Introduction
Ankylosing spondylitis (AS), a subtype of
spondyloarthritis (Online Mendelian Inheritance in Man, ref no.
106300; https://www.omim.org/entry/106300), is a progressive
chronic disease characterized by inflammatory lower back pain, and
is occasionally accompanied by peripheral arthritis, enthesis,
iritis, spinal deformity and ankyloses (1,2). AS
is highly heritable (>90%) and demonstrates an estimated
prevalence of 0.1–0.4% in the Caucasian population and 0.2–0.54% in
the Chinese population (3,4).
Previous studies have indicated that AS is strongly
associated with the human leukocyte antigen-B27 gene (3,5), a
haplotype of the major histocompatibility complex (MHC). However,
additional studies have suggested that non-MHC genes may be
involved (6–8). Recently, according to the first
genome-wide association study of AS in a Caucasian population,
endoplasmic reticulum aminopeptidase 1 (ERAP1; also known as
ARTS1), located on chromosome 5q15, has been demonstrated to
serve an important role in the risk of developing AS (7). Subsequently, a number of European and
Asian studies have attempted to replicate this study using
different single nucleotide polymorphisms (SNPs), and associations
have been reported in different populations (1,9–17).
However, a number of case-control studies have failed to report
this association using the same genetic markers (4,18–20).
Potential rationales for these inconsistent results include ethnic
differences between populations, the heterogeneity of AS and
inadequate statistical power in certain studies. These
inconsistencies may be overcome by performing a meta-analysis,
which provides a quantitative approach for combining different
independent studies and may maximize the overall statistical power
(21,22).
There are >13 known AS-associated SNPs that span
the ERAP1 gene locus, including rs3734016, rs26653, rs27895,
rs2287987, rs27434, rs30187, rs10050860, rs17482078, rs27044,
rs1065407, rs27980, rs7711564 and rs27037, which have been used as
genetic markers in multiple association studies (1,7,9–20). A
total of 8 SNPs (rs3734016, rs26653, rs27895, rs2287987, rs30187,
rs10050860, rs17482078 and rs27044) are non-synonymous
substitutions in the coding region of the ERAP1 gene, which
implies that the corresponding amino acid substitutions exhibit
functional effects (9,11).
In the present study, a two-stage bioinformatics
analysis was performed in order to investigate the role of common
genetic variants of ERAP1 in AS. In the first stage, a
classical meta-analysis was used to assess all of the AS-associated
SNPs in ERAP1, using all published case-control association
studies. In the second stage, the functional effects of these
genetic variants of ERAP1 were investigated using protein
structure analysis.
Materials and methods
Literature search
To identify studies for inclusion in the
meta-analysis, PubMed (http://www.ncbi.nlm.nih.gov), Scopus (http://www.scopus.com) and Embase (http://www.elsevier.com/online-tools/embase) citations
up to June 2016 were queried with the following search terms:
‘ERAP1’, ‘endoplasmic reticulum aminopeptidase 1’, ‘ARTS1’,
‘Ankylosing spondylitis’ and ‘AS’. The retrieved abstracts were
read to identify studies that examined the association between a
polymorphism in the ERAP1 gene locus and AS. Studies of this
type were subsequently read in full to assess their appropriateness
for inclusion in the meta-analysis. All references cited in these
studies were reviewed to identify additional studies not indexed by
PubMed, Scopus and Embase.
Inclusion criteria, exclusion criteria
and data extraction
Only studies that tested ≥1 polymorphism within the
ERAP1 gene locus were included in the current meta-analysis.
In addition, studies that met all of the following criteria
were included: i) Publication in a peer-reviewed journal; ii)
publication in English; iii) presentation of original data on
genotype and/or alleles in case and control samples; iv)
independence from other studies (i.e. studies that included and
re-analyzed a previously published data set were not regarded as
independent, and in such cases, only the study that had published
the primary data set was included in the meta-analysis); and v)
presence of sufficient data to calculate an effect size (23). For each included study, the
following data were extracted by two independent investigators
using standard forms: First author; journal; year of publication;
study design; ethnicity of the subjects; sample size; phenotype
information; genotype and allele distribution of subjects with and
without AS (24).
Statistical analysis
Population-based studies were collected and
subdivided into European and Asian ethnic populations. Data
regarding the genotype and/or allele distributions are summarized
in Tables I–IV. The genotype and/or allele
frequencies in these studies were analyzed using the EpiInfo™
program version 7.2 (Centers for Disease Control and Prevention,
Atlanta, GA, USA; http://www.cdc.gov/epiinfo), and P<0.05 was
considered to indicate a statistically significant difference.
Prior to the pooling procedure, Cochran's χ2-based
Q-statistic, which was considered significant at P<0.10, was
performed to assess the heterogeneity within the group of odds
ratios (ORs). The extent of the inconsistency across the studies
was quantified using the I2 statistic, and
I2>50% was considered to be a large heterogeneity
value among studies (21). The
natural logarithms of the OR estimates were determined using
random-effect or fixed-effect models, depending on the
heterogeneity among studies. The significance of the pooled ORs was
determined using the Z-test. An ancillary procedure for funnel plot
asymmetry was additionally used to qualitatively assess the
evidence for publication bias. The above statistical analyses were
performed using the RevMan software program (version 5.2;
http://www.cochrane.org/revman)
(22).
 | Table I.Summary of the association studies
published up to June 2016 investigating the SNPs in ERAP1
and AS. |
Table I.
Summary of the association studies
published up to June 2016 investigating the SNPs in ERAP1
and AS.
| Author, year | Country | Ethnicity | Cases | Controls | No. of studied
SNPs | Number of positive
SNPsa | Detailed data | Quality control of
current meta-analysis | (Refs.) |
|---|
| WTCCC, 2007 | UK | European | 922 | 1,500 | 7 | 5 | Yes | Included |
(7) |
|
| USA | European | 471 | 625 | 4 | 4 | No | Excluded |
(7) |
| Davidson SI, et
al, 2009 | China | Asian | 527 | 945 | 33 | 7 | Yes | Included | (20) |
| Zvyagin IV, et
al, 2010 | Russia | European | 84 | 77 | 5 | 3 | Yes | Included |
(9) |
| Pazar B, et
al, 2010 | Hungary | European | 297 | 200 | 5 | 4 | Yes | Included | (10) |
| Szczypiorska M,
et al, 2011 | Spain | European | 300 | 300 | 8 | 5 | Yes | Included | (11) |
| Bang SY, et
al, 2011 | Korea | Asian | 1,164 | 752 | 2 | 2 | Yes | Included | (12) |
| Lin Z, et
al, 2012 | China | Asian | 1,837 | 4,231 | 2 | 2 | Yes | Included |
(1) |
| Mahmoudi M, et
al, 2012 | Iran | Asian | 387 | 316 | 4 | 2 | Yes | Included | (13) |
| Wu W, et al,
2012 | China | Asian | 328 | 627 | 1 | 1 | Yes | Included | (16) |
| Cinar M, et
al, 2013 | Turkey | European | 150 | 150 | 10 | 1 | Yes | Included | (18) |
| Cherciu M, et
al, 2013 | Romania | European | 137 | 139 | 2 | 1 | Yes | Included | (17) |
| Zhang Z, et
al, 2014 | China | Asian | 602 | 619 | 4 | 1 | Yes | Included | (19) |
| Chen C, et
al, 2015 | China | Asian | 368 | 460 | 6 | 4 | Yes | Included | (14) |
| Wang J, et
al, 2015 | China | Asian | 100 | 100 | 2 | 1 | Yes | Included | (15) |
| Maksymowych WP,
et al, 2009 | Canada | European | 992 | 1,437 | 6 | 3 | No | Excluded | (8) |
| Pimentel-Santos FM,
et al, 2009 | Portugal | European | 358 | 285 | 5 | 2 | No | Excluded | (27) |
| Harvey D, et
al, 2009 | UK | European | 730 | 1,021 | 4 | 4 | No | Excluded | (28) |
|
| UK | European | 1,604 | 1,021 | 23 | 11 | No | Excluded | (28) |
| Choi CB, et
al, 2010 | Korea | Asian | 872 | 403 | 5 | 2 | No | Excluded | (29) |
| Reveille JD, et
al, 2010 | Mixed | European | 2,053 | 5,140 | 2 | 2 | No | Excluded | (30) |
|
| UK | European | 898 | 1,518 | 2 | 2 | No | Excluded | (30) |
| Li C, et al,
2010 | China | Asian | 471 | 456 | 6 | 2 | No | Excluded | (31) |
| TASC, 2011 | Mixed | European | 1,787 | 4,800 | 2 | 1 | No | Excluded | (34) |
|
| Mixed | European | 3,023 | 8,779 | 2 | 1 | No | Excluded | (34) |
|
| Mixed | European | 2,111 | 4,483 | 2 | 2 | No | Excluded | (34) |
| Wang CM, et
al, 2012 | China | Asian | 797 | 1,150 | 4 | 4 | No | Excluded | (32) |
| Kadi A, et
al, 2013 | France | European | 180 | 384 | 3 | 1 | No | Excluded |
(2) |
|
| Belgium | European | 256 | 248 | 3 | 2 | No | Excluded |
(2) |
| Liu Y, et
al, 2015 | China | Asian | 707 | 837 | 8 | 5 | No | Excluded | (33) |
 | Table IV.Fixed-effects and random-effects
model summary OR and 95% CI values for 11 ERAP1 SNPs
associated with AS risk. |
Table IV.
Fixed-effects and random-effects
model summary OR and 95% CI values for 11 ERAP1 SNPs
associated with AS risk.
|
| Heterogeneity
test | Fixed-effects
model | Random-effects
model |
|---|
|
|
|
|
|
|---|
| SNPs (minor allele)
and analysis (included studies/included samples) | I2
(%) | P-value | OR (95% CI) | Z-test | P-value | OR (95% CI) | Z-test | P-value |
|---|
| rs3734016 (A) |
|
|
|
|
|
|
|
|
|
Combined (2/2) | 18 |
2.70×10−1 | 0.91
(0.77–1.08) | 1.11 |
2.70×10−1 |
|
|
|
| rs26653 (C) |
|
|
|
|
|
|
|
|
|
European (2/2) | 0 |
3.20×10−1 | 1.41
(1.16–1.71) | 3.44 |
6.0×10−4a |
|
|
|
| rs27895 (A) |
|
|
|
|
|
|
|
|
|
European (2/2) | 0 |
9.10×10−1 | 1.14
(0.93–1.39) | 1.22 |
2.20×10−1 |
|
|
|
| rs2287987 (C) |
|
|
|
|
|
|
|
|
| Combined (6/6) | 23 |
2.60×10−1 | 0.71
(0.64–0.79) | 6.00 |
<1.00×10−5a |
|
|
|
|
European (5/5) | 35 |
1.90×10−1 | 0.70
(0.63–0.79) | 5.91 |
<1.00×10−5 |
|
|
|
| rs27434 (G) |
|
|
|
|
|
|
|
|
| Asian
(7/7) | 92 |
<1.0×10−5a |
|
|
| 0.76
(0.62–0.93) | 2.62 |
9.00×10−3a |
| rs10050860 (T) |
|
|
|
|
|
|
|
|
| Combined (6/6) | 88 |
<1.0×10−5a |
|
|
| 0.53
(0.36–0.78) | 3.24 |
1.00×10−3a |
|
European (5/5) | 11 |
3.4×10−1 | 0.71
(0.63–0.80) | 5.80 |
<1.00×10−5a |
|
|
|
| rs27044 (G) |
|
|
|
|
|
|
|
|
| Combined (8/8) | 97 |
<1.0×10−5a |
|
|
| 1.06
(0.65–1.71) | 0.22 |
8.20×10−1 |
| Asian
(2/2) | 99 |
<1.0×10−5a |
|
|
| 0.53
(0.13–2.18) | 0.88 |
3.80×10−1 |
|
European (6/6) | 0 |
4.90×10−1 | 1.35
(1.23–1.49) | 6.24 |
<1.00×10−5a |
|
|
|
| rs1065407 (C) |
|
|
|
|
|
|
|
|
| Asian
(2/2) | 96 |
<1.0×10−5a |
|
|
| 0.61
(0.18–2.02) | 0.81 |
4.20×10−1 |
| rs27980 (C) |
|
|
|
|
|
|
|
|
| Combined (3/3) | 29 |
2.50×10−1 | 0.88
(0.79–0.98) | 2.36 |
2.00×10−2a |
|
|
|
| Asian
(2/2) | 61 |
1.10×10−1 | 0.87
(0.78–0.98) | 2.40 |
2.00×10−2a |
|
|
|
| rs7711564 (G) |
|
|
|
|
|
|
|
|
| Combined (3/3) | 0 |
5.70×10−1 | 0.79
(0.69–0.91) | 3.29 |
1.00×10−3a |
|
|
|
| Asian
(2/2) | 0 |
3.40×10−1 | 0.78
(0.68–0.91) | 3.23 |
1.00×10−3a |
|
|
|
| rs27037 (T) |
|
|
|
|
|
|
|
|
| Combined (4/4) | 2 |
3.80×10−1 | 1.22
(1.12–1.33) | 4.53 |
<1.00×10−5aa |
|
|
|
| Asian
(3/3) | 33 |
2.20×10−1 | 1.22
(1.11–1.33) | 4.34 |
<1.00×10−4aa |
|
|
|
Structural and functional
analysis
The functional effects of the non-synonymous
variants of ERAP1 were analyzed using the PolyPhen-2
software program (http://genetics.bwh.harvard.edu/pph2/), which is an
automatic tool for predicting the possible effects of an amino acid
substitution on the structure and function of a human protein. This
prediction is based on a number of features contained in the
sequence, as well as phylogenetic and structural information
characterizing the substitution (25). Further structural analysis was
performed with the molecular visualization software PyMOL (version
1.5.0.4; Schrödinger, Inc., Portland, OR, USA), on the basis of the
2YD0 (http://www.rcsb.org/pdb/explore.do?structureId=2yd0)
and 3MDJ (http://www.rcsb.org/pdb/explore/literature.do?structureId=3MDJ)
Protein Data Bank (PDB) structures (26).
Results
Available studies
In total, ≥89 studies were identified by the
combined search. The reviews and studies written in languages other
than English were excluded, leaving 63 studies. An additional 39
references that did not clearly meet the criteria or were not SNP
association studies were further excluded. Therefore, a total of 24
studies remained. Nine additional references were excluded due to
the fact that they did not supply the original data regarding
genotypes and/or alleles in their samples (2,8,27–33),
and one study was excluded due to the analysis of the same samples
as previous studies (Table I)
(34). Ultimately, a total of 14
studies contributed to available data regarding the following 13
identified SNPs in ERAP1 associated with AS: Rs3734016,
rs26653, rs27895, rs2287987, rs27434, rs30187, rs10050860,
rs17482078, rs27044, rs1065407, rs27980, rs7711564 and rs27037
(Table II) (1,7,9–20).
 | Table II.Details of the AS-associated SNPs in
ERAP1 identified in the association studies. |
Table II.
Details of the AS-associated SNPs in
ERAP1 identified in the association studies.
|
|
|
| Genotypes for
cases | Genotypes for
controls |
| Minor allele:major
allele |
|
|
|---|
|
|
|
|
|
|
|
|
|
|
|---|
| SNP (minor
allele/major allele), genomic position (bp) | Country | Case/control | 11 | 12 | 22 | 11 | 12 | 22 | P-value | Cases | Controls | P-value | (Refs.) |
|---|
| rs3734016(A/G),
96,803,761 |
|
|
|
|
|
|
|
|
|
|
|
|
|
| WTCCC,
2007 | UK | 922/1,500 | 0 | 93 | 829 | 4 | 137 | 1,325 |
2.40×10−1 | 93:1,751 | 145:2,787 |
8.80×10−1 |
(7) |
|
Davidson SI, et al,
2009 | China | 527/945 | 6 | 126 | 358 | 24 | 228 | 593 |
1.25×10−1 | 138:842 | 276:1,414 |
1.22×10−1 | (20) |
| rs26653(C/G),
96,803,547 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Szczypiorska M, et al,
2011 | Spain | 300/300 | 40 | 130 | 119 | 25 | 125 | 138 |
8.40×10−2 | 210:368 | 175:401 |
3.20×10−2a | (11) |
| Cinar
M, et al, 2013 | Turkey | 150/150 | 36 | 78 | 36 | 21 | 73 | 56 |
1.50×10−2a | 150:150 | 115:185 |
4.00×10−3a | (18) |
| rs27895(A/G),
96,793,840 |
|
|
|
|
|
|
|
|
|
|
|
|
|
| WTCCC,
2007 | UK | 922/1,500 | 3 | 132 | 787 | 6 | 184 | 1,276 |
4.44×10−1 | 138:1,706 | 196:2,736 |
2.92×10−1 |
(7) |
|
Szczypiorska M, et al,
2011 | Spain | 300/300 | 1 | 36 | 258 | 2 | 29 | 265 |
5.55×10−1 | 38:552 | 33:559 |
5.31×10−1 | (11) |
| rs2287987(C/T),
96,793,832 |
|
|
|
|
|
|
|
|
|
|
|
|
|
| WTCCC,
2007 | UK | 922/1,500 | 16 | 293 | 611 | 83 | 481 | 893 |
6.86×10−6a | 325:1,515 | 647:2,267 |
1.57×10−4a |
(7) |
| Zvyagin
IV, et al, 2010 | Russia | 84/77 | 1 | 13 | 70 | 3 | 26 | 44 |
5.00×10−3a | 15:153 | 32:114 |
1.30×10−3a |
(9) |
| Pazar
B, et al, 2010 | Hungary | 297/200 | 4 | 93 | 200 | 14 | 72 | 114 |
1.00×10−3a | 101:493 | 100:300 |
2.10×10−3a | (10) |
|
Szczypiorska M, et al,
2011 | Spain | 300/300 | 6 | 83 | 204 | 10 | 104 | 182 |
1.00×10−1 | 95:491 | 124:468 |
3.70×10−2a | (11) |
| Cinar
M, et al, 2013 | Turkey | 150/150 | 2 | 22 | 126 | 1 | 38 | 111 |
6.20×10−2 | 26:274 | 40:260 |
6.80×10−2 | (18) |
| Chen C,
et al, 2015 | China | 368/460 | NA | NA | NA | NA | NA | NA | NA | 47:689 | 73:847 |
2.27×10−1 | (14) |
| rs27434(G/A),
96,793,809 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Davidson SI, et al,
2009 | China | 527/945 | 99 | 259 | 121 | 206 | 418 | 195 |
1.84×10−1 | 457:501 | 830:808 |
1.45×10−1 | (20) |
| Bang
SY, et al, 2011 | Korea | 1,164/752 | 198 | 626 | 295 | 216 | 336 | 167 |
<1.00×10−8a | 1,022:1,216 | 768:670 |
4.60×10−6a | (12) |
| Lin Z,
et al, 2012 | China | 1,837/4,231 | 331 | 980 | 526 | 941 | 2,126 | 1,163 |
9.60×10−4a | 1,642:2,032 | 4,008:4,452 |
6.50×10−3a |
(1) |
|
Mahmoudi M, et al,
2012 | Iran | 387/316 | 138 | 188 | 55 | 147 | 141 | 28 |
7.00×10−3a | 464:298 | 435:197 |
2.10×10−3a | (13) |
| Zhang
Z, et al, 2014 | China | 602/619 | 165 | 311 | 125 | 157 | 287 | 165 |
3.60×10−2a | 641:561 | 601:617 |
5.00×10−2 | (19) |
| Chen C,
et al, 2015 | China | 368/460 | NA | NA | NA | NA | NA | NA | NA | 230:506 | 476:444 |
<1.0×10−8a | (14) |
| Wang J,
et al, 2015 | China | 100/100 | 22 | 49 | 29 | 37 | 42 | 21 |
6.00×10−2 | 93:107 | 116:84 |
2.10×10−2a | (15) |
| rs30187(T/C),
96,788,627 |
|
|
|
|
|
|
|
|
|
|
|
|
|
| WTCCC,
2007 | UK | 922/1,500 | 134 | 472 | 316 | 177 | 627 | 662 |
9.50×10−7a | 740:1104 | 981:1,951 |
2.90×10−6a | (7) |
| Zvyagin
IV, et al, 2010 | Russia | 84/77 | 7 | 48 | 29 | 9 | 33 | 35 |
1.93×10−1 | 62:106 | 51:103 |
4.78×10−1 | (9) |
| Pazar
B, et al, 2010 | Hungary | 297/200 | 45 | 136 | 116 | 26 | 76 | 98 |
8.90×10−2 | 226:368 | 128:272 |
5.10×10−2 | (10) |
|
Szczypiorska M, et al,
2011 | Spain | 300/300 | 57 | 162 | 74 | 46 | 144 | 99 |
5.40×10−2 | 276:310 | 236:342 |
3.10×10−2a | (11) |
| Lin Z,
et al, 2012 | China | 1,837/4,231 | 528 | 979 | 330 | 1,164 | 2,130 | 937 |
1.00×10−3a | 2,035:1,639 | 4,458:4,004 |
6.00×10−3a |
(1) |
|
Mahmoudi M, et al,
2012 | Iran | 387/316 | 103 | 190 | 88 | 45 | 163 | 104 |
6.00×10−5a | 396:366 | 253:371 |
2.30×10−5a | (13) |
| Cinar
M, et al, 2013 | Turkey | 150/150 | 36 | 85 | 29 | 29 | 103 | 18 |
8.00×10−2 | 157:143 | 161:139 |
7.44×10−1 | (18) |
| Cherciu
M, et al, 2013 | Romania | 137/139 | 14 | 80 | 43 | 15 | 59 | 65 |
2.20×10−2a | 108:166 | 89:189 |
7.00×10−2 | (17) |
| Chen C,
et al, 2015 | China | 368/460 | NA | NA | NA | NA | NA | NA | NA | 412:324 | 446:474 |
2.00×10−3a | (14) |
| rs10050860(T/C),
96,786,506 |
|
|
|
|
|
|
|
|
|
|
|
|
|
| WTCCC,
2007 | UK | 922/1,500 | 17 | 296 | 609 | 86 | 489 | 891 |
6.11×10−6a | 330:1514 | 661:2,271 |
1.15×10−4a |
(7) |
| Zvyagin
IV, et al, 2010 | Russia | 84/77 | 0 | 18 | 66 | 5 | 26 | 46 |
8.00×10−3a | 18:140 | 36:118 |
2.40×10−3a |
(9) |
| Pazar
B, et al, 2010 | Hungary | 297/200 | 4 | 87 | 206 | 14 | 64 | 122 |
2.00×10−3a | 95:499 | 92:308 |
5.60×10−3a | (10) |
|
Szczypiorska M, et
al,2011 | Spain | 300/300 | 6 | 82 | 205 | 11 | 100 | 180 |
8.80×10−2 | 94:492 | 122:460 |
3.00×10−2a | (11) |
| Cinar
M, et al, 2013 | Turkey | 150/150 | 5 | 27 | 118 | 3 | 44 | 103 |
6.10×10−2 | 37:263 | 50:250 |
1.32×10−1 | (18) |
| Chen C,
et al, 2015 | China | 368/460 | NA | NA | NA | NA | NA | NA | NA | 37:699 | 186:734 |
<1.00×10−7a | (14) |
| rs17482078(T/C),
96,783,162 |
|
|
|
|
|
|
|
|
|
|
|
|
|
| WTCCC,
2007 | UK | 922/1,500 | 16 | 285 | 621 | 78 | 476 | 912 |
2.40×10−5a | 317:1,527 | 632:2,300 |
2.30×10−4a |
(7) |
| Zvyagin
IV, et al, 2010 | Russia | 84, 77 | 2 | 17 | 65 | 3 | 27 | 47 |
7.90×10−2 | 21:147 | 33:121 |
3.20×10−2a |
(9) |
| Pazar
B, et al, 2010 | Hungary | 297/200 | 5 | 85 | 207 | 13 | 58 | 129 |
1.70×10−2a | 95:499 | 84:316 |
4.40×10−2a | (10) |
|
Szczypiorska M, et al,
2011 | Spain | 300/300 | 7 | 82 | 206 | 12 | 100 | 185 |
1.21×10−1 | 96:494 | 124:470 |
4.20×10−2a | (11) |
| Cinar
M, et al, 2013 | Turkey | 150/150 | 2 | 23 | 125 | 1 | 39 | 110 |
6.70×10−2 | 27:273 | 41:259 |
7.20×10−2 | (18) |
| rs27044(G/C),
96,783,148 |
|
|
|
|
|
|
|
|
|
|
|
|
|
| WTCCC,
2007 | UK | 922/1,500 | 94 | 432 | 395 | 119 | 553 | 793 |
6.20×10−7a | 620:1,222 | 791:2,139 |
9.00×10−7a |
(7) |
| Zvyagin
IV, et al, 2010 | Russia | 84/77 | 7 | 42 | 35 | 9 | 26 | 42 |
1.13×10−1 | 56:112 | 44:110 |
3.57×10−1 | (9) |
| Pazar
B, et al, 2010 | Hungary | 297/200 | 27 | 136 | 134 | 14 | 60 | 126 |
4.30×10−4a | 190:404 | 88:312 |
5.90×10−4a | (10) |
|
Szczypiorska M, et al,
2011 | Spain | 300/300 | 34 | 149 | 109 | 35 | 127 | 132 |
1.38×10−1 | 217:367 | 197:391 |
1.91×10−1 | (11) |
| Wu W,
et al, 2012 | China | 328/627 | 48 | 156 | 178 | 285 | 252 | 90 |
<1.00×10−7a | 252:512 | 822:432 |
<1.00×10−7a | (16) |
| Cinar
M, et al, 2013 | Turkey | 150/150 | 19 | 67 | 64 | 12 | 71 | 67 |
4.14×10−1 | 105:195 | 95:205 |
3.87×10−1 | (18) |
| Cherciu
M, et al, 2013 | Romania | 137/139 | 9 | 66 | 62 | 7 | 50 | 82 |
7.40×10−2 | 84:190 | 64:214 |
4.30×10−2a | (17) |
| Chen C,
et al, 2015 | China | 368/460 | NA | NA | NA | NA | NA | NA | NA | 361:375 | 431:489 |
3.73×10−1 | (14) |
| rs1065407(C/A),
96,776,379 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Davidson SI, et al,
2009 | China | 527/945 | 0 | 52 | 439 | 3 | 74 | 767 |
2.33×10−1 | 52:930 | 80:1,608 |
5.23×10−1 | (20) |
| Chen C,
et al, 2015 | China | 368/460 | NA | NA | NA | NA | NA | NA | NA | 52:684 | 172:748 |
<1.00×10−7a | (14) |
| rs27980(C/A),
96,762,191 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Davidson SI, et al,
2009 | China | 527/945 | 87 | 252 | 152 | 211 | 407 | 227 |
8.00×10−3a | 426:556 | 829:861 |
4.60×10−3a | (20) |
| Cinar
M, et al, 2013 | Turkey | 150/150 | 11 | 68 | 71 | 13 | 67 | 70 |
9.13×10−1 | 90:210 | 93:207 |
7.90×10−1 | (18) |
| Zhang
Z, et al, 2014 | China | 602/619 | 125 | 294 | 179 | 140 | 293 | 182 |
7.27×10−1 | 544:652 | 573:657 |
5.87×10−1 | (19) |
| rs7711564(G/C),
96,760,515 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Davidson SI, et al,
2009 | China | 527/945 | 88 | 249 | 150 | 209 | 405 | 226 |
1.40×10−2a | 425:549 | 823:857 |
7.70×10−3a | (20) |
| Cinar
M, et al, 2013 | Turkey | 150/150 | 4 | 60 | 86 | 10 | 56 | 84 |
2.55×10−1 | 68:232 | 76:224 |
4.45×10−1 | (18) |
| Wang J,
et al, 2015 | China | 100/100 | 28 | 59 | 13 | 41 | 53 | 6 |
6.90×10−2 | 115:85 | 135:65 |
3.90×10−2a | (15) |
| rs27037(T/G),
96,758,990 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Davidson SI, et al,
2009 | China | 527/945 | 97 | 258 | 135 | 139 | 402 | 285 |
2.80×10−2a | 452:528 | 680:972 |
1.30×10−2a | (20) |
| Bang
SY, et al, 2011 | Korea | 1,164/752 | 142 | 578 | 403 | 89 | 280 | 343 |
3.20×10−7a | 862:1,384 | 458:966 |
1.30×10−4a | (12) |
| Cinar
M, et al, 2013 | Turkey | 150/150 | 5 | 77 | 68 | 1 | 71 | 78 |
1.66×10−1 | 87:213 | 73:227 |
1.97×10−1 | (18) |
| Zhang
Z, et al, 2014 | China | 602/619 | 99 | 295 | 201 | 94 | 296 | 224 |
5.83×10−1 | 493:697 | 484:744 |
3.13×10−1 | (19) |
Meta-analysis
A total of 9 studies, which included 7 studies
involving subjects of European-descent and two studies involving
subject of Asian descent, contributed 4,469 cases and 7,324
controls for the analysis of the association between the
ERAP1 SNP rs30187 and AS. Using a random-effects model, a
significant difference was identified between the patients and
controls for the T-allele of rs30187 (subtotal OR=1.27; 95%
CI=1.15–1.40; Z=4.73; P<1.0×10−5) in all of the
samples combined, and significant evidence of between-study
heterogeneity was identified among the group of allele-wise ORs
(P=0.03; I2=54%; Table
III). In addition, studies were analyzed separately by
ethnicity (European and Asian) to limit the ethnic heterogeneity.
The fixed-effects model was used for the European studies (P=0.53;
I2=0%), and the random-effects model was used for the
Asian studies (P=0.004, I2=82%) according to their
heterogeneity tests. A statistically significant summary OR was
identified in European studies (subtotal OR=1.29; 95% CI=1.17–1.41;
Z=5.48; P<1.0×10−5) and Asian studies (subtotal
OR=1.31; 95% CI=1.06–1.63; Z=2.45; P=0.01; Table III).
 | Table III.Fixed- and random-effects model
summary OR and 95% CI values for the ERAP1 SNPs, rs30187 and
rs17482078, associated with AS risk. |
Table III.
Fixed- and random-effects model
summary OR and 95% CI values for the ERAP1 SNPs, rs30187 and
rs17482078, associated with AS risk.
|
|
|
| Heterogeneity
test | Fixed-effects
model | Random-effects
model |
|---|
|
|
|
|
|
|
|
|---|
| SNPs (minor allele)
and analysis (included studies/included samples) | I2
(%) | P-value | OR (95% CI) | Z-test | P-value | OR (95% CI) | Z-test | P-value |
|---|
| rs30187 (T) |
|
|
|
|
|
|
|
|
|
Combined (9/9) | 54 |
3.0×10−2 |
|
|
| 1.27
(1.15–1.40) | 4.73 |
<1.00×10−5a |
| Asian
(3/3) | 82 |
4.0×10−3 |
|
|
| 1.31
(1.06–1.63) | 2.45 |
1.00×10−2a |
|
European (6/6) | 0 |
5.3×10−1 | 1.29
(1.17–1.41) | 5.48 |
<1.00×10−5a |
|
|
|
| rs17482078 (T) |
|
|
|
|
|
|
|
|
|
European (5/5) | 0 |
7.80×10−1 | 0.73
(0.65–0.82) | 5.25 |
<1.00×10−5a |
|
|
|
The results of the publication bias tests for the
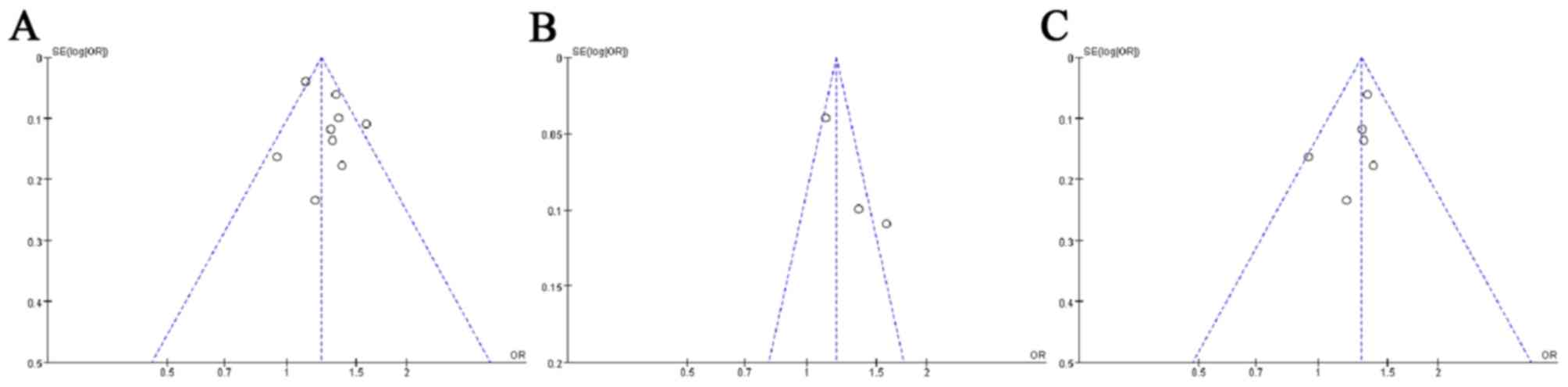
rs30187 SNP are presented in Fig.
1. The results demonstrated that no publication bias existed in
this group, as the shapes of the funnel plots did not reveal any
obvious asymmetry.
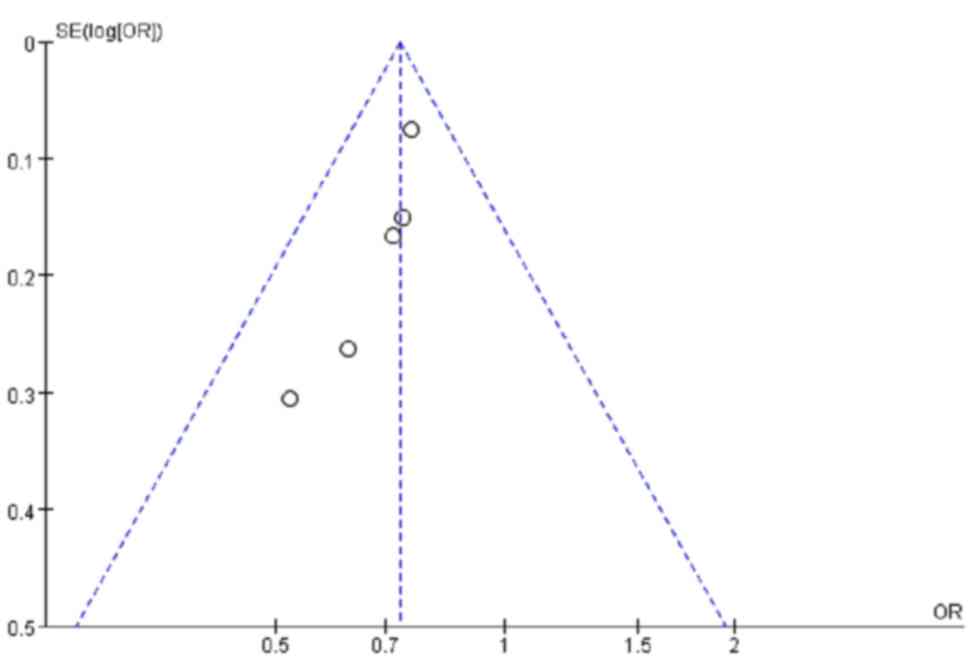
A total of 5 studies involving European populations
contributed 1,748 cases and 2,190 controls for the analysis of the
association between the ERAP1 SNP rs17482078 and AS. For the
T-allele of rs17482078, no significant heterogeneity was detected
(P=0.78; I2=0%). The fixed-effects summary OR was 0.73
(95% CI=0.65–0.82), and a significant association was observed
(Z=5.25; P<1.0×10−5; Table III). There was no evidence of
publication bias in these 5 studies (Fig. 2).
Further meta-analyses were performed using the
random-effects or fixed-effects models for rs3734016, rs26653,
rs27895, rs2287987, rs27434, rs10050860, rs27044, rs1065407,
rs27980, rs7711564 and rs27037 SNPs, according to the results of
the heterogeneity test. The summary ORs for 8 SNPs (rs26653,
rs2287987, rs27434, rs10050860, rs27044, rs27980, rs7711564 and
rs27037) were statistically significant in the combined, European
studies and Asian studies (Table
IV). There was no evidence of publication bias for these SNPs
in their associated studies (data not shown).
Structural and functional
analysis
A total of 9 SNPs (rs3734016, rs26653, rs27895,
rs2287987, rs27434, rs30187, rs10050860, rs17482078 and rs27044)
lead to a genetic variation within the coding region of the
ERAP1 gene. The PolyPhen-2 software program was used to
predict the structural and functional effects of these variations
on ERAP1, and the results are presented in Table V. For rs30187 and rs17482078, the
results of the functional prediction analysis suggested that these
mutations were potentially damaging (scores 0.998 and 0.759,
respectively). No predictions outside of the benign score range
were identified for the remaining 6 SNPs (rs3734016, rs26653,
rs27895, rs2287987, rs10050860 and rs27044). As the rs27434 SNP
generates a synonymous substitution in exon 6, the PolyPhen-2
software was unable to analyze it (Table V).
 | Table V.Predicted effects of the identified
SNPs on ERAP1 protein function. |
Table V.
Predicted effects of the identified
SNPs on ERAP1 protein function.
| ERAP1 SNP | Genomic coordinates
(bp) | Amino acid sequence
alteration or gene location | PolyPhen-2
phenotype prediction | Score |
|---|
| rs3734016 | 96,803,761 | E56K | Benign | 0.008 |
| rs26653 | 96,803,547 | R127P | Benign | 0.000 |
| rs27895 | 96,793,840 | G346D | Benign | 0.016 |
| rs2287987 | 96,793,832 | M349V | Benign | 0.203 |
| rs27434 | 96,793,809 | A356A | – | – |
| rs30187 | 96,788,627 | K528R | Probably
damaging | 0.998 |
| rs10050860 | 96,786,506 | D575N | Benign | 0.000 |
| rs17482078 | 96,783,162 | R725Q | Probably
damaging | 0.759 |
| rs27044 | 96,783,148 | Q730E | Benign | 0.038 |
| rs1065407 | 96,776,379 | Intron | – | – |
| rs27980 | 96,762,191 | 3′-UTR | – | – |
| rs7711564 | 96,760,515 | Proximal to the
3′-end of ERAP1 | – | – |
| rs27037 | 96,758,990 | Proximal to the
3′-end of ERAP1 | – | – |
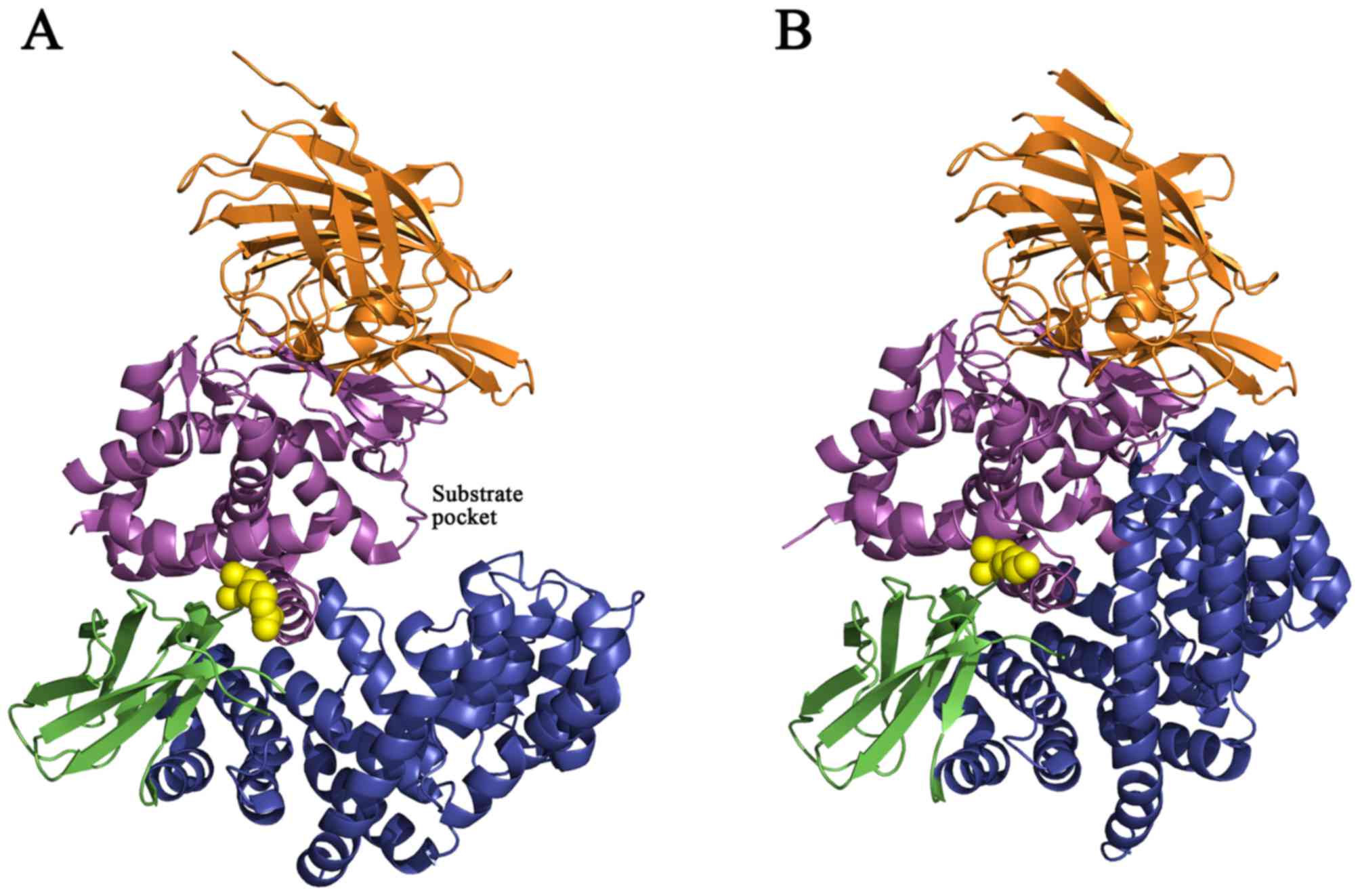
The crystal structure of ERAP1 revealed four protein
domains and a large cavity between domains II and IV (Figs. 3 and 4) (35).
Domain I (residues 46–254; brown region, Figs. 4 and 5) is an all-β-sheet domain that docks
above the thermolysin domain, caps the active site and provides
binding sites for the N-terminus of a substrate peptide. Domain II
(residues 255–529; purple region, Figs. 3 and 4) is the catalytic domain that possesses
a zinc atom, the exo-peptidase specific G-A-M-E-N motif and the
canonical zinc-binding motif (H-E-X-X-H-X18-E) on a
thermolysin-like αβ fold. Domain III (residues 530–614; green
region, Figs. 3 and 4) is composed of two β-sheets that forms
a β-sandwich domain between domains II and IV. Domain IV (residues
615–940; blue region, Figs. 3 and
4) consisted solely of α-helices
and displayed a bowl-like shape. In the closed state, domain IV
arches over the catalytic domain and forms a large central cavity
that completely obstructs the active site (35,36).
ERAP1 is a multifunctional enzyme involved in
cleaving peptides to an optimal length for presentation by MHC
class I molecules. The crystal structures of ERAP1 display open and
close states, and this enzyme is inactive in its open form
(36). The rs30187 SNP (K528R;
yellow circles, Fig. 3) is located
near the entrance of the substrate pocket and may affect
substrate-binding affinity with the enzyme and reduce ERAP1
aminopeptidase activity toward a synthetic peptide substrate
(Fig. 3) (11,37).
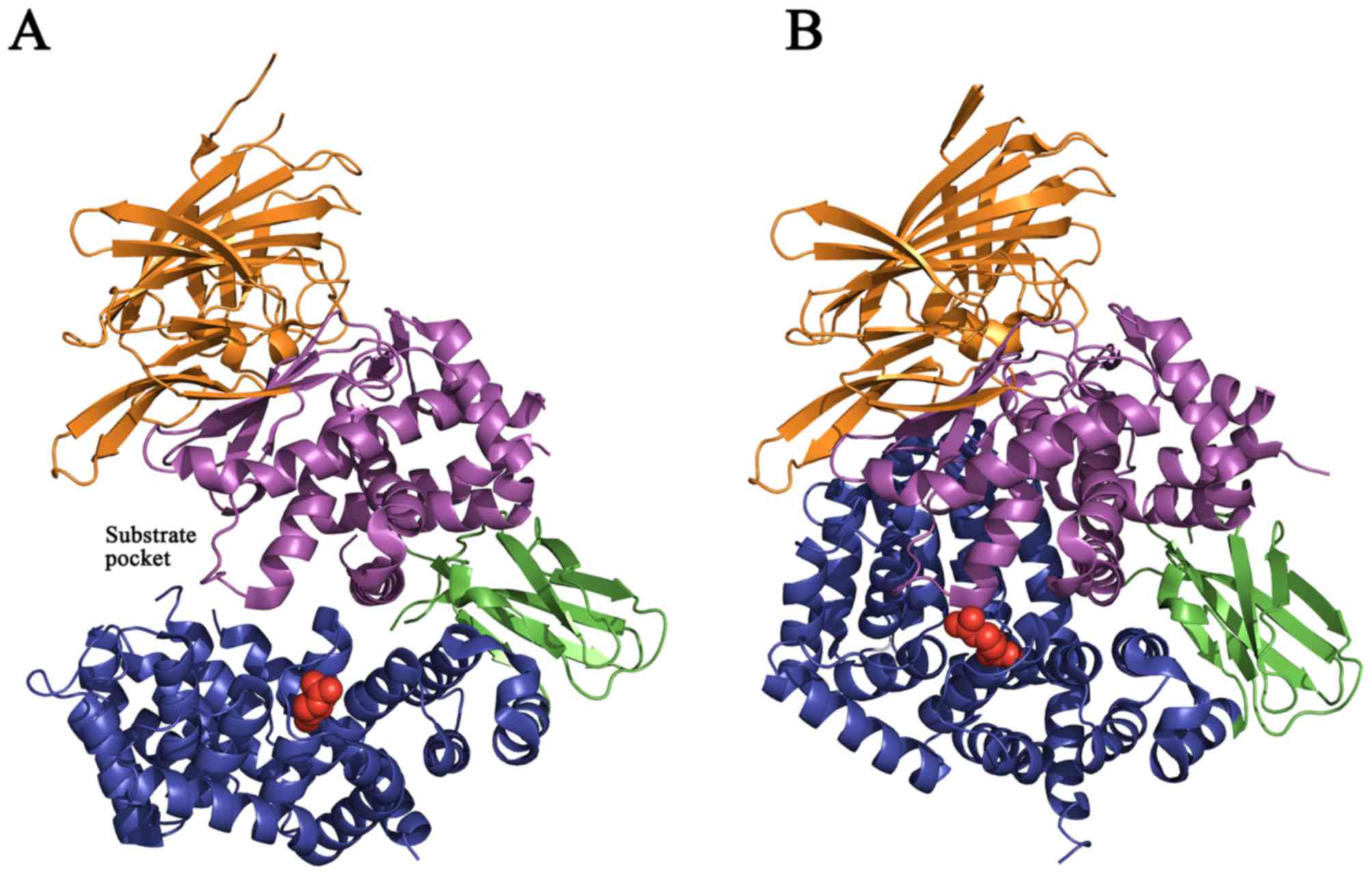
In addition, the rs17482078 SNP (R725Q; red circles, Fig. 4) is located on the inner surface of
the C-terminal cavity and may affect the substrate sequence or
length specificity (35).
Protein structure prediction analysis provided
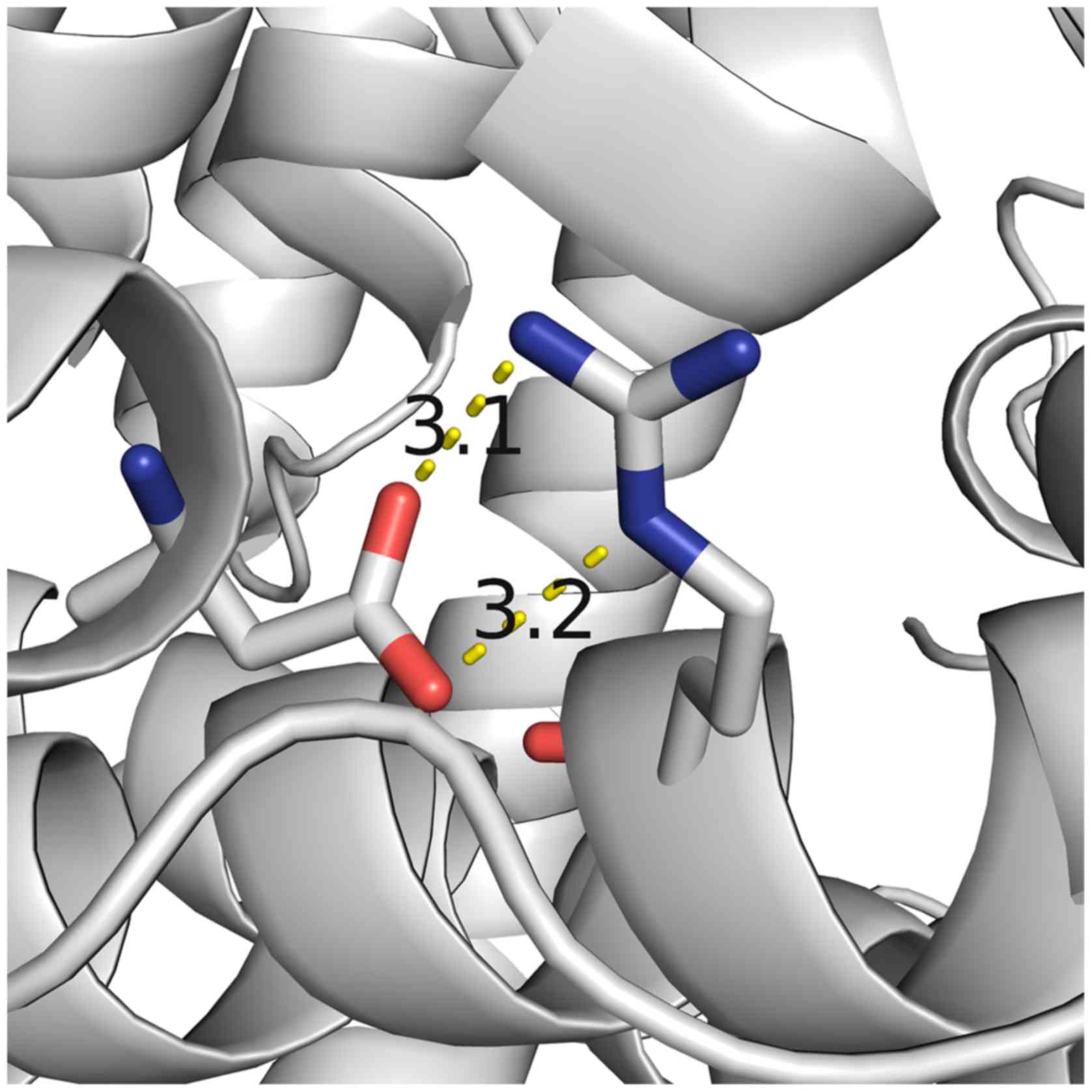
evidence for the functional role of amino acid residue R725. Two
hydrogen bonds were observed to form between R725 and D766 residues
with distances of 3.1 Å and 3.2 Å in the closed form of ERAP1
(Fig. 5). Based on the
aforementioned results and the molecular modeling structure of
ERAP1, the R725Q SNP may therefore affect the stability of the
C-terminus of ERAP1 in its active state.
Discussion
In the present study, a two-stage bioinformatic
analysis was performed to investigate the association between 13
SNPs in the ERAP1 locus and AS using ethnically diverse
independent samples from 14 previously published studies (24). The functional effects of
non-synonymous variations were analyzed with protein structure
prediction analysis software, and the crystal structure of ERAP1
was examined using the PDB database.
For the rs30187 SNP, the p-value in the combined
population was <0.00001, suggesting an unequivocal association
with AS. When the samples were stratified by ethnicity (European
and Asian), the p-values for the association tests for this SNP
remained significant (<0.00001 and 0.02, respectively), thus
providing additional evidence for the association of this SNP with
AS in the two populations (22).
For the rs17482078 SNP, a significant association was observed in
the European population (P<0.00001), therefore suggesting an
association of this SNP with AS. In addition, 11 additional SNPs
(rs3734016, rs26653, rs27895, rs2287987, rs27434, rs10050860,
rs27044, rs1065407, rs27980, rs7711564 and rs27037) were
investigated to determine their association with AS. The summary
ORs for 8 SNPs (rs26653, rs2287987, rs27434, rs10050860, rs27044,
rs27980, rs7711564, and rs27037) were statistically significant in
the combined, European and Asian studies when using the
random-effects or fixed-effects models. However, prior to the
meta-analysis, 10 studies investigating ERAP1 and AS were
excluded from the final statistical analysis due to limited
available data. All of the excluded studies also demonstrated
significant associations in the above 13 SNPs between the case and
control populations (data not shown). These results provided
further evidence of an association between ERAP1 and AS.
Therefore, the results of the present study suggest that
ERAP1 may be an important susceptibility gene for AS, which
is consistent with the results from a number of previously
published studies (1,7,10,12,14,18,20,27,30,32,38–40).
The ERAP1 gene is located on chromosome 5q15
and is translated into two isoforms comprising 941 and 948 amino
acids, which are generated via alternative splicing. The active
site of ERAP1 spans 375 amino acids (41). Although ERAP1 does not contain any
obvious endoplasmic reticulum (ER) retention motifs, as identified
in additional ER resident proteins, it is known to localize within
the ER; however, exon 10 may serve a role in ER retention (42). ERAP1 is known to have two major
functions. Firstly, it is involved in cleaving peptides to the
optimal length for MHC class I presentation, and secondly, it
cleaves different cell surface cytokine receptors, including tumor
necrosis factor receptor 1, interleukin (IL)-1 receptor II and IL-6
receptor α (43).
A previous meta-analysis investigated the potential
molecular mechanisms underlying the effect of different genetic
variants of ERAP 1in the development of AS (40). Goto et al (37) demonstrated that a K528R
substitution resulted in reduced ERAP1 enzymatic activity by
reducing the hydrolysis of the bioactive hormones, angiotensin II
and kallidin (37,42). In addition, these authors
identified little difference between the activities of the N575 and
E730 mutants when compared with the wild-type (WT), and their
results were consistent with the PolyPhen-2 prediction analysis
performed in the present study (42). The potentially damaging
substitution at R528 (rs30187) has been well documented, and
similar results among studies have indicated a 30–40% reduction in
enzymatic activity compared with that of the WT (34–37,44).
K528R is located near the entrance of the substrate pocket, and may
contribute to substrate-binding affinity, thus leading to reduced
ERAP1 aminopeptidase activity with a synthetic peptide substrate
(37).
It is hypothesized that Q725 (rs17482078) is an
additional potentially damaging substitution, and various in
vitro studies have suggested that it decreases the enzymatic
activity by 40% compared with that of the WT (34,37).
The protein structure analysis performed in the present study,
suggested that this substitution may result the disruption of two
hydrogen bonds between R725 and D766 in the active state of ERAP1
and affect the stability of the C-terminus. Based on these
alterations, a decrease in the enzymatic activity induced by the
substitution of R to Q at position 725 may be expected. However, in
the present study, the results of the meta-analysis at stage 1
suggested the opposite, as the minor allele of rs17482078 (Q725)
was observed to theoretically decrease the risk of AS in the
case-control studies involving different populations. Due to the
limited understanding of the association between the structure and
function of ERAP1, a simple interpretation of the role of
rs17482078 is therefore not possible at this stage. Therefore, as
further studies emerge, the current findings may be updated and
more reliable estimates of the role of this SNP may be
obtained.
In conclusion, the results of the two-stage
bioinformatics analysis performed in the current study suggested
that ERAP1 may present an important susceptibility gene for
AS, and revealed two functional SNPs (rs30187 and rs17482078) that
may decrease the enzymatic activity of ERAP1 by affecting its
protein structure. Therefore, future studies investigating the role
of these ERAP1 variants in influencing protein structure are
warranted.
Acknowledgements
The authors of the present study are indebted to all
of the individuals who have participated in, or helped with, the
research. The present study was supported by the National Natural
Science Foundation of China (grant nos. 31371298, 81301151 and
81272023), the Program for New Century Excellent Talents in
University (grant no. NCET-13-0452) and the Key Project of
International Scientific Cooperation of Shaanxi Province (grant no.
S2013KW25-02).
References
|
1
|
Lin Z, Bei JX, Shen M, Li Q, Liao Z, Zhang
Y, Lv Q, Wei Q, Low HQ, Guo YM, et al: A genome-wide association
study in Han Chinese identifies new susceptibility loci for
ankylosing spondylitis. Nat Genet. 44:73–77. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kadi A, Izac B, Said-Nahal R, Leboime A,
Van Praet L, de Vlam K, Elewaut D, Chiocchia G and Breban M:
Investigating the genetic association between ERAP1 and
spondyloarthritis. Ann Rheum Dis. 72:608–613. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Braun J, Bollow M, Remlinger G, Eggens U,
Rudwaleit M, Distler A and Sieper J: Prevalence of
spondylarthropathies in HLA-B27 positive and negative blood donors.
Arthritis Rheum. 41:58–67. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cai G, Xin L, Wang L, Fan D, Liu L, Hu Y,
Ding N, Xu S, Xia G, Jin X, et al: Associations between ERAP1
polymorphisms and ankylosing spondylitis susceptibility: An updated
meta-analysis. Mod Rheumatol. 25:453–461. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Brewerton DA, Hart FD, Nicholls A, Caffrey
M, James DC and Sturrock RD: Ankylosing spondylitis and HL-A 27.
Lancet. 1:904–907. 1973. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
van der Linden S, Valkenburg H and Cats A:
The risk of developing ankylosing spondylitis in HLA-B27 positive
individuals: A family and population study. Br J Rheumatol. 22 4
Suppl 2:S18–S19. 1983. View Article : Google Scholar
|
|
7
|
Wellcome Trust Case Control Consortium1;
Australo-Anglo-American Spondylitis Consortium (TASC), ; Burton PR,
Clayton DG, Cardon LR, Craddock N, Deloukas P, Duncanson A,
Kwiatkowski DP, McCarthy MI, et al: Association scan of 14,500
nonsynonymous SNPs in four diseases identifies autoimmunity
variants. Nat Genet. 39:1329–1337. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Maksymowych WP, Inman RD, Gladman DD,
Reeve JP, Pope A and Rahman P: Association of a specific
ERAP1/ARTS1 haplotype with disease susceptibility in ankylosing
spondylitis. Arthritis Rheum. 60:1317–1323. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zvyagin IV, Dorodnykh VY, Mamedov IZ,
Staroverov DB, Bochkova AG, Rebrikov DV and Lebedev YB: Association
of ERAP1 allelic variants with risk of ankylosing spondylitis. Acta
Naturae. 2:72–77. 2010.PubMed/NCBI
|
|
10
|
Pazár B, Sáfrány E, Gergely P, Szántó S,
Szekanecz Z and Poór G: Association of ARTS1 gene polymorphisms
with ankylosing spondylitis in the Hungarian population: The
rs27044 variant is associated with HLA-B*2705 subtype in Hungarian
patients with ankylosing spondylitis. J Rheumatol. 37:379–384.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Szczypiorska M, Sánchez A, Bartolomé N,
Arteta D, Sanz J, Brito E, Fernández P, Collantes E, Martínez A,
Tejedor D, et al: ERAP1 polymorphisms and haplotypes are associated
with ankylosing spondylitis susceptibility and functional severity
in a Spanish population. Rheumatology (Oxford). 50:1969–1975. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bang SY, Kim TH, Lee B, Kwon E, Choi SH,
Lee KS, Shim SC, Pope A, Rahman P, Reveille JD and Inman RD:
Genetic studies of ankylosing spondylitis in Koreans confirm
associations with ERAP1 and 2p15 reported in white patients. J
Rheumatol. 38:322–324. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mahmoudi M, Jamshidi AR, Amirzargar AA,
Farhadi E, Nourijelyani K, Fallahi S, Oraei M, Noori S and Nicknam
MH: Association between endoplasmic reticulum aminopeptidase-1
(ERAP-1) and susceptibility to ankylosing spondylitis in Iran. Iran
J Allergy Asthma Immunol. 11:294–300. 2012.PubMed/NCBI
|
|
14
|
Chen C and Zhang X: ERAP1 variants are
associated with ankylosing spondylitis in East Asian population: A
new Chinese case-control study and meta-analysis of published
series. Int J Immunogenet. 42:168–173. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang J, Li H, Wang J and Gao X:
Association between ERAP1 gene polymorphisms and ankylosing
spondylitis susceptibility in Han population. Int J Clin Exp
Pathol. 8:11641–11646. 2015.PubMed/NCBI
|
|
16
|
Wu W, Ding Y, Chen Y, Hua Z, Liu H, Wang H
and Jiao G: Susceptibility to ankylosing spondylitis: Evidence for
the role of ERAP1, TGFb1 and TLR9 gene polymorphisms. Rheumatol
Int. 32:2517–2521. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cherciu M, Popa LO, Bojinca M, Dutescu MI,
Bojinca V, Bara C and Popa OM: Functional variants of ERAP1 gene
are associated with HLA-B27 positive spondyloarthritis. Tissue
Antigens. 82:192–196. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cinar M, Akar H, Yilmaz S, Simsek I,
Karkucak M, Sagkan RI, Pekel A, Erdem H, Avci IY, Acikel C, et al:
A polymorphism in ERAP1 is associated with susceptibility to
ankylosing spondylitis in a Turkish population. Rheumatol Int.
33:2851–2858. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang Z, Dai D, Yu K, Yuan F, Jin J, Ding
L, Hao Y, Liang F, Liu N, Zhao X, et al: Association of HLA-B27 and
ERAP1 with ankylosing spondylitis susceptibility in Beijing Han
Chinese. Tissue Antigens. 83:324–329. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Davidson SI, Wu X, Liu Y, Wei M, Danoy PA,
Thomas G, Cai Q, Sun L, Duncan E, Wang N, et al: Association of
ERAP1, but not IL23R, with ankylosing spondylitis in a Han Chinese
population. Arthritis Rheum. 60:3263–3268. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chapman K, Takahashi A, Meulenbelt I,
Watson C, Rodriguez-Lopez J, Egli R, Tsezou A, Malizos KN,
Kloppenburg M, Shi D, et al: A meta-analysis of European and Asian
cohorts reveals a global role of a functional SNP in the 5UTR of
GDF5 with osteoarthritis susceptibility. Hum Mol Genet.
17:1497–1504. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang R, Yao J, Xu P, Ji B, Luck JV, Chin
B, Lu S, Kelsoe JR and Ma J: A comprehensive meta-analysis of
association between genetic variants of GDF5 and osteoarthritis of
the knee, hip and hand. Inflamm Res. 64:405–414. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu M, St Clair D and He L: Testing for
genetic association between the ZDHHC8 gene locus and
susceptibility to schizophrenia: An integrated analysis of multiple
datasets. Am J Med Genet B Neuropsychiatr Genet. 153B:1–1275. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nie F, Wang X, Zhao P, Yang H, Zhu W, Zhao
Y, Chen B, Valenzuela RK, Zhang R, Gallitano AL and Ma J: Genetic
analysis of SNPs in CACNA1C and ANK3 gene with schizophrenia: A
comprehensive meta-analysis. Am J Med Genet B Neuropsychiatr Genet.
168:637–648. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Adzhubei IA, Schmidt S, Peshkin L,
Ramensky VE, Gerasimova A, Bork P, Kondrashov AS and Sunyaev SR: A
method and server for predicting damaging missense mutations.
Nature methods. 7:248–249. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Berman HM, Westbrook J, Feng Z, Gilliland
G, Bhat TN, Weissig H, Shindyalov IN and Bourne PE: The protein
data bank. Nucleic Acids Res. 28:235–242. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pimentel-Santos FM, Ligeiro D, Matos M,
Mourão AF, Sousa E, Pinto P, Ribeiro A, Sousa M, Barcelos A,
Godinho F, et al: Association of IL23R and ERAP1 genes with
ankylosing spondylitis in a Portuguese population. Clin Exp
Rheumatol. 27:800–806. 2009.PubMed/NCBI
|
|
28
|
Harvey D, Pointon JJ, Evans DM, Karaderi
T, Farrar C, Appleton LH, Sturrock RD, Stone MA, Oppermann U, Brown
MA and Wordsworth BP: Investigating the genetic association between
ERAP1 and ankylosing spondylitis. Hum Mol Genet. 18:4204–4212.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Choi CB, Kim TH, Jun JB, Lee HS, Shim SC,
Lee B, Pope A, Uddin M, Rahman P and Inman RD: ARTS1 polymorphisms
are associated with ankylosing spondylitis in Koreans. Ann Rheum
Dis. 69:582–584. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Australo-Anglo-American Spondyloarthritis
Consortium (TASC), ; Reveille JD, Sims AM, Danoy P, Evans DM, Leo
P, Pointon JJ, Jin R, Zhou X, Bradbury LA, et al: Genome-wide
association study of ankylosing spondylitis identifies non-MHC
susceptibility loci. Nat Genet. 42:123–127. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li C, Lin Z, Xie Y, Guo Z, Huang J, Wei Q,
Li QX, Wang X, Cao S, Liao Z, et al: ERAP1 is associated with
ankylosing spondylitis in Han Chinese. J Rheumatol. 38:317–321.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang CM, Ho HH, Chang SW, Wu YJ, Lin JC,
Chang PY, Wu J and Chen JY: ERAP1 genetic variations associated
with HLA-B27 interaction and disease severity of syndesmophytes
formation in Taiwanese ankylosing spondylitis. Arthritis Res Ther.
14:R1252012. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu Y, Li L, Shi S, Chen X, Gao J, Zhu M
and Yuan J: Association study of ankylosing spondylitis and
polymorphisms in ERAP1 gene in Zhejiang Han Chinese population.
Rheumatol Int. 36:243–248. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Evans DM, Spencer CC, Pointon JJ, Su Z,
Harvey D, Kochan G, Oppermann U, Dilthey A, Pirinen M, Stone MA, et
al: Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis
implicates peptide handling in the mechanism for HLA-B27 in disease
susceptibility. Nat Genet. 43:761–767. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Nguyen TT, Chang SC, Evnouchidou I, York
IA, Zikos C, Rock KL, Goldberg AL, Stratikos E and Stern LJ:
Structural basis for antigenic peptide precursor processing by the
endoplasmic reticulum aminopeptidase ERAP1. Nat Struct Mol Biol.
18:604–613. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kochan G, Krojer T, Harvey D, Fischer R,
Chen L, Vollmar M, von Delft F, Kavanagh KL, Brown MA, Bowness P,
et al: Crystal structures of the endoplasmic reticulum
aminopeptidase-1 (ERAP1) reveal the molecular basis for N-terminal
peptide trimming. Proc Natl Acad Sci USA. 108:7745–7750. 2011;
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Goto Y, Hattori A, Ishii Y and Tsujimoto
M: Reduced activity of the hypertension-associated Lys528Arg mutant
of human adipocyte-derived leucine aminopeptidase
(A-LAP)/ER-aminopeptidase-1. FEBS Lett. 580:1833–1838. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Robinson PC and Brown MA: Genetics of
ankylosing spondylitis. Mol Immunol. 57:2–11. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lee YH, Choi SJ, Ji JD and Song GG:
Associations between ERAP1 polymorphisms and ankylosing spondylitis
susceptibility: A meta-analysis. Inflamm Res. 60:999–1003. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chen R, Yao L, Meng T and Xu W: The
association between seven ERAP1 polymorphisms and ankylosing
spondylitis susceptibility: A meta-analysis involving 8,530 cases
and 12,449 controls. Rheumatol Int. 32:909–914. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yousaf N, Low WY, Onipinla A, Mein C,
Caulfield M, Munroe PB and Chernajovsky Y: Differences between
disease-associated endoplasmic reticulum aminopeptidase 1 (ERAP1)
isoforms in cellular expression, interactions with tumour necrosis
factor receptor 1 (TNF-R1) and regulation by cytokines. Clin Exp
Immunol. 180:289–304. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Reeves E, Elliott T, James E and Edwards
CJ: ERAP1 in the pathogenesis of ankylosing spondylitis. Immunol
Res. 60:257–269. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Haroon N: Endoplasmic reticulum
aminopeptidase 1 and interleukin-23 receptor in ankylosing
spondylitis. Curr Rheumatol Rep. 14:383–389. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Evnouchidou I, Kamal RP, Seregin SS, Goto
Y, Tsujimoto M, Hattori A, Voulgari PV, Drosos AA, Amalfitano A,
York IA and Stratikos E: Cutting Edge: Coding single nucleotide
polymorphisms of endoplasmic reticulum aminopeptidase 1 can affect
antigenic peptide generation in vitro by influencing basic
enzymatic properties of the enzyme. J Immunol. 186:1909–1913. 2011.
View Article : Google Scholar : PubMed/NCBI
|