|
1
|
Taylor FB Jr, Kinasewitz GT and Lupu F:
Pathophysiology, staging and therapy of severe sepsis in baboon
models. J Cell Mol Med. 16:672–682. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
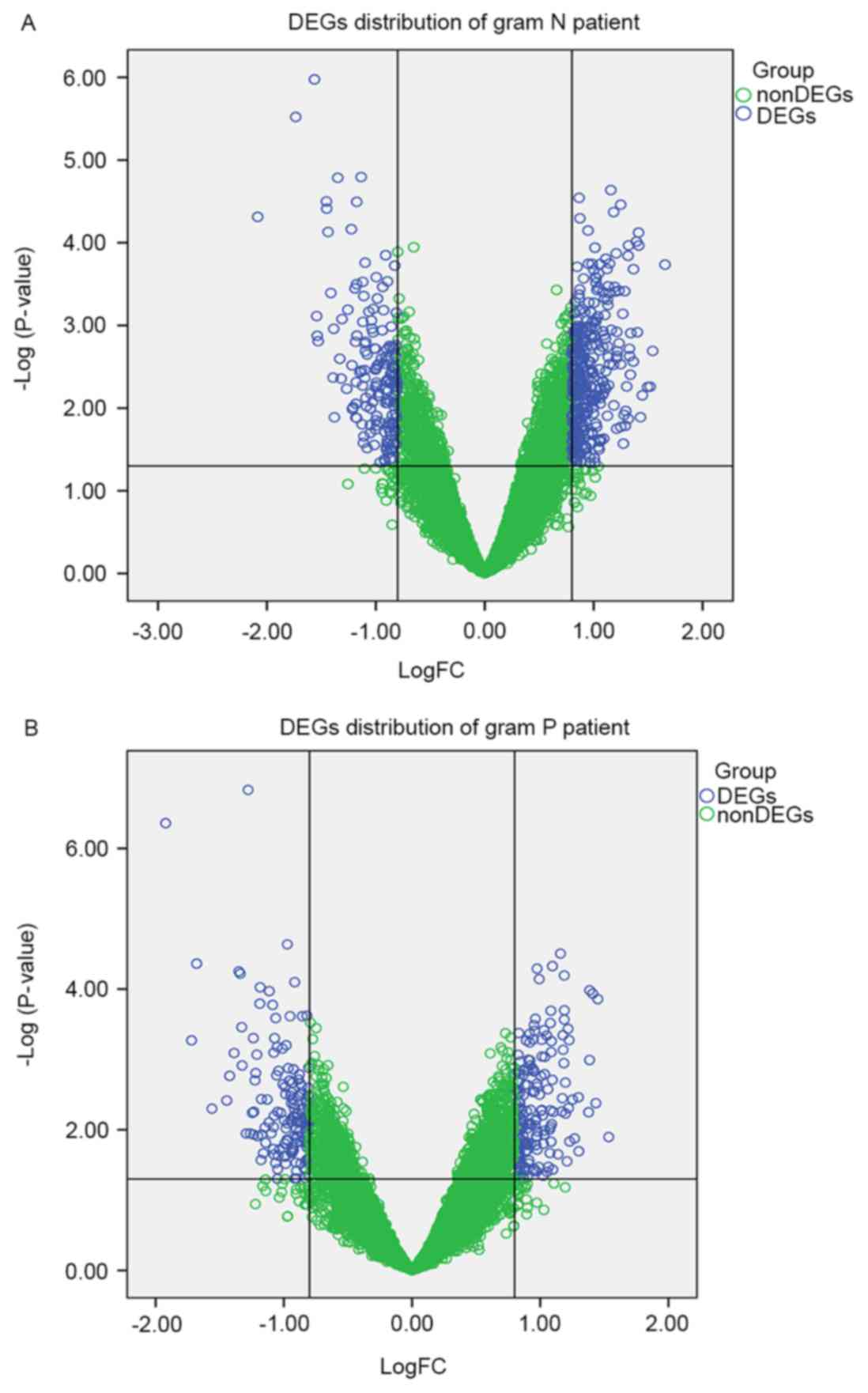
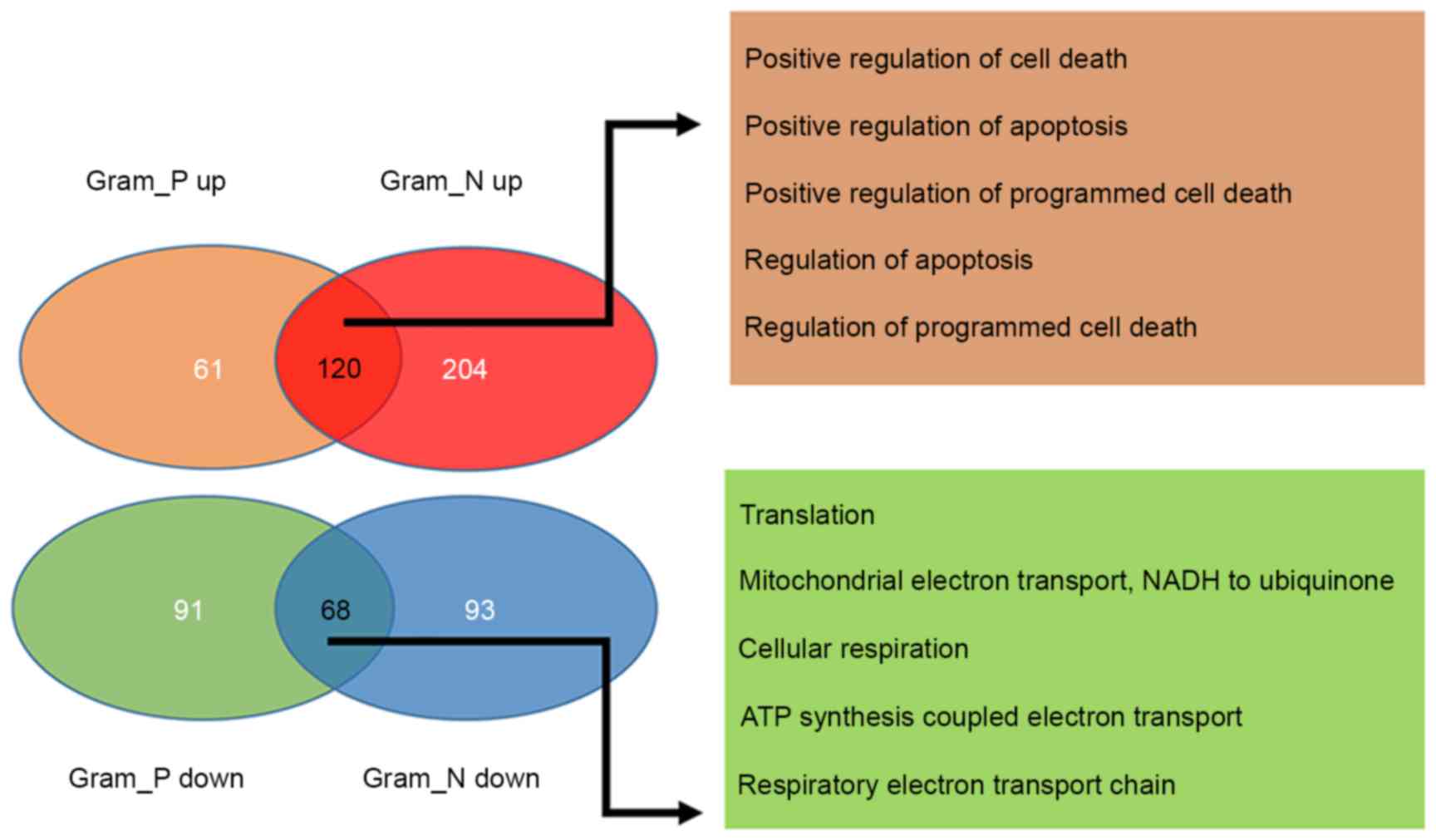
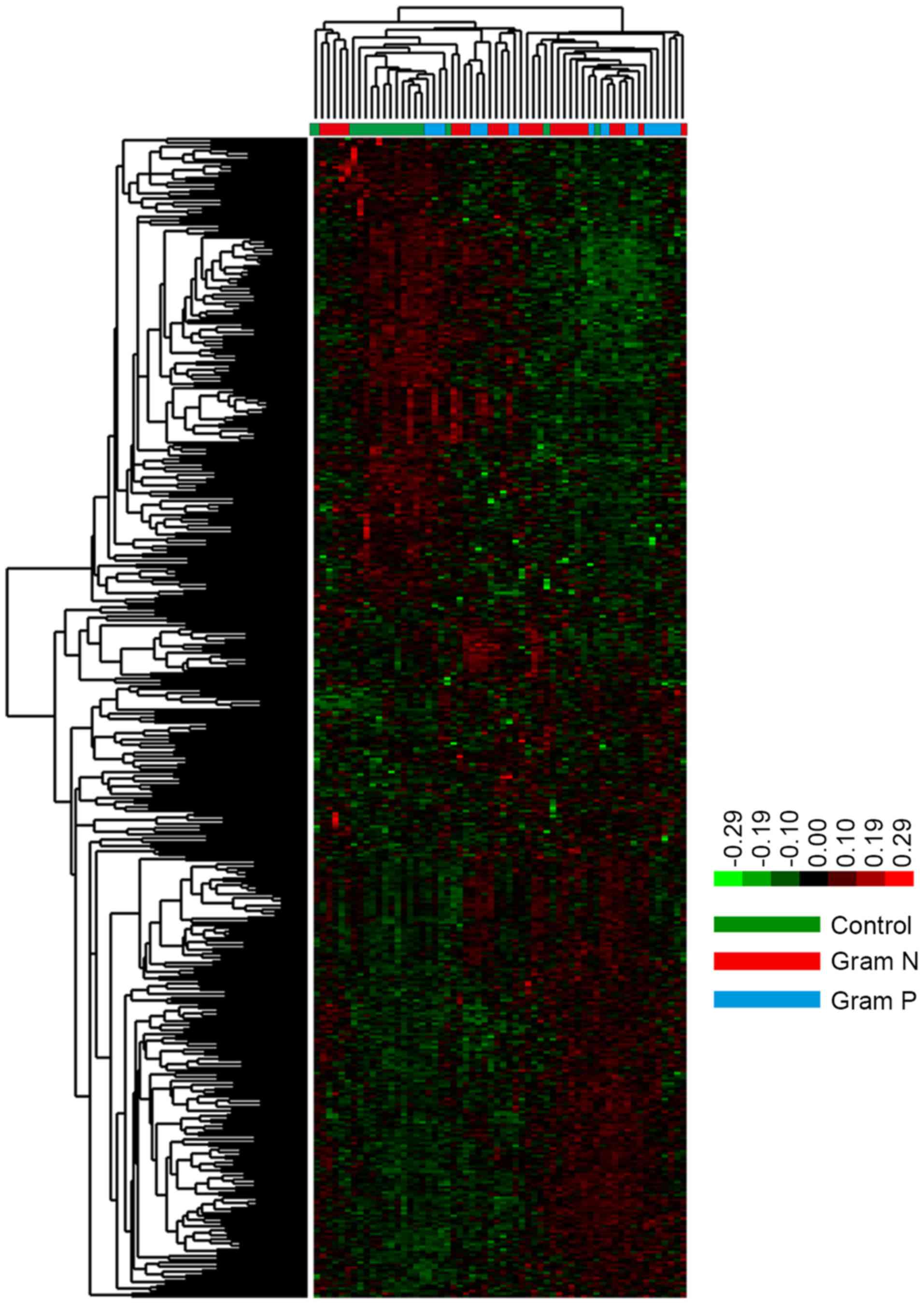
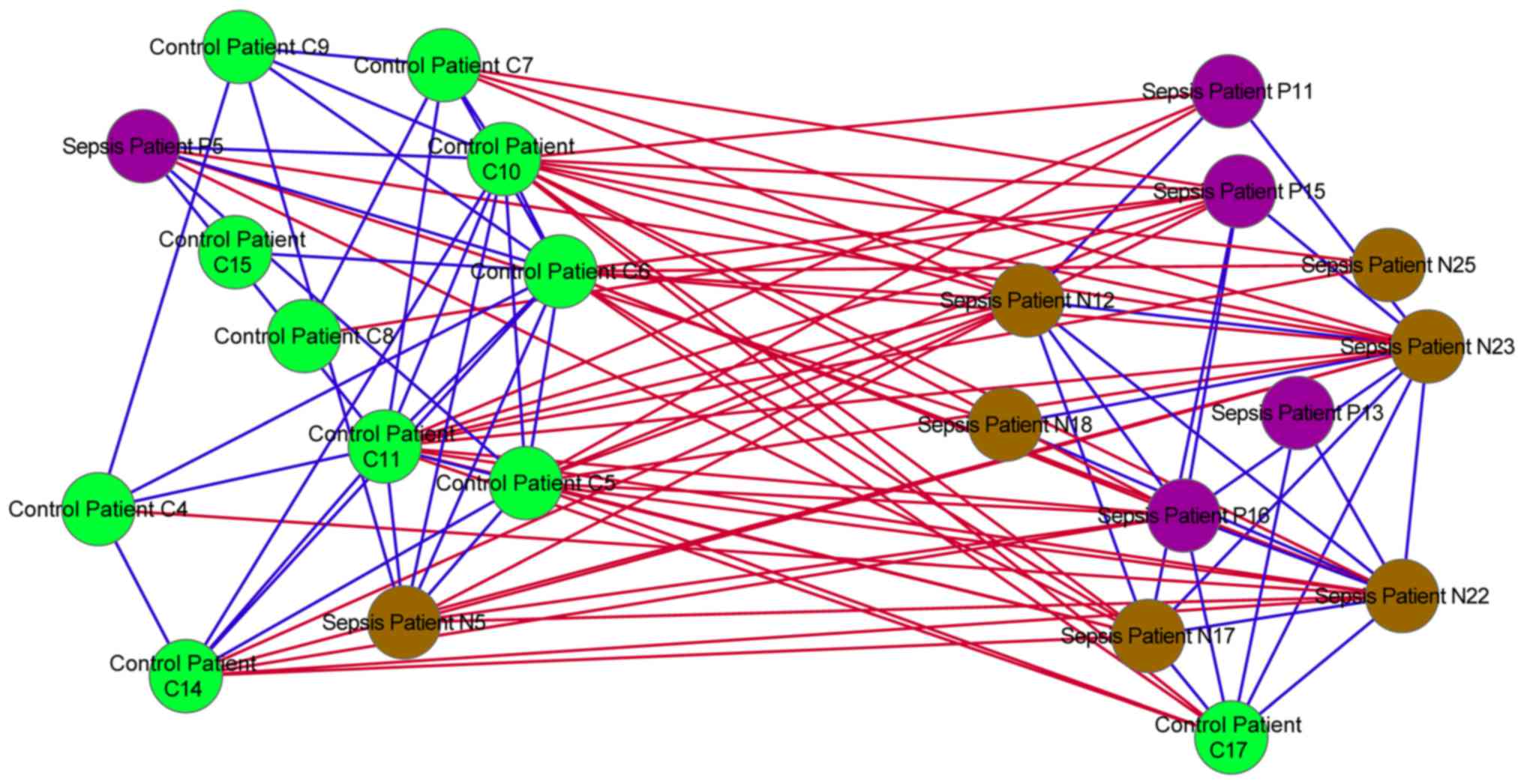
2
|
Brodská H, Malíčková K, Adámková V,
Benáková H, Šťastná MM and Zima T: Significantly higher
procalcitonin levels could differentiate Gram-negative sepsis from
Gram-positive and fungal sepsis. Clin Exp Med. 13:165–170. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Deutschman C and Tracey K: Sepsis: Current
dogma and new perspectives. Immunity. 40:463–475. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lyle NH, Pena OM, Boyd JH and Hancock REW:
Barriers to the effective treatment of sepsis: Antimicrobial
agents, sepsis definitions, and host-directed therapies. Ann N Y
Acad Sci. 1323:101–114. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hotchkiss RS and Karl IE: The
pathophysiology and treatment of sepsis. N Engl J Med. 348:138–150.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Koh EM, Lee SG, Kim CK, Kim M, Yong D, Lee
K, Kim JM, Kim DS and Chong Y: Microorganisms isolated from blood
cultures and their antimicrobial susceptibility patterns at a
university hospital during 1994–2003. Korean J Lab Med. 27:265–275.
2007.(In Korean). View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Glik J, Kawecki M, Gaździk T and Nowak M:
The impact of the types of microorganisms isolated from blood and
wounds on the results of treatment in burn patients with sepsis.
Pol Przegl Chir. 84:6–16. 2012.PubMed/NCBI
|
|
8
|
Tang BM, McLean AS, Dawes IW, Huang SJ,
Cowley MJ and Lin RC: Gene-expression profiling of gram-positive
and gram-negative sepsis in critically ill patients. Crit Care Med.
36:1125–1128. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mahabeleshwar GH, Qureshi MA, Takami Y,
Sharma N, Lingrel JB and Jain MK: A myeloid hypoxia-inducible
factor 1α-Krüppel-like factor 2 pathway regulates gram-positive
endotoxin-mediated sepsis. J Biol Chem. 287:1448–1457. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Giamarellos-Bourboulis EJ, van de Veerdonk
FL, Mouktaroudi M, Raftogiannis M, Antonopoulou A, Joosten LA,
Pickkers P, Savva A, Georgitsi M, van der Meer JW and Netea MG:
Inhibition of caspase-1 activation in Gram-negative sepsis and
experimental endotoxemia. Crit Care. 15:R272011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kager LM, Weehuizen TA, Wiersinga WJ,
Roelofs JJ, Meijers JC, Dondorp AM, van't Veer C and van der Poll
T: Endogenous α2-antiplasmin is protective during severe
gram-negative sepsis (melioidosis). Am J Respir Crit Care Med.
188:967–975. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kager LM, Wiersinga WJ, Roelofs JJ, de
Boer OJ, Weiler H, van't Veer C and van der Poll T: A
thrombomodulin mutation that impairs active protein C generation is
detrimental in severe pneumonia-derived gram-negative sepsis
(melioidosis). PLoS Negl Trop Dis. 8:e28192014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Afsari B, Geman D and Fertig EJ: Learning
dysregulated pathways in cancers from differential variability
analysis. Cancer Inform. 13:(Suppl 5). S61–S67. 2014.
|
|
14
|
Calandra T, Cohen J, et al: International
Sepsis Forum Definition of Infection in the ICU Consensus
Conference: The international sepsis forum consensus conference on
definitions of infection in the intensive care unit. Crit Care Med.
33:1538–1548. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Murie C, Barette C, Lafanechère L and
Nadon R: Control-plate regression (CPR) normalization for
high-throughput screens with many active features. J Biomol Screen.
19:661–671. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen H and Boutros PC: VennDiagram: A
package for the generation of highly-customizable Venn and Euler
diagrams in R. BMC Bioinformatics. 12:352011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gene Ontology Consortium, . Blake JA,
Dolan M, Drabkin H, Hill DP, Li N, Sitnikov D, Bridges S, Burgess
S, Buza T, et al: Gene ontology annotations and resources. Nucleic
Acids Res. 41:(Database issue). D530–D535. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
da Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2008. View Article : Google Scholar
|
|
20
|
Zhou J, Dong X, Zhou Q, Wang H, Qian Y,
Tian W, Ma D and Li X: microRNA expression profiling of heart
tissue during fetal development. Int J Mol Med. 33:1250–1260.
2014.PubMed/NCBI
|
|
21
|
Zhai Y, Tchieu J and Saier MH Jr: A
web-based tree view (TV) program for the visualization of
phylogenetic trees. J Mol Microbiol Biotechnol. 4:69–70.
2002.PubMed/NCBI
|
|
22
|
Weaver B and Wuensch KL: SPSS and SAS
programs for comparing Pearson correlations and OLS regression
coefficients. Behav Res Methods. 45:880–895. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Demchak B, Hull T, Reich M, Liefeld T,
Smoot M, Ideker T and Mesirov JP: Cytoscape: The network
visualization tool for GenomeSpace workflows. F1000Res.
3:1512014.PubMed/NCBI
|
|
24
|
Kotera M, Moriya Y, Tokimatsu T, Kanehisa
M and Goto S: KEGG and GenomeNet, New Developments. Metagenomic
Analysis. 329–339. 2015.
|
|
25
|
Liberti L, Lavor C, Maculan N and
Mucherino A: Euclidean distance geometry and applications.
Quantitative Biol. 56:3–69. 2012.
|
|
26
|
Bódai T, Altmann EG and Endler A:
Stochastic perturbations in open chaotic systems: Random versus
noisy maps. Phys Rev E Stat Nonlin Soft Matter Phys. 87:0429022013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Schmidt MV, Paulus P, Kuhn AM, Weigert A,
Morbitzer V, Zacharowski K, Kempf VA, Brüne B and von Knethen A:
Peroxisome proliferator-activated receptor γ-induced T cell
apoptosis reduces survival during polymicrobial sepsis. Am J Respir
Crit Care Med. 184:64–74. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chang K, Svabek C, Vazquez-Guillamet C,
Sato B, Rasche D, Wilson S, Robbins P, Ulbrandt N, Suzich J, Green
J, et al: Targeting the programmed cell death 1: Programmed cell
death ligand 1 pathway reverses T cell exhaustion in patients with
sepsis. Crit Care. 18:R32014. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Härter L, Mica L, Stocker R, Trentz O and
Keel M: Mcl-1 correlates with reduced apoptosis in neutrophils from
patients with sepsis. J Am Coll Surg. 197:964–973. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Galley H: Oxidative stress and
mitochondrial dysfunction in sepsis. Br J Anaesth. 107:57–64. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lee I and Hüttemann M: Energy crisis: The
role of oxidative phosphorylation in acute inflammation and sepsis.
Biochim Biophys Acta. 1842:1579–1586. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Petruzzella V, Vergari R, Puzziferri I,
Boffoli D, Lamantea E, Zeviani M and Papa S: A nonsense mutation in
the NDUFS4 gene encoding the 18 kDa (AQDQ) subunit of complex I
abolishes assembly and activity of the complex in a patient with
Leigh-like syndrome. Hum Mol Genet. 10:529–535. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ugalde C, Hinttala R, Timal S, Smeets R,
Rodenburg RJ, Uusimaa J, van Heuvel LP, Nijtmans LG, Majamaa K and
Smeitink JA: Mutated ND2 impairs mitochondrial complex I assembly
and leads to Leigh syndrome. Mol Genet Metab. 90:10–14. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bottley A, Phillips NM, Webb TE, Willis AE
and Spriggs KA: eIF4A inhibition allows translational regulation of
mRNAs encoding proteins involved in Alzheimer's disease. PLoS One.
5:pii: e13030. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dröse S and Brandt U: Molecular mechanisms
of superoxide production by the mitochondrial respiratory
chainMitochondrial Oxidative Phosphorylation. Springer; pp.
145–169. 2012, View Article : Google Scholar
|
|
36
|
Van Raamsdonk JM and Hekimi S: Superoxide
dismutase is dispensable for normal animal lifespan. Proc Natl Acad
Sci USA. 109:5785–5790. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Schulte J, Struck J, Köhrle J and Müller
B: Circulating levels of peroxiredoxin 4 as a novel biomarker of
oxidative stress in patients with sepsis. Shock. 35:460–465. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Owens KM, Kulawiec M, Desouki MM,
Vanniarajan A and Singh KK: Impaired OXPHOS complex III in breast
cancer. PLoS One. 6:e238462011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Modena P, Testi MA, Facchinetti F,
Mezzanzanica D, Radice MT, Pilotti S and Sozzi G: UQCRH gene
encoding mitochondrial Hinge protein is interrupted by a
translocation in a soft-tissue sarcoma and epigenetically
inactivated in some cancer cell lines. Oncogene. 22:4586–4593.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li Y, Pei J, Xia H, Ke H, Wang H and Tao
W: Lats2, a putative tumor suppressor, inhibits G1/S transition.
Oncogene. 22:4398–4405. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yang QH, Liu DW, Long Y, Liu HZ, Chai WZ
and Wang XT: Acute renal failure during sepsis: Potential role of
cell cycle regulation. J Infect. 58:459–464. 2009. View Article : Google Scholar : PubMed/NCBI
|