat Prod
Commun. 11:739–740. 2016.
|
|
7
|
Sathiyaraj G, Srinivasan S, Subramanium S,
Kim YJ, Kim YJ, Kwon WS and Yang DC: Polygalacturonase inhibiting
protein: Isolation, developmental regulation and pathogen related
expression in Panax ginseng C.A. Meyer. Mol Biol Rep. 37:3445–3454.
2010. View Article : Google Scholar
|
|
8
|
Han JY, Kim HJ, Kwon YS and Choi YE: The
Cyt P450 enzyme CYP716A47 catalyzes the formation of
protopanaxadiol from dammarenediol-II duringginsenoside
biosynthesis in Panax ginseng. Plant Cell Physiol. 52:2062–2073.
2011. View Article : Google Scholar
|
|
9
|
Bustin SA: Quantification of mRNA using
real-time reverse transcription PCR (RT-PCR): Trends and problems.
J Mol Endocrinol. 29:23–39. 2002. View Article : Google Scholar
|
|
10
|
Thellin O, Zorzi W, Lakaye B, De Borman B,
Coumans B, Hennen G, Grisar T, Igout A and Heinen E: Housekeeping
genes as internal standards: Use and limits. J Biotechnol.
75:291–295. 1999. View Article : Google Scholar
|
|
11
|
Bustin SA: Absolute quantification of mRNA
using real-time reverse transcription polymerase chain reaction
assays. J Mol Endocrinol. 25:169–193. 2000. View Article : Google Scholar
|
|
12
|
Schmittgen TD and Zakrajsek BA: Effect of
experimental treatment on housekeeping gene expression: Validation
by real-time, quantitative RT-PCR. J Biochem Biophys Methods.
46:69–81. 2000. View Article : Google Scholar
|
|
13
|
Lee PD, Sladek R, Greenwood CM and Hudson
TJ: Control genes and variability: Absence of ubiquitous reference
transcripts in diverse mammalian expression studies. Genome Res.
12:292–297. 2002. View Article : Google Scholar :
|
|
14
|
Huang Z, Lin J, Cheng Z, Xu M, Guo M,
Huang X, Yang Z and Zheng J: Production of oleanane-type sapogenin
in transgenic rice via expression of β-amyrin synthase gene from
Panax japonicas C. A. Mey. BMC Biotechnol. 15:452015. View Article : Google Scholar :
|
|
15
|
Lim W, Shim MK, Kim S and Lee Y: Red
ginseng represses hypoxia-induced cyclooxygenase-2 through sirtuin1
activation. Phytomedicine. 22:597–604. 2015. View Article : Google Scholar
|
|
16
|
Oh GS, Yoon J, Lee GG, Oh WK and Kim SW:
20 (S)-protopanaxatriol inhibits liver X receptor α-mediated
expression of lipogenic genes in hepatocytes. J Pharmacol Sci.
128:71–77. 2015. View Article : Google Scholar
|
|
17
|
Qi J, Sun P, Liao D, Sun T, Zhu J and Li
X: Transcriptomic analysis of american ginseng seeds during the
dormancy release process by RNA-Seq. PLoS One. 10:e01185582015.
View Article : Google Scholar :
|
|
18
|
Zhu L, Li J, Xing N, Han D, Kuang H and Ge
P: American ginseng regulates gene expression to protect against
premature ovarian failure in rats. Biomed Res Int. 2015:7671242015.
View Article : Google Scholar :
|
|
19
|
Liu J, Wang Q, Sun M, Zhu L, Yang M and
Zhao Y: Selection of reference genes for quantitative real-time PCR
normalization in Panax ginseng at different stages of growth and in
different organs. PLoS One. 9:e1121772014. View Article : Google Scholar :
|
|
20
|
Mortazavi A, Williams BA, McCue K,
Schaeffer L and Wold B: Mapping and quantifying mammalian
transcriptomes by RNA-Seq. Nat Methods. 5:621–628. 2008. View Article : Google Scholar
|
|
21
|
Wang Z, Gerstein M and Snyder M: RNA-Seq:
A revolutionary tool for transcriptomics. Nat Rev Genet. 10:57–63.
2009. View
Article : Google Scholar :
|
|
22
|
Macrae T, Sargeant T, Lemieux S, Hébert J,
Deneault E and Sauvageau G: RNA-Seq reveals spliceosome and
proteasome genes as most consistent transcripts in human cancer
cells. PLoS One. 8:e728842013. View Article : Google Scholar :
|
|
23
|
Cankorur-Cetinkaya A, Dereli E, Eraslan S,
Karabekmez E, Dikicioglu D and Kirdar B: A novel strategy for
selection and validation of reference genes in dynamic
multidimensional experimental design in yeast. PLoS One.
7:e383512012. View Article : Google Scholar :
|
|
24
|
de Jonge HJ, Fehrmann RS, de Bont ES,
Hofstra RM, Gerbens F, Kamps WA, de Vries EG, van der Zee AG, te
Meerman GJ and ter Elst A: Evidence based selection of housekeeping
genes. PLoS One. 2:e8982007. View Article : Google Scholar :
|
|
25
|
Vandesompele J, De Preter K, Pattyn F,
Poppe B, Van Roy N, De Paepe A and Speleman F: Accurate
normalization of real-time quantitative RT-PCR data by geometric
averaging of multiple internal control genes. Genome Biol.
3:research0034. 2002. View Article : Google Scholar :
|
|
26
|
Andersen CL, Jensen JL and Ørntoft TF:
Normalization of real-time quantitative reverse transcription-PCR
data: A model-based variance estimation approach to identify genes
suited for normalization, applied to bladder and colon cancer data
sets. Cancer Res. 64:5245–5250. 2004. View Article : Google Scholar
|
|
27
|
Pfaffl MW, Tichopad A, Prgomet C and
Neuvians TP: Determination of stable housekeeping genes,
differentially regulated target genes and sample integrity:
BestKeeper--Excel-based tool using pair-wise correlations.
Biotechnol Lett. 26:509–515. 2004. View Article : Google Scholar
|
|
28
|
Yao B, Zhao Y, Zhang H, Zhang M, Liu M,
Liu H and Li J: Sequencing and de novo analysis of the Chinese Sika
deer antler-tip transcriptome during the ossification stage using
Illumina RNA-Seq technology. Biotechnol Lett. 34:813–822. 2012.
View Article : Google Scholar
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
30
|
Rozen S and Skaletsky H: Primer3 on the
WWW for general users and for biologist programmers. Methods Mol
Biol. 132:365–386. 2000.
|
|
31
|
Bustin SA, Benes V, Garson JA, Hellemans
J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL,
et al: The MIQE guidelines: Minimum information for publication of
quantitative real-time PCR experiments. Clin Chem. 55:611–622.
2009. View Article : Google Scholar
|
|
32
|
Pombo-Suarez M, Calaza M, Gomez-Reino JJ
and Gonzalez A: Reference genes for normalization of gene
expression studies in human osteoarthritic articular cartilage. BMC
Mol Biol. 9:172008. View Article : Google Scholar :
|
|
33
|
Warzybok A and Migocka M: Reliable
reference genes for normalization of gene expression in cucumber
grown under different nitrogen nutrition. PLoS One. 8:e728872013.
View Article : Google Scholar :
|
|
34
|
Gibbs GM, Roelants K and O'Bryan MK: The
CAP superfamily: Cysteine-rich secretory proteins, antigen 5, and
pathogenesis-related 1 proteins-roles in reproduction, cancer and
immune defense. Endocr Rev. 29:865–897. 2008. View Article : Google Scholar
|
|
35
|
Sels J, Mathys J, De Coninck BM, Cammue BP
and De Bolle MF: Plant pathogenesis-related (PR) proteins: A focus
on PR peptides. Plant Physiol Biochem. 46:941–950. 2008. View Article : Google Scholar
|
|
36
|
Etschmann B, Wilcken B, Stoevesand K, von
der Schulenburg A and Sterner-Kock A: Selection of reference genes
for quantitative real-time PCR analysis in canine mammary tumors
using the GeNorm algorithm. Vet Pathol. 43:934–942. 2006.
View Article : Google Scholar
|
|
37
|
Huis R, Hawkins S and Neutelings G:
Selection of reference genes for quantitative gene expression
normalization in flax (Linum usitatissimum L.). BMC Plant Biol.
10:712010. View Article : Google Scholar :
|
|
38
|
Hamalainen HK, Tubman JC, Vikman S, Kyrölä
T, Ylikoski E, Warrington JA and Lahesmaa R: Identification and
validation of endogenous reference genes for expression profiling
of T helper cell differentiation by quantitative real-time RT-PCR.
Anal Biochem. 299:63–70. 2001. View Article : Google Scholar
|
|
39
|
Oshlack A, Robinson MD and Young MD: From
RNA-seq reads to differential expression results. Genome Biol.
11:2202010. View Article : Google Scholar :
|
|
40
|
Mane SP, Evans C, Cooper KL, Crasta OR,
Folkerts O, Hutchison SK, Harkins TT, Thierry-Mieg D, Thierry-Mieg
J and Jensen RV: Transcriptome sequencing of the microarray quality
control (MAQC) RNA reference samples using next generation
sequencing. BMC Genomics. 10:2642009. View Article : Google Scholar :
|
|
41
|
Wilhelm BT and Landry JR:
RNA-Seq-quantitative measurement of expression through massively
parallel RNA-sequencing. Methods. 48:249–257. 2009. View Article : Google Scholar
|
|
42
|
Shendure J: The beginning of the end for
microarrays? Nat Methods. 5:585–587. 2008. View Article : Google Scholar
|
|
43
|
Jian B, Liu B, Bi Y, Hou W, Wu C and Han
T: Validation of internal control for gene expression study in
soybean by quantitative real-time PCR. BMC Mol Biol. 9:592008.
View Article : Google Scholar :
|
|
44
|
Muraoka M, Kawakita M and Ishida N:
Molecular characterization of human UDP-glucuronic
acid/UDP-N-acetylgalactosamine transporter, a novel nucleotide
sugar transporter with dual substrate specificity. FEBS Lett.
495:87–93. 2001. View Article : Google Scholar
|
|
45
|
Bayliss R, Ribbeck K, Akin D, Kent HM,
Feldherr CM, Görlich D and Stewart M: Interaction between NTF2 and
xFxFG-containing nucleoporins is required to mediate nuclear import
of RanGDP. J Mol Biol. 293:579–593. 1999. View Article : Google Scholar
|
|
46
|
Goepfert S, Vidoudez C, Rezzonico E,
Hiltunen JK and Poirier Y: Molecular identification and
characterization of the Arabidopsis delta(3,5),
delta(2,4)-dienoyl-coenzyme A isomerase, a peroxisomal enzyme
participating in the beta-oxidation cycle of unsaturated fatty
acids. Plant Physiol. 138:1947–1956. 2005. View Article : Google Scholar :
|
|
47
|
Czechowski T, Stitt M, Altmann T, Udvardi
MK and Scheible WR: Genome-wide identification and testing of
superior reference genes for transcript normalization in
Arabidopsis. Plant Physiol. 139:5–17. 2005. View Article : Google Scholar :
|
|
48
|
Goidin D, Mamessier A, Staquet MJ, Schmitt
D and Berthier-Vergnes O: Ribosomal 18S RNA prevails over
glyceraldehyde-3-phosphate dehydrogenase and beta-actin genes as
internal standard for quantitative comparison of mRNA levels in
invasive and noninvasive human melanoma cell subpopulations. Anal
Biochem. 295:17–21. 2001. View Article : Google Scholar
|
|
49
|
Selvey S, Thompson EW, Matthaei K, Lea RA,
Irving MG and Griffiths LR: Beta-actin: An unsuitable internal
control for RT-PCR. Mol Cell Probes. 15:307–311. 2001. View Article : Google Scholar
|
|
50
|
Warrington JA, Nair A, Mahadevappa M and
Tsyganskaya M: Comparison of human adult and fetal expression and
identification of 535 housekeeping/maintenance genes. Physiol
Genomics. 2:143–147. 2000.
|