Introduction
Low back pain is mainly caused by degenerative disc
disease (DDD) (1). A number of
pathological factors may be responsible for DDD, such as apoptosis,
inflammation and cartilage matrix degradation (2–4);
metabolic exchange reduction, which results in nutrition
insufficiency and waste accumulation, is considered to be the most
important of these mechanisms (5).
The intervertebral disc (IVD) is an avascular organ, and metabolic
exchange primarily depends on the diffusion of cartilage endplate
(CEP) in mature IVDs (6). The CEP
is a horizontal, thin layer of hyaline cartilage, which separates
the IVD from the vertebrae. Blood vessels in adjacent vertebrae end
at the IVD-vertebra interface without reaching the inner component
of the IVD (7). CEP is the most
important channel of metabolic substance exchange, and its
degeneration is considered to be the origination of DDD (8).
Our previous study reported the existence of
CEP-derived stem cells (CESCs), which exhibited the ability for
both chondrogenic and osteogenic differentiation (9). This differentiation potential enable
CESCs to serve important roles in the restoration and regeneration
of cartilage, and the differentiation status of CESCs may be
responsible for CEP degeneration.
As an avascular organ, the IVD remains in a
physiologically hypoxic microenvironment (10). Hypoxia has been reported to
regulate the biological activities of mesenchymal stem cells
(MSCs), such as apoptosis, proliferation and differentiation
(11,12), which indicated that physiological
hypoxia may serve vital roles in regulating the physiological
functions of CESCs.
Data from our previous study on the effects of
hypoxia on the biological activities of CESCs indicated that
alternative splicing (AS) may serve as a ‘middle link’ between
them, through which hypoxia may regulate the biological activities
of CESCs (9). This assumption may
be due to two reasons. First, several AS events may be
hypoxia-inducible; for example, inhibitory PAS domain protein
generated unique splicing variants of exon 6 and exon 3 exclusion
in a hypoxic microenvironment, which subsequently formed a new
negative-feedback modulation mode of adaptive responses to ischemia
(13). In addition, a spliced
variant of tyrosine kinase receptor A (TrkA), TrkA-III, was
demonstrated to be induced by hypoxia (14), and the ability of this spliced
variant to form stress-resistant and angiogenic isoforms suggested
its potential roles in the protective effects against hypoxia
(14). Second, the roles of AS in
stem cell fate also have been reported previously. For example, the
variant of transcription initiation factor TFIID subunit 4 (TAF4)
that lacks the human TAF4-TAF homology domain has been demonstrated
to lead to the promotion of chondrogenic differentiation in human
MSCs (15). A conserved embryonic
stem cell-specific AS event was previously identified that altered
the DNA-binding activity of forkhead box P1, which indicated that
the pluripotency and reprogramming of embryonic stem cells may be
regulated (16).
High-throughput screening is a powerful technology
for investigating the changes in the transcription profile and AS
genome-wide. A previous study used exon microarrays to examine the
gene expression profiles and AS events in human umbilical vein
endothelial cells under hypoxic conditions (17). Another study used
microarray-screening technology to identify a hypoxia-induced
splice variant, laminin subunit α3, which was closely associated
with poor prognosis in patients with head and neck cancers
(18). Our previous studies
revealed gene expression profiles and AS events during the process
of chondrogenesis in CESCs under hypoxia and normoxia (19,20);
however, high-throughput analysis of the general regulatory effects
of hypoxia on the various aspects of CESCs biological function
(without the induction of differentiation) has not yet been
reported. In the present study, isolated CESCs were cultured in
normal growth medium under normoxic and hypoxic conditions. RNA was
extracted, purified and analyzed using the Affymetrix Human
Transcriptome Array 2.0 System, and a comparative analysis of gene
expressions and AS events between the normoxic and hypoxic groups
was conducted. Gene Ontology (GO) and Kyoto Encyclopedia of Genes
and Genomes (KEGG) pathway analysis were used for the functional
enrichment of genes of interest to explain the possible mechanisms
of transcription and alternative splicing. Data from the present
study may aid in identifying the roles of the physiological hypoxic
microenvironment on the fate of CESCs, which may be beneficial to
the understanding the pathological mechanisms of DDD and may lead
to the development of new therapies.
Materials and methods
Patients and subjects
CEP samples were obtained from three patients that
underwent discectomy operations at the Xinqiao Hospital, Third
Military Medical University (Chongqing, China; Table I). All procedures in the present
study were approved by the Ethics Committee of Xinqiao Hospital,
Third Military Medical University and were conducted in accordance
with the Declaration of Helsinki; written informed consent was
obtained from each patient.
 | Table I.Patient information. |
Table I.
Patient information.
| Case | Sex | Age (years) | Diagnosis | Degenerated disc
level | Surgery |
|---|
| 1 | Male | 50 |
Spondylolisthesis | L4-L5 | TLIF |
| 2 | Male | 55 |
Spondylolisthesis | L4-L5 | TLIF |
| 3 | Female | 52 |
Spondylolisthesis | L4-L5 | TLIF |
CESC isolation and culture
CEP samples were cut into pieces and digested in
Dulbecco's modified Eagle's medium (DMEM)/Ham's F12 medium
(Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) containing
1% fetal calf serum (FCS; Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) and 0.2% collagenase II (Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany) for 12 h at 37°C. The suspension was
passed through a cell filter (70 µm) and centrifuged at 110 × g for
5 min at room temperature. Following removal of the supernatant,
the pellet was resuspended in 3 ml DMEM/F12 containing 10% FCS and
1% penicillin-streptomycin (Hyclone; GE Healthcare Life Sciences).
The cells were then cultured in a 25-cm2 cell-culture
flask at 37°C and 5% CO2 for 6 weeks. Cells were
subjected to agarose selection following the first passage of
expansion, as previously described (9). Briefly, Costar culture dishes
(Corning Inc., Corning, NY, USA) were pre-coated with 1% low
melting-point agarose, and a mixture containing DMEM/F12 (0.5 ml),
2% low melting-point agarose (0.5 ml) and 5×104
suspended CEP cells in culture medium (1 ml) was transferred to the
culture dishes and incubated in humidified atmosphere in 5%
CO2 and 37°C. The culture medium was changed twice per
week and, 6 weeks later, cell aggregates were carefully aspirated
with a sterile Pasteur pipette and cultured in a 25 cm2
cell culture flask in 5% CO2 and 37°C for 2 weeks to
reach 80–100% confluence. CESCs which were sub-cultured to the
third passage, after ~6 weeks of culture, were used.
Oxygen deprivation
For hypoxic culture conditions, CESCs at a density
of 2.5×104/ml were cultured in a 1% O2
microenvironment at 37°C. For normoxic culture conditions, CESCs
were cultured in a 21% O2 microenvironment at 37°C. The
culture medium was changed twice per week over a 21-day period.
GeneChip human transcriptome array
(HTA) 2.0
Total RNA was extracted from cells
(~5×104) using TRIzol (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. RNA was
purified with an RNeasy kit (Qiagen GmbH, Hilden, Germany)
according to the manufacturer's protocol. The RNA purity was
determined by the ratio of optical density (OD) value in 260 nm/OD
value in 280 nm using a NanoDrop spectrophotometer (NanoDrop
Technologies; Thermo Fisher Scientific, Inc., Wilmington, DE, USA).
RNA quality was assessed by 1% agarose gel electrophoresis. The
samples with a bright band of ribosomal 28S/18S RNA at a ratio
>1.5:1 were used for the microarray analysis. For gene
expression profiling, RNA was hybridized using the GeneChip HTA 2.0
system (Affymetrix; Thermo Fisher Scientific, Inc.). A total of 100
ng RNA was used to generate biotinylated and amplified sense-strand
cDNA from the whole expressed genome using GeneChip WT PLUS Reagent
kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) according
to the manufacturer's protocol. GeneChip HTA 2.0 cartridges were
hybridized in a 45°C incubator (GeneChip Hybridization Oven 640;
Thermo Fisher Scientific, Inc.) for 16 h under rotation at 60 rpm.
After the hybridization, the microarrays were stained using the
GeneChip Fluidics Station 450 (Thermo Fisher Scientific, Inc.) and
screened using the GeneChip Scanner 3000 7G (Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. Gene
probes of the HTA 2.0 target exons and junctions and are able to
detect gene expression and AS events simultaneously. Microarray
screening was performed by CapitalBio Corporation (Beijing, China)
according to the manufacturer's protocol. Image signals of the
probes were obtained and saved as DAT files. The GeneChip Command
Console (AGCC) software version 3.2 (Affymetrix; Thermo Fisher
Scientific, Inc.) was used to convert the image signals (DAT files)
into digital signals (CEL files), which were converted to CHP files
for probeset signal integration, quantile normalization and
background correction through the robust multichip analysis (RMA)
algorithm using the Affymetrix Expression Console software version
1.3 (Affymetrix; Thermo Fisher Scientific, Inc.). The CHP files
were then analyzed by Affymetrix Transcriptome Analysis Console
software version 3.0 (Affymetrix; Thermo Fisher Scientific, Inc.)
to determine the differentially expressed genes (DEGs) and
alternatively spliced genes (ASGs). GO terms and related signaling
pathway enrichments were determined using web tools, such as DAVID
(https://david.ncifcrf.gov), Molecule
Annotation System (http://bioinfo.capitalbio.com/mas3) and KEGG
(http://www.genome.jp/kegg. The workflow
of the present study procedures was as previously described
(20). The microarray data have
been submitted to the Gene Expression Omnibus (accession no.
GSE84484).
Criteria for the detection of DEGs and
ASGs
Data from the cell samples cultured under normoxia
were used as a control to calculate the fold change in gene
expression. The default filter criteria were set as fold change ≤-2
or ≥2 (linear) for significant DEGs. For ASG identification, the
splicing index (SI) calculation mode was used to analyze the level
of exon inclusion/exclusion. SI indicates the ratio of the signal
intensity of an exon normalized to that of the target gene between
two groups. SI values were calculated using the following
formulas:
Equation1:[NI(i,j)A]=(exonisignalintensityinconditionA)/(genejsignalintensityinconditionA)
Equation2:SI(X,Y)=log2{[NI(X,Y)H/NI(X,Y)N]}
Where NI(i,j)A represents the normalized
intensity (NI) of the ith exon signal normalized to that
of the jth gene signal under condition A, N represents
the normoxic culture condition and H represents the hypoxic culture
condition. The default filter criteria were defined as SI (linear)
≤-2 or ≥2.
DEG validation by reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells
(~5×104 cells/ml) using TRIzol reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. RNA was purified with an RNeasy kit (Qiagen GmbH, Hilden,
Germany) according to the manufacturer's protocol. The RNA purity
was determined by the ratio of optical density (OD) value in 260
nm/OD value in 280 nm using a NanoDrop spectrophotometer (NanoDrop
Technologies; Thermo Fisher Scientific, Inc.). RNA quality was
assessed by 1% agarose gel electrophoresis. The samples with a
bright band of ribosomal 28S/18S RNA at a ratio >1.5:1 were
used. RNA was reverse transcribed into cDNA with the PrimeScript RT
Master Mix kit (cat no. RR047A; Takara Bio, Inc., Otsu Japan). qPCR
was performed with the ABI 7900H Real-Time PCR system and the SYBR
Premix Ex Taq II kit (cat no. RR820A; Takara Bio, Inc.) with the
following conditions: At 95°C for 30 sec, then 40 cycles at 95°C
for 5 sec and at 60°C for 34 sec; a dissociation curve analysis was
performed in a temperature range of 60–95°C. The mRNA expression
levels of each gene were normalized to the level of β-actin
expression and analyzed using the 2−∆∆Cq method
(21). Data were analyzed using
StepOne software version 2.3 (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The primers used for qPCR are listed in Table II-A.
 | Table II.List of primers used for quantitative
and semi-quantitative PCR. |
Table II.
List of primers used for quantitative
and semi-quantitative PCR.
| A, Quantitative PCR
primers for DEG validation |
|---|
|
|---|
| Gene | Primer sequence
(5′→3′) |
|---|
| VCAM1 | F:
GGACCACATCTACGCTGACA |
| VCAM1 | R:
TTGACTGTGATCGGCTTCCC |
| COMP | F:
CTTCAGGGCCTTCCAGACAG |
| COMP | R:
CGCATAGTCGTCATCCGTGA |
| FGF9 | F:
CGGCTACAACGCTCCGC |
| FGF9 | R:
TGGCGGCGACAAATCTCC |
| IRAK3 | F:
AGGATTTCCGCGGTTGTGTA |
| IRAK3 | R:
ACTCAACACTGCTCCCAGGC |
| CFD | F:
GCGGTGAGGAGGCCTGG |
| CFD | R:
GAACCTGCACCTTCCCGTC |
| TFRC | F:
CCAGGCTATAAACCGCCG |
| TFRC | R:
CCAGGCTGAACCGGGTATATG |
| PGAM2 | F:
TAGCAAGGAGCGTCGGTACG |
| PGAM2 | R:
GCCTGGTCTGACATCCCTTC |
| CAPN6 | F:
CTTGGGGCAGGTTACTCTGG |
| CAPN6 | R:
CTAAGACCTGGCACCTGACG |
| β-actin | F:
CAACCGGGAAGGAAATGAATGG |
| β-actin | R:
GCCCAATACGACCAAATCAGAG |
|
| B,
Semi-quantitative PCR primers for ASGs validation |
|
| Gene | Primer sequence
(5′→3′) |
|
| GFRA2 | F:
GGACTCGGGATCTTCATCG |
| GFRA2 | R:
AGGTCACCGGCTCATAGGG |
| CADM1 | F:
AGTCGATGATGAAATGCC |
| CADM1 | R:
CAGAATGATGAGCAAGCA |
| NR4A3 | F:
CCGCTCCTCCTACACTCT |
| NR4A3 | R:
TCCGTGGTGTATTCCGAG |
| RXFP1 | F:
AGTGCTCCCTTGGCTATT |
| RXFP1 | R:
AGAAACCGATGGAACAGC |
| NDUFC1 | F:
AACGAGAACAACGGAGGC |
| NDUFC1 | R:
AGTTGGCAACAGAACCAG |
| TMEM106B | F:
GTTATCCTACCCCTCCCC |
| TMEM106B | R:
CCAGTCCATTCCTCATGT |
| FN1 | F:
TTATAGAATTACCACAACCC |
| FN1 | R:
TTCACAGGTGAGTAACGC |
| β-actin | F:
CAACCGGGAAGGAAATGAATGG |
| β-actin | R:
GCCCAATACGACCAAATCAGAG |
ASG validation by semi-quantitative
RT-PCR
Total RNA extraction and reverse transcription of
cDNA were carried out as aforementioned. For semi-quantitative PCR,
cDNA (2 µl) was combined with Premix Taq (cat no. RR901A; Takara
Bio, Inc.) and primers that were designed flanking the
constitutively expressed exons; primers are listed in Table II-B. PCR thermocycling conditions
were as follows: At 94°C for 30 sec, then 30 cycles at 94°C for 30
sec, at 55°C for 30 sec and at 72°C for 60 sec. PCR products were
resolved on a 1.5% agarose gel, using a DNA ladder (Thermo Fisher
Scientific, Inc.) and gold view nucleic acid gel stain regent
(Beijing Solarbio Science & Technology Co., Ltd., Beijing,
China). Gels were imaged using a fluorescence imaging analysis
system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The mRNA
expression levels of each ASG were normalized to β-actin.
Differences of ASG between two groups were reflected in the ratio
of exon-inclusion fragment expression level vs. β-actin expression
level/exon-exclusion fragment expression level vs. β-actin
expression level. The ASGs used for validation were in accordance
with the following selection criteria: i) The whole exon
exclusion/inclusion was included; ii) relatively higher SI absolute
value was included; iii) the first or the last AS exon was
excluded.
Statistical analysis
The data of 3 independent experiments were expressed
as the mean ± standard deviation. Independent-samples t-test was
used to determine the significance between groups. P<0.05 was
considered to indicate a statistically significant difference.
Statistical analysis was performed using SPSS software version 19.0
(IBM Corp., Armonk, NY, USA).
Results
Detection, validation and functional
analysis of DEGs under normoxic and hypoxic conditions in
CESCs
HTA 2.0 data analysis identified a total of 448
DEGs, of which 330 (73%) were upregulated and 118 (27%) were
downregulated. Based on these results, 10 DEGs were chosen for
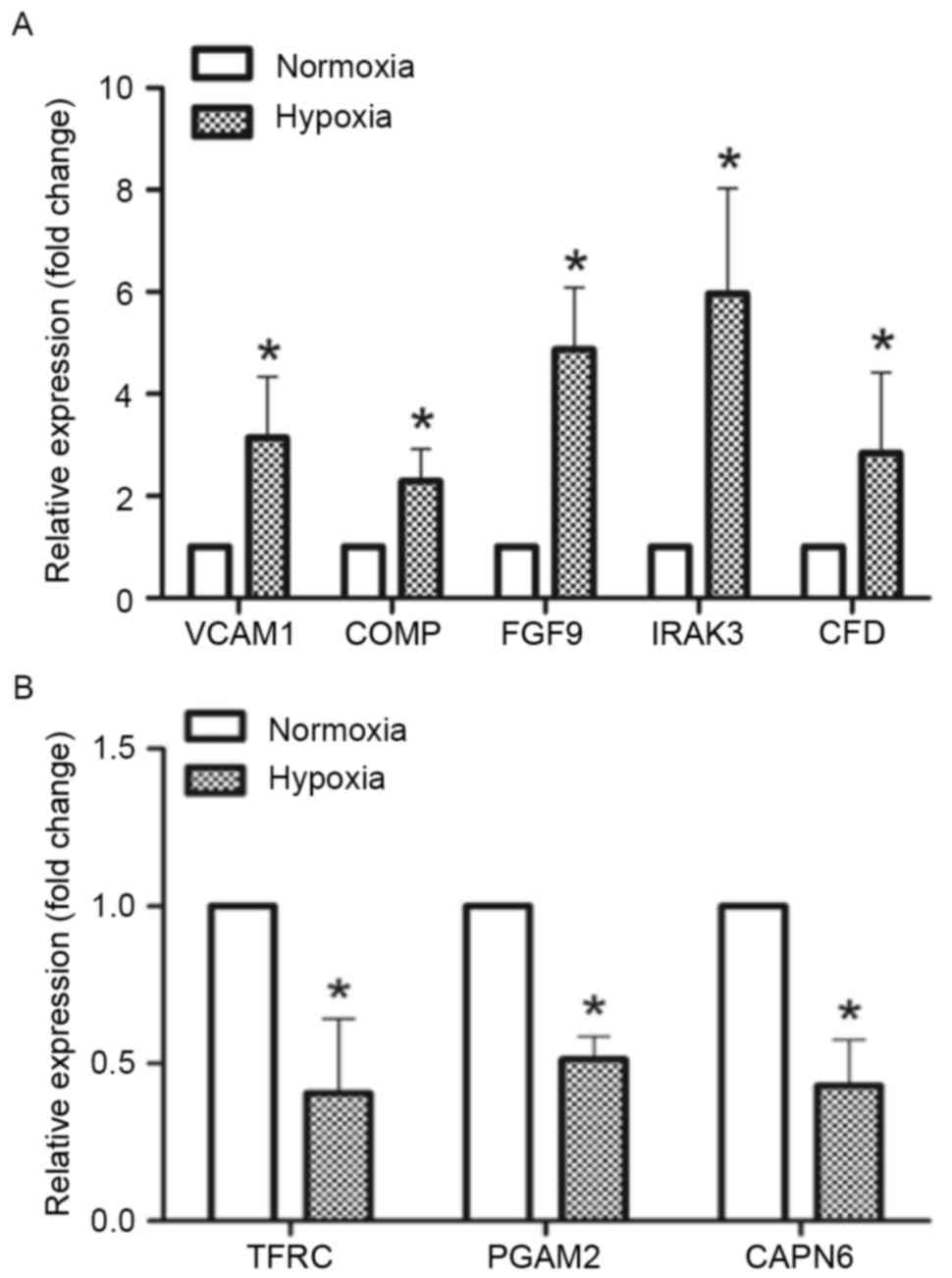
validation by RT-qPCR, and 8 of these were validated successfully.
Since the GeneChip signal increases linearly as target gene
expression increases, but the qPCR signal increases exponentially
as target gene expression increases, the ratio calculated by
GeneChip and qPCR was different. Thus, a similar tendency for
increase or decrease in the results from both methods was
considered to indicate successful validation of the DEGs. In the
present study, the expression of 5 genes [vascular cell adhesion
molecule 1, cartilage oligomeric matrix protein, fibroblast growth
factor 9, interleukin 1 receptor-associated kinase 3 (IRAK3) and
complement factor D (CFD)] was upregulated in the hypoxia group
compared with the normoxia group. Conversely, the expression of 3
genes (transferrin receptor, phosphoglycerate mutase 2 and calpain
6) was downregulated in the hypoxia group compared with the
normoxia group (Fig. 1).
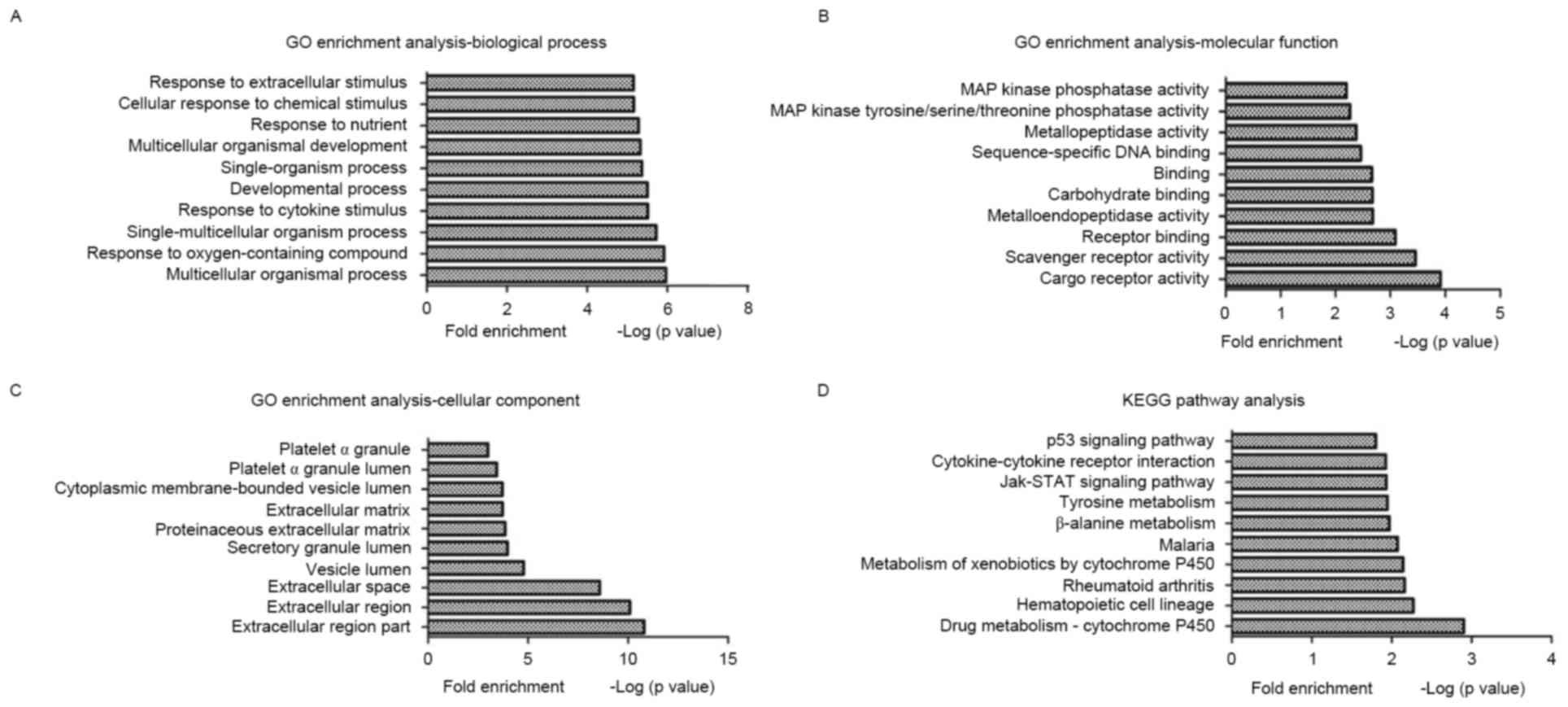
Enriched biological processes, molecular functions
and cellular components in CESCs were identified by GO analysis of
the DEGs under normoxia and hypoxia. GO terms were highly enriched
in CESCs under hypoxia, such as multicellular organismal process,
cargo receptor activity and extracellular region part; the top 10
GO terms from each category are indicated in Fig. 2A-C.
The KEGG mapping tool was used to identify the
enriched functional signaling pathways for the identified DEGs
under hypoxia in CESCs. The results revealed that several signaling
pathways were significantly enriched, such as drug
metabolism-cytochrome P450, hematopoietic cell lineage and
rheumatoid arthritis; the top 10 KEGG pathways are in Fig. 2D.
Detection, validation and functional
analysis of ASGs under normoxic and hypoxic conditions in
CESCs
Genome-wide AS analysis between the normoxic group
and hypoxic group identified 9,475 alternatively spliced exons,
which belonged to 1,808 ASGs. In addition, 3,677 (39%) of the
alternatively spliced exons with an SI value ≥2 were defined as
‘exon inclusion’ events, whereas the remaining 5,798 (61%) exons
were considered as ‘exon exclusion’ events (with an SI value ≤-2).
Under hypoxic conditions, each ASG had an average of 5.2
(9,475/1,808) alternatively spliced exons, which confirmed the
occurrence of multiple alternative splicing events in the same
gene. For example, nine alternatively spliced exons were identified
for nuclear receptor subfamily 4 group A member 3 (NR4A3), which
suggested that AS regulation was complex. In addition, 114 of the
1,808 ASGs were also identified as DEGs, indicating an underlying
link between AS and differential gene expression. A total of 10
ASGs were chosen for validation by semi-quantitative PCR analysis,
of which 7 were successfully validated. Successful validation was
confirmed when the HTA 2.0 array results and semi-quantitative PCR
demonstrated that an exon was more likely to be spliced or
preserved under hypoxic conditions compared with under normoxic
conditions. In the present study, AS events were observed and
validated in GDNF family receptor α2, cell adhesion molecule 1,
NR4A3, relaxin/insulin-like family peptide receptor 1,
NADH:ubiquinione oxidoreductase subunit C1, transmembrane protein
106B and fibronectin 1 under hypoxic conditions compared with under
normoxic conditions (Fig. 3).
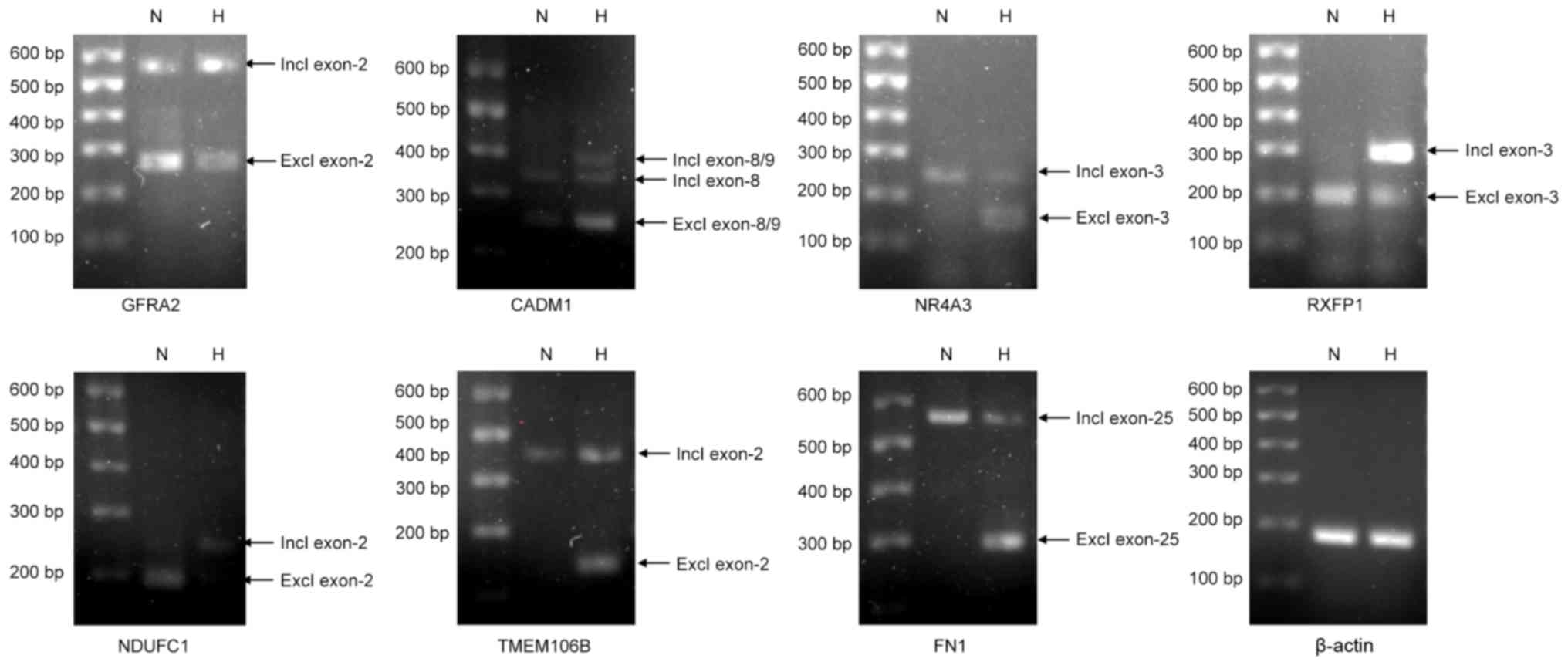 | Figure 3.Validation of the ASGs identified for
CESCs under normoxic and hypoxic conditions. CESCs were exposed to
normoxic or hypoxic conditions for 21 days. A total of 10 ASGs were
selected for further validation by RT-PCR, of which 7 were
successfully validated. β-actin was used as an internal control.
ASG, alternatively spliced gene; CADM1, cell adhesion molecule 1;
CESC, cartilage endplate-derived stem-cells; excl, exclusion; FN1,
fibronectin 1; GFRA2, GDNF family receptor α2; H, hypoxia; incl,
inclusion; N, normoxia; NDUFC1, NADH:ubiquinione oxidoreductase
subunit C1; NR4A3, nuclear receptor subfamily 4 group A member 3;
RT-PCR, reverse transcription-polymerase chain reaction; RXFP1,
relaxin/insulin-like family peptide receptor 1; TMEM106B,
transmembrane protein 106B. |
GO term enrichment analysis of ASGs in CESCs under
normoxic and hypoxic conditions was conducted to determine the
enriched biological processes, molecular functions and cellular
components. The results revealed enrichment in GO terms, such as
antigen processing and presentation of peptide antigen via major
histocompatibility complex (MHC) class I, protein binding and
cytoplasm; the top 10 GO terms for ASGs under hypoxic conditions in
CESCs are presented in Table
III.
 | Table III.List of the top 10 GO terms
identified for alternatively spliced genes under hypoxic conditions
in cartilage endplate stem cells. |
Table III.
List of the top 10 GO terms
identified for alternatively spliced genes under hypoxic conditions
in cartilage endplate stem cells.
| A, Biological
process |
|---|
|
|---|
| GO term | Count | P-value |
|---|
| GO:0002474 antigen
processing and presentation of peptide antigen via MHC class I | 60 |
1.97×10−119 |
| GO:0044419
interspecies interaction between organisms | 82 |
2.15×10−116 |
| GO:0006355
regulation of transcription, DNA-dependent | 145 |
7.89×10−110 |
| GO:0006955 immune
response | 101 |
1.72×10−103 |
| GO:0019882 antigen
processing and presentation | 61 |
5.82×10−100 |
| GO:0006350
transcription | 122 |
4.81×10−79 |
| GO:0007155 cell
adhesion | 74 |
2.23×10−70 |
| GO:0007165 signal
transduction | 128 |
3.81×10−68 |
| GO:0055114
oxidation reduction | 57 |
7.07×10−54 |
| GO:0006468 protein
amino acid phosphorylation | 52 |
1.62×10−44 |
|
| B, Molecular
function |
|
| GO term | Count | P-value |
|
| GO:0005515 protein
binding | 496 | 0 |
| GO:0008270 zinc ion
binding | 222 |
2.30×10−211 |
| GO:0046872 metal
ion binding | 237 |
1.14×10−174 |
| GO:0000166
nucleotide binding | 173 |
3.72×10−155 |
| GO:0005524 ATP
binding | 136 |
9.80×10−133 |
| GO:0005509 calcium
ion binding | 103 |
9.50×10−105 |
| GO:0016740
transferase activity | 114 |
1.62×10−91 |
| GO:0003700
transcription factor activity | 79 |
1.40×10−70 |
| GO:0008233
peptidase activity | 64 |
2.37×10−65 |
| GO:0016787
hydrolase activity | 93 |
1.10×10−55 |
|
| C, Cellular
component |
|
| GO term | Count | P-value |
|
| GO:0005737
cytoplasm | 430 | 0 |
| GO:0005634
nucleus | 445 | 0 |
| GO:0016020
membrane | 411 |
1.05×10−308 |
| GO:0016021 integral
to membrane | 355 |
2.29×10−286 |
| GO:0005886 plasma
membrane | 217 |
1.30×10−162 |
| GO:0005576
extracellular region | 176 |
1.46×10−157 |
| GO:0042612 MHC
class I protein complex | 60 |
1.39×10−115 |
| GO:0005887 integral
to plasma membrane | 107 |
5.12×10−99 |
| GO:0005829
cytosol | 99 |
8.65×10−98 |
| GO:0005794 Golgi
apparatus | 77 |
5.77×10−72 |
The 1,808 identified ASGs were further examined by
KEGG pathway analysis to identify the enriched functional pathways.
These results revealed that several vital pathways were
significantly enriched, such as focal adhesion, cell adhesion
molecules and axon guidance; a list of the top 10 KEGG pathways
identified for ASGs under hypoxic conditions in CESCs is presented
in Table IV.
 | Table IV.List of the top 10 enriched signaling
pathways identified by Kyoto Encyclopedia of Genes and Genomes for
alternatively spliced genes under hypoxic conditions in cartilage
endplate stem cells. |
Table IV.
List of the top 10 enriched signaling
pathways identified by Kyoto Encyclopedia of Genes and Genomes for
alternatively spliced genes under hypoxic conditions in cartilage
endplate stem cells.
| Pathway | Count | P-value |
|---|
| Focal adhesion | 37 |
1.01×10−22 |
| Cell adhesion
molecules | 28 |
2.60×10−19 |
| Axon guidance | 26 |
1.56×10−17 |
| MAPK signaling
pathway | 35 |
1.44×10−16 |
| ECM-receptor
interaction | 21 |
1.57×10−16 |
| Type I diabetes
mellitus | 15 |
2.00×10−14 |
| Small cell lung
cancer | 19 |
5.00×10−14 |
| Wnt signaling
pathway | 23 |
5.78×10−13 |
| Calcium signaling
pathway | 25 |
6.98×10−13 |
| Allograft
rejection | 13 |
1.06×10−12 |
Discussion
The CEP is an avascular organ, and the oxygen
tension within it is as low as 1% (22). A previous study reported that in
healthy juveniles and adolescents with no history of low back pain,
the CEP was free of blood vessels. Conversely, in adults and
seniors with a history of DDD, defects in the CEP were noted
through which blood vessels could be observed (23). Blood vessel invasion was also noted
in painful IVD and osteoarthritis model (24,25).
In degenerated CEP, the destruction of the avascular
microenvironment may lead to the destruction of the physiological
hypoxic microenvironment of CESCs; therefore, an improved
understanding of the effects of the hypoxic microenvironment on
CESCs may aid in our understanding of CEP degeneration.
MSC differentiation under hypoxic conditions has
previously been investigated: Bone marrow cells at an oxygen
concentration of 4–7%, adipocytes at 3.8–9.6% O2 and
muscle tissue at a concentration of 1–10% (26–29).
In addition, several studies have focused on the regulatory effects
of hypoxia on various biological processes in MSCs, including cell
cycle, survival, proliferation, differentiation, morphology,
immunophenotype and cytogenetics (11,12,30,31).
However, the effects of hypoxia on stem cells have not been
examined on a genome-wide scale. In the present study, gene
expression profiling and AS events in CESCs under hypoxia were
examined with the Affymetrix HTA 2.0 system, and the GO terms and
KEGG pathways were analyzed with relevant web-based bioinformatics
tools.
A total of 448 DEGs were identified between CESCs in
the normoxic group and CESCs in the hypoxic group. GO term and KEGG
pathway analyses of the DEGs revealed that hypoxia may influence
CESC fate in a number of aspects of biological processes, molecular
functions, cellular components and signaling pathways, which was
consistent with a previous study (20). Several of the identified genes were
chosen for validation by PCR, including IRAK3 and CFD. Members of
the IRAK protein family have been reported to be necessary
components of toll/interleukin-receptor immune signaling pathways
(32,33). IRAK3 is mainly expressed in
macrophages and monocytes, and negatively regulates the toll-like
receptor signaling (34). The CFD
protein is an essential component of the alternative complement
pathway, and has been reported to serve a role in humoral immunity
against infectious agents (35,36).
CFD cleaves CFB, following CFB binding to complement component 3b,
thereby forming the active complement component 3 convertase
(37,38). IRAK3 and CFD are vital components
of immune signaling pathways, which indicated their potential
necessary roles in the hypoxia-regulated fate of CESCs.
Due to the avascular properties and low
immunogenicity of IVD cells, the roles of the immune signaling
pathway in IVD cells had not previously been investigated. However,
a number of immune signals have been recently demonstrated to
exhibit non-immune effects. For example, although the complement
pathway is known to serve essential roles in immune surveillance,
it has been reported to be involved in apoptotic waste clearance,
stem cell differentiation and recruitment of stem cells (39). Several markers of the complement
cascades have been demonstrated to be expressed in both MSCs and
osteoblasts, such as C5a anaphylatoxin chemotactic receptor (C5aR),
C5, C3aR and C3, in addition to cell-surface markers CD59, CD55 and
CD46 (40,41). As hypoxia is an important activator
of the immune response (42,43),
immune signaling may serve an essential role in the CESC response
to the hypoxic microenvironment.
The present study aimed to obtain a global view of
AS events that occur in CESCs under hypoxic conditions. A total of
9,475 alternatively spliced exons belonging to 1,808 ASGs were
detected and analyzed for GO term enrichment. Consistent with the
DEG analysis results, both antigen processing/presentation of
peptide antigen via MHC class I and immune response were also
significantly enriched and ranked within the top 10 GO terms of
biological processes. MHC class I protein complex was also
significantly enriched and ranked within the top 10 GO terms of
molecular function, indicating an abundant occurrence of AS events
in the immune signaling pathways. A previous study demonstrated
that AS may regulate the immune response by generating different
protein isoforms (44); therefore,
it is reasonable to consider that AS is highly involved in the
immune signaling pathway in CESCs under hypoxia.
The validated AS events from the present study may
be considered as examples to highlight the function of AS on immune
pathways. For example, glial cell line-derived neurotrophic factor
(GDNF) family receptor α2 (GFRA2) is a
glycosylphosphatidylinositol-linked surface receptor for both GDNF
and neurturin (NTN), and mediates RET receptor tyrosine kinase
activation. GFRA2 is a highly-expressed ligand-binding chain in B
cells, T cells and monocytes. The RET/GFRA2 signals have been
reported to be essential for immune cell development and the
protective immunity of central nervous system (45,46).
A GFRA2 isoform that lacks exon 2 was revealed to be expressed at
low levels in lipopolysaccharide (LPS) + interferon γ
(IFN-γ)-activated peripheral blood mononuclear cells (PBMCs),
resting monocytes and neuroblastoma cells. Expression of the
full-length GFRA2 isoform was observed in resting and (LPS +
IFN-γ)-activated PBMCs, resting and (LPS + IFN-γ)-activated
monocytes and B cells, particularly following activation with
Staphylococcus aureus Cowan I (SAC) (46). The lack of exon 2 resulted in the
loss of N-glycosylation sites, which may have influenced the LPS,
IFN-γ and SAC-mediated RET/GFRA2 immune signals (46). In the present study, the ratio of
variant lacking exon 2/full-length variant was lower in the hypoxia
exposed group compared with that in normoxia exposed group, which
indicated that the RET/GFRA2 immune signals may be involved in
CESCs under hypoxia.
In addition, GO term analysis revealed enrichment in
the biological process of DNA-dependent regulation of
transcription, the molecular function of nucleotide binding and
transcription factor activity. A number of vital regulatory
molecules are transcription factor, such as SRY box 9 and
runt-related transcription factor 2, the functions of which are to
bind the promoter region to facilitate or suppress transcription
(47,48). Additionally, the cellular component
terms of cytoplasm and nucleus were significantly enriched, which
indicated an abundant and frequent occurrence of cytoplasm/nucleus
translocation by which many important molecules, such as nuclear
factor κB (49) and AKT (50), serve their regulatory roles. These
results indicated a relationship between AS events and nuclear
signal transduction in CESCs under hypoxia.
KEGG analysis of ASGs identified several AS-related
signaling pathways in CESCs under hypoxic conditions, such as focal
adhesion, cell adhesion molecules and extracellular matrix
(ECM)-receptor interaction. Focal adhesion with the cytoskeleton
may influence stem cell morphology, mechanical nature and the
direction of differentiation (51). In the process of focal adhesion,
signal transduction of matrix-cell adhesion is based on the
interaction between stem cells and ECM-receptors (52). The results of KEGG analysis
indicated that AS may serve regulatory roles through these
mentioned signaling pathways in CESCs under hypoxic conditions.
In conclusion, the present study demonstrated the
differentially expressed genes and AS events in CESCs under hypoxia
on a genome-wide scale, followed by GO and KEGG analysis. These
results provided preliminary insight into the mechanisms of
hypoxia-regulated CESC differentiation at the level of gene
expression and alternative splicing, which may aid in our
understanding of CEP degeneration processes.
Acknowledgements
We sincerely thank Mr Yi Zha for assistance with
microarray data analysis. This work was supported by the National
Natural Science Foundation of China (grant nos. 81472076, 81271982,
81401801 and 81272028).
Glossary
Abbreviations
Abbreviations:
|
AS
|
alternative splicing
|
|
ASG
|
alternatively spliced gene
|
|
CEP
|
cartilage endplate
|
|
CESC
|
cartilage endplate-derived stem
cell
|
|
DDD
|
degenerative disc disease
|
|
DEG
|
differentially expressed gene
|
|
IVD
|
intervertebral disc
|
|
MSC
|
mesenchymal-derived stem cell
|
References
|
1
|
Andersson GB: Epidemiological features of
chronic low-back pain. Lancet. 354:581–585. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Le Maitre CL, Freemont AJ and Hoyland JA:
Accelerated cellular senescence in degenerate intervertebral discs:
A possible role in the pathogenesis of intervertebral disc
degeneration. Arthritis Res Ther. 9:R452007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Peng B, Hao J, Hou S, Wu W, Jiang D, Fu X
and Yang Y: Possible pathogenesis of painful intervertebral disc
degeneration. Spine (Phila Pa 1976). 31:560–566. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang AM, Cao P, Yee A, Chan D and Wu EX:
Detection of extracellular matrix degradation in intervertebral
disc degeneration by diffusion magnetic resonance spectroscopy.
Magn Reson Med. 73:1703–1712. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Buckwalter JA: Aging and degeneration of
the human intervertebral disc. Spine (Phila Pa 1976). 20:1307–1314.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Holm S, Maroudas A, Urban JP, Selstam G
and Nachemson A: Nutrition of the intervertebral disc: Solute
transport and metabolism. Connect Tissue Res. 8:101–119. 1981.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Raj PP: Intervertebral disc:
Anatomy-physiology-pathophysiology-treatment. Pain Pract. 8:18–44.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li FC, Zhang N, Chen WS and Chen QX:
Endplate degeneration may be the origination of the vacuum
phenomenon in intervertebral discs. Med Hypotheses. 75:169–171.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu LT, Huang B, Li CQ, Zhuang Y, Wang J
and Zhou Y: Characteristics of stem cells derived from the
degenerated human intervertebral disc cartilage endplate. PLoS One.
6:e262852011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Boskey AL: Signaling in response to
hypoxia and normoxia in the intervertebral disc. Arthritis Rheum.
58:3637–3639. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Carmeliet P, Dor Y, Herbert JM, Fukumura
D, Brusselmans K, Dewerchin M, Neeman M, Bono F, Abramovitch R,
Maxwell P, et al: Role of HIF-1alpha in hypoxia-mediated apoptosis,
cell proliferation and tumour angiogenesis. Nature. 394:485–490.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Merceron C, Vinatier C, Portron S, Masson
M, Amiaud J, Guigand L, Chérel Y, Weiss P and Guicheux J:
Differential effects of hypoxia on osteochondrogenic potential of
human adipose-derived stem cells. Am J Physiol Cell Physiol.
298:C355–C364. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Makino Y, Kanopka A, Wilson WJ, Tanaka H
and Poellinger L: Inhibitory PAS domain protein (IPAS) is a
hypoxia-inducible splicing variant of the hypoxia-inducible
factor-3alpha locus. J Biol Chem. 277:32405–32408. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tacconelli A, Farina AR, Cappabianca L,
Desantis G, Tessitore A, Vetuschi A, Sferra R, Rucci N, Argenti B,
Screpanti I, et al: TrkA alternative splicing: A regulated
tumor-promoting switch in human neuroblastoma. Cancer Cell.
6:347–360. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kazantseva J, Kivil A, Tints K, Kazantseva
A, Neuman T and Palm K: Alternative splicing targeting the
hTAF4-TAFH domain of TAF4 represses proliferation and accelerates
chondrogenic differentiation of human mesenchymal stem cells. PLoS
One. 8:e747992013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gabut M, Samavarchi-Tehrani P, Wang X,
Slobodeniuc V, O'Hanlon D, Sung HK, Alvarez M, Talukder S, Pan Q,
Mazzoni EO, et al: An alternative splicing switch regulates
embryonic stem cell pluripotency and reprogramming. Cell.
147:132–146. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hang X, Li P, Li Z, Qu W, Yu Y, Li H, Shen
Z, Zheng H, Gao Y, Wu Y, et al: Transcription and splicing
regulation in human umbilical vein endothelial cells under hypoxic
stress conditions by exon array. BMC Genomics. 10:1262009.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Moller-Levet CS, Betts GN, Harris AL,
Homer JJ, West CM and Miller CJ: Exon array analysis of head and
neck cancers identifies a hypoxia related splice variant of LAMA3
associated with a poor prognosis. PLoS Comput Biol. 5:e10005712009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shang J, Fan X, Shangguan L, Liu H and
Zhou Y: Global Gene expression profiling and alternative splicing
events during the chondrogenic differentiation of human cartilage
endplate-derived stem cells. Biomed Res Int. 2015:6049722015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yao Y, Shang J, Song W, Deng Q, Liu H and
Zhou Y: Global profiling of the gene expression and alternative
splicing events during hypoxia-regulated chondrogenic
differentiation in human cartilage endplate-derived stem cells.
Genomics. 107:170–177. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lee DC, Adams CS, Albert TJ, Shapiro IM,
Evans SM and Koch CJ: In situ oxygen utilization in the rat
intervertebral disc. J Anat. 210:294–303. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nerlich AG, Schaaf R, Wälchli B and Boos
N: Temporo-spatial distribution of blood vessels in human lumbar
intervertebral discs. Eur Spine J. 16:547–555. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Freemont AJ, Watkins A, Le Maitre C, Baird
P, Jeziorska M, Knight MT, Ross ER, O'Brien JP and Hoyland JA:
Nerve growth factor expression and innervation of the painful
intervertebral disc. J Pathol. 197:286–292. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Walsh DA, McWilliams DF, Turley MJ, Dixon
MR, Fransès RE, Mapp PI and Wilson D: Angiogenesis and nerve growth
factor at the osteochondral junction in rheumatoid arthritis and
osteoarthritis. Rheumatology (Oxford). 49:1852–1861. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Amorin B, Alegretti AP, Valim VS, Silva
AM, Silva MA, Sehn F and Silla L: Characteristics of mesenchymal
stem cells under hypoxia. Cell Bio. 2:11–19. 2013.
|
|
27
|
Schiller ZA, Schiele NR, Sims JK, Lee K
and Kuo CK: Adipogenesis of adipose-derived stem cells may be
regulated via the cytoskeleton at physiological oxygen levels in
vitro. Stem Cell Res Ther. 4:792013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Redshaw Z and Loughna PT: Oxygen
concentration modulates the differentiation of muscle stem cells
toward myogenic and adipogenic fates. Differentiation. 84:193–202.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
D'Ippolito G, Diabira S, Howard GA, Roos
BA and Schiller PC: Low oxygen tension inhibits osteogenic
differentiation and enhances stemness of human MIAMI cells. Bone.
39:513–522. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dos Santos F, Andrade PZ, Boura JS,
Abecasis MM, da Silva CL and Cabral JM: Ex vivo expansion of human
mesenchymal stem cells: A more effective cell proliferation
kinetics and metabolism under hypoxia. J Cell Physiol. 223:27–35.
2010.PubMed/NCBI
|
|
31
|
Potier E, Ferreira E, Meunier A, Sedel L,
Logeart-Avramoglou D and Petite H: Prolonged hypoxia concomitant
with serum deprivation induces massive human mesenchymal stem cell
death. Tissue Eng. 13:1325–1331. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kobayashi K, Hernandez LD, Galán JE,
Janeway CA Jr, Medzhitov R and Flavell RA: IRAK-M is a negative
regulator of Toll-like receptor signaling. Cell. 110:191–202. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fernandes-Alnemri T, Kang S, Anderson C,
Sagara J, Fitzgerald KA and Alnemri ES: Cutting edge: TLR signaling
licenses IRAK1 for rapid activation of the NLRP3 inflammasome. J
Immunol. 191:3995–3999. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cohen P: The TLR and IL-1 signalling
network at a glance. J Cell Sci. 127:2383–2390. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sprong T, Roos D, Weemaes C, Neeleman C,
Geesing CL, Mollnes TE and van Deuren M: Deficient alternative
complement pathway activation due to factor D deficiency by 2 novel
mutations in the complement factor D gene in a family with
meningococcal infections. Blood. 107:4865–4870. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Rohrer B, Guo Y, Kunchithapautham K and
Gilkeson GS: Eliminating complement factor D reduces photoreceptor
susceptibility to light-induced damage. Invest Ophthalmol Vis Sci.
48:5282–5289. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Anderson DH, Radeke MJ, Gallo NB, Chapin
EA, Johnson PT, Curletti CR, Hancox LS, Hu J, Ebright JN, Malek G,
et al: The pivotal role of the complement system in aging and
age-related macular degeneration: Hypothesis re-visited. Prog Retin
Eye Res. 29:95–112. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zeng J, Chen Y, Tong Z, Zhou X, Zhao C,
Wang K, Hughes G, Kasuga D, Bedell M, Lee C, et al: Lack of
association of CFD polymorphisms with advanced age-related macular
degeneration. Mol Vis. 16:2273–2278. 2010.PubMed/NCBI
|
|
39
|
Schraufstatter IU, Khaldoyanidi SK and
DiScipio RG: Complement activation in the context of stem cells and
tissue repair. World J Stem Cells. 7:1090–1108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lee DS, Yi TG, Lee HJ, Kim SN, Park S,
Jeon MS and Song SU: Mesenchymal stem cells infected with
Mycoplasma arginini secrete complement C3 to regulate
immunoglobulin production in B lymphocytes. Cell Death Dis.
5:e11922014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Soland MA, Bego M, Colletti E, Zanjani ED,
St Jeor S, Porada CD and Almeida-Porada G: Mesenchymal stem cells
engineered to inhibit complement-mediated damage. PLoS One.
8:e604612013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Collard CD, Väkevä A, Morrissey MA, Agah
A, Rollins SA, Reenstra WR, Buras JA, Meri S and Stahl GL:
Complement activation after oxidative stress: Role of the lectin
complement pathway. Am J Pathol. 156:1549–1556. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cowell RM, Plane JM and Silverstein FS:
Complement activation contributes to hypoxic-ischemic brain injury
in neonatal rats. J Neurosci. 23:9459–9468. 2003.PubMed/NCBI
|
|
44
|
Olszowski T, Poziomkowska-Gęsicka I,
Jensenius JC and Adler G: Lectin pathway of complement activation
in a Polish woman with MASP-2 deficiency. Immunobiology.
219:261–262. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Almeida AR, Arroz-Madeira S,
Fonseca-Pereira D, Ribeiro H, Lasrado R, Pachnis V and
Veiga-Fernandes H: RET/GFRα signals are dispensable for thymic T
cell development in vivo. PloS One. 7:e529492012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Vargas-Leal V, Bruno R, Derfuss T,
Krumbholz M, Hohlfeld R and Meinl E: Expression and function of
glial cell line-derived neurotrophic factor family ligands and
their receptors on human immune cells. J Immunol. 175:2301–2308.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu CF and Lefebvre V: The transcription
factors SOX9 and SOX5/SOX6 cooperate genome-wide through
super-enhancers to drive chondrogenesis. Nucleic Acids Res.
43:8183–8203. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Choung HW, Lee DS, Lee HK, Shon WJ and
Park JC: Preameloblast-derived factors mediate osteoblast
differentiation of human bone marrow mesenchymal stem cells by
Runx2-Osterix-BSP signaling. Tissue Eng Part A. 22:93–102. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Haddad JJ: Endotoxin-mediated regulation
of nuclear factor-kappaB nuclear translocation and activation in
the hippocampus of the central nervous system: Modulation by
intracerebroventricular treatment with thymulin and the
immunomodulatory role of the IkappaB-alpha/pIkappaB-alpha pathway.
Neuroscience. 164:1509–1520. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Nguyen TL Xuan, Choi JW, Lee SB, Ye K, Woo
SD, Lee KH and Ahn JY: Akt phosphorylation is essential for nuclear
translocation and retention in NGF-stimulated PC12 cells. Biochem
Biophys Res Commun. 349:789–798. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Mathieu PS and Loboa EG: Cytoskeletal and
focal adhesion influences on mesenchymal stem cell shape,
mechanical properties and differentiation down osteogenic,
adipogenic and chondrogenic pathways. Tissue Eng Part B Rev.
18:436–444. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lee HJ, Jang M, Kim H, Kwak W, Park W,
Hwang JY, Lee CK, Jang GW, Park MN, Kim HC, et al: Comparative
transcriptome analysis of adipose tissues reveals that ECM-Receptor
interaction is involved in the depot-specific adipogenesis in
cattle. PLoS One. 8:e662672013. View Article : Google Scholar : PubMed/NCBI
|