Introduction
Tumor incidence is associated with impaired
antitumor immunity in the patients. Effectively stimulating the
immune system to kill tumor cells is a critical factor for the
success or failure of antitumor immunotherapy (1). Efforts have been made to construct
antitumor vaccines, aiming to induce antitumor immunity using tumor
cells, genes that express tumor antigens or tumor-associated
proteins and peptides (2). Tumor
cell lysates (TCLs) have been reported to induce antitumor
immunity, since all antigens and signal transduction factors that
are present in tumor cells exist in the TCL (3,4);
thus, they may have potential to be used in antitumor vaccines. In
a dendritic cell (DC)-based antitumor study, lung cancer TCL was
used to activate mouse DCs in vitro; subsequently, the
activated DCs were transfused back into the mice to treat the tumor
(5). In addition, our previous
study combined Lewis lung cancer TCL with heat shock protein 65 to
activate immunocytes in mice; the results demonstrated that the
activated immunocytes induced anti-lung cancer immunity in
vivo (6). Although TCLs have
been used in antitumor immunotherapy, the key proteins involved in
antitumor immunity remain unknown. Receptor for activated C kinase
1 (RACK1) is expressed in numerous tumor types, including lung
cancer, colorectal cancer and hepatocellular carcinoma (7). This factor is able to activate the
Sonic hedgehog signaling pathway, which promotes tumor cell
proliferation and inhibits apoptosis (8). Similarly, catenin β-like 1 (CTNNBL1)
is a protein that has been detected in colorectal cancer,
osteosarcoma and B lymphocytes (9–12).
Although the function of this protein has yet to be determined, it
has been identified as a putative regulator of the canonical Wnt
signaling pathway using RNA interference technology, where it acts
upstream of, or in parallel to β-catenin, particularly in
colorectal cancer cell proliferation (9). RACK1 and CTNNBL1 are expressed in
numerous tumor cell types to transduce the Sonic hedgehog cell
proliferation signaling pathway; therefore, it is hypothesized that
these proteins must also exist in TCLs. In the present study, RACK1
and CTNNBL1 were detected in TCL prepared from Lewis lung cancer
cells, and their roles in the activation of mouse splenocytes were
determined.
Materials and methods
Animals and cell lines
Female C57BL/6 mice (n=9; age, 6–8 weeks; weight,
18–22 g) were purchased from Anhui Medical University Laboratory
Animal Center (Hefei, China) and were maintained in microisolator
cages under pathogen-free conditions, at a temperature of 20–26°C
under a 12-h light/dark cycle. Food and water was autoclaved before
feeding and provided ad libitum. The mice were evenly divided into
3 groups; a PBS group, whole TCL group and TCL
RACK1-/CTNNBL1-group. Experimental manipulation of the mice was
undertaken in accordance with the National Institutes of Health
Guide for the Care and Use of Laboratory Animals (https://www.ncbi.nlm.nih.gov/books/NBK54050), with the
approval of the Scientific Investigation Board of Science and
Technology of Anhui Province and the animal experiments of the
present study were also approved by the Ethics Committee of Wannan
Medical College. The mouse Lewis lung cancer cell line was
purchased from Wuhan Boster Biological Technology, Ltd. (Wuhan,
China). The cells were cultured in high-glucose Dulbecco's modified
Eagle's medium (Hyclone; GE Healthcare Life Sciences, Logan, UT,
USA) supplemented with 10% fetal calf serum (FCS; Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA), 100 U/ml penicillin and
100 µg/ml streptomycin (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany). The present study was approved by the Ethics Committee of
Wannan Medical College (Wuhu, China).
Preparation of TCL
To prepare the TCL, cultured Lewis cells were lysed
using a freeze-thaw cycle in PBS solution between −70°C and 37°C.
After five cycles, the prepared TCL was stored at −70°C until
further use. The TCL was observed under a microscope (Olympus
Corporation, Tokyo, Japan) using trypan blue staining
(Sigma-Aldrich; Merck KGaA) to ensure all of the cells were
lysed.
Preparation of mouse type II alveolar
epithelial cell lysate
To prepare the lysate, cultured type II alveolar
epithelial cells were lysed using a freeze-thaw cycle in PBS
solution between −70°C and 37°C. After five cycles, the prepared
lysate was stored at −70°C until further use. The lysate was
observed under a microscope (Olympus Corporation, Tokyo, Japan)
using trypan blue staining (Sigma-Aldrich; Merck KGaA) to ensure
all of the cells were lysed.
Isobaric tags for relative and
absolute quantitation (iTRAQ) analysis
TCL was prepared from Lewis lung cancer cells or
mouse type II alveolar epithelial cell using the above method.
Protein concentration of TCL was determined using a bicinchoninic
acid (BCA) assay using bovine serum albumin as standard (BCA Assay
kit; Beyotime Institute of Biotechnology, Shanghai, China).
Subsequently, 100 µg TCL protein was digested with trypsin (Promega
Corporation, Madison, WI, USA) overnight at 37°C. Digested samples
were labeled using the 8-plex iTRAQ kit (Applied Biosystems; Thermo
Fisher Scientific Inc.) according to the manufacturer's protocol.
iTRAQ-labeled samples were diluted to 100 µl with 20 mM
HCOONH4, 2M NaOH (pH 10) prior to high-performance
liquid chromatography on a Gemini-NX 3u C18 110A (150×2.00 mm)
column (Phenomenex, Torrance, CA, USA). Peptides were separated
using linear gradient elution; the mobile phases were comprised as
follows: Mobile phase A, 20 mM HCOONH4 and 2M NaOH;
mobile phase B, 20 mM HCOONH4, 2M NaOH and 80%
acetonitrile (ACN). The gradient was increased from 5 to 40% mobile
phase B in 30 min at a flow rate of 0.2 ml/min. The UV detector was
set at 214/280 nm, and fractions were collected every 1 min. In
total, 24 fractions were collected and dried by vacuum centrifuge
in 12,000 × g overnight at 4°C. Acidified with 50%
CF3COOH, the peptides were separated using linear
gradient elution; the mobile phases were comprised as follows:
Mobile phase A, 5% ACN and 0.1% formic acid (FA); mobile phase B,
80% ACN and 0.1% FA. The gradient was increased from 5 to 90%
mobile phase B in 50 min at a flow rate of 300 nl/min. The tandem
mass spectrometry (MS/MS) analysis was performed using a Q Exative
system (Thermo Fisher Scientific, Inc.) in Information-Dependent
Mode. Briefly, MS spectra were acquired across the mass range of
350–1800 m/z in high resolution mode (>70,000 full width at half
maximum) using a 40 msec accumulation time per spectrum. A maximum
of 20 precursors per cycle were chosen for fragmentation from each
MS spectrum with 60 msec minimum accumulation time for each
precursor and dynamic exclusion for 20 sec. Tandem mass spectra
were recorded in high sensitivity mode (resolution>17,500) with
rolling collision energy on and iTRAQ reagent collision energy
adjustment on. For protein identification, the MS/MS spectra were
processed using ProteinPilot software version 5.0 (Sciex,
Framingham, MA, USA). Differentially abundant proteins were
examined using QuickGO (https://www.ebi.ac.uk/QuickGO/) for Gene Ontology
annotation and enrichment analysis.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from mouse Lewis lung cancer
cell whole TCL and reverse transcribed into cDNA using an RNAprep
pure kit (Tiangen Biotech Co. Ltd., Beijing, China) and FastQuant
RT kit (with gDNase; Tiangen Biotech Co. Ltd.), respectively. Oligo
dT mixed with total RNA was and placed in a warm bath at 70°C for10
min, and then into ice-water mixture. dNTP\RTase\Rnase free
H2O was added to prepare the reverse transcription
reaction system. The system was warmed at 42°C for 1 h, and then
was heated to 70°C for10 min to produce cDNA. Subsequently, qPCR
was performed to detect the mRNA expression levels of RACK1,
CTNNBL1, Cullin 3, B-cell lymphoma 2-associated transcription
factor 1 (BCLAF1), ribosomal protein S6 (RPS6), superoxide
dismutase 1 (SOD1), high mobility group box 1 (HMGB1), inhibitor of
growth family member 1 (ING1), ERCC excision repair 3, TFIIH core
complex helicase subunit (ERCC3) and GAPDH in mouse Lewis lung
cancer cell TCL using a SuperReal PreMix Plus kit with SYBR Green
(Tiangen Biotech Co. Ltd.). Thermocycling conditions were 30 sec at
95°C, 2 step PCR at 95°C for 0.5 sec and at 60°C for 30 sec, for 40
cycles. Following dissociation was 1 cycle at 95°C for 15 sec, at
60°C for 30 sec and at 95°C for 15 sec successively. The qPCR data
were analyzed with the ABI Step One PCR system (Applied Biosystems;
Thermo Fisher Scientific Inc.). Relative gene expression was
quantified using the 2−ΔΔCq (13) and normalized to GAPDH (14). All qPCR primer sequences are
presented in Table I.
 | Table I.Primer sequences for quantitative
polymerase chain reaction. |
Table I.
Primer sequences for quantitative
polymerase chain reaction.
| Gene | Forward primer
(5′-3′) | Reverse primer
(5′-3′) |
|---|
| BCLAF1 |
TGAGACGACCTTATGGGTACA |
ATCTGCTTCGGGATCTTTGAG |
| RPS6 |
TGTTACTCCTCGTGTCCTGC |
CCAAAAGTTTAGCGTATTCTGC |
| SOD1 |
TGTACCAGTGCAGGACCTCAT |
GCCCAAGTCATCTTGTTTCTC |
| HMGB1 |
GCAGATGACAAGCAGCCCTAT |
GCTCTTTTCAGCCTTGACCAC |
| ING1 |
CAGGCAGATAAGCCGAATAAC |
AGGAGACCTGGTTGCACAGAC |
| ERCC3 |
CCCGATGTCTCCCGAGTTCTA |
GGTGTTGATTTTGGGGTTGTG |
| CTNNBL1 |
TTGGCTTACGGACCATCTTTC |
TGTTTCTCCCCTTCAATCTTCT |
| RACK1 |
TGAAGCAAGAAGTTATCAGCAC |
CTCGCACCAAGTTGTCTGTAT |
| GAPDH |
TGGTGAAGGTCGGTGTGAAC |
GCTCCTGGAAGATGGTGATGG |
Immunoprecipitation (IP)
Protein concentration of the TCL prepared from Lewis
lung cancer cells was measured using the BCA protein assay kit.
Subsequently, IP was conducted using a Pierce Classic IP kit (cat.
no. 26146; Pierce; Thermo Fisher Scientific, Inc.). For 1 mg
lysate, 80 µl Control Agarose Resin slurry was added to preclear
the lysate. Subsequently, anti-RACK1 (cat. no. 5432; dilution,
1:50; Cell Signaling Technology, Inc., Danvers, MA, USA) was added
to the precleared cell lysate in a microcentrifuge tube; the
antibody/lysate solution was diluted to 300–600 µl with PBS. The
solution was incubated overnight at 4°C to form the immune complex
in the spin column. To capture the immune complex, the
antibody/lysate sample was added to Protein A/G Plus Agarose in the
spin column (supplied in the IP kit), and the column was incubated
with gentle end-over-end shaking for 1 h at 4°C. Subsequently, the
sample was centrifuged 100 × g for 1 min and the flow-through was
saved. The spin column containing the resin was transferred into a
new collection tube and 2X reducing sample buffer was added and
incubated at 100°C for 5–10 min. The samples were further
centrifuged to collect the eluate. Finally, the eluate and
flow-through were separated by 10% SDS-PAGE and analyzed by western
blotting. First round IP can remove RACK1 from the TCL. To remove
CTNNBL1, the flow-through of first round IP was collected and then
used to process second round of IP with anti-CTNNBL1 at 5 µg/mg of
lysate (cat. no. ab95170; Abcam, Cambridge, MA, USA). Besides
antibody, other steps of second round of IP is the same as the
first round.
Flow cytometric analysis
C57/BL6 mice (3 per group) were used for spleen
isolation. The spleen was grinded, and depleted of red blood cells
using a red blood cell lysis buffer (Tiangen Biotech Co. Ltd.).
Then, the spleen cells were centrifuged at 500 × g for 5 min at
room temperature, counted and cocultured at a density of
1×106/ml with either TCL prepared from Lewis lung cancer
cells (1×106), TCL with RACK1 and CTNNBL1 removed or PBS
for 48 h at 37°C. Subsequently, the cells were collected, washed
and resuspended (1,000 cells) in PBS supplemented with 1%
heat-inactivated FCS. Thereafter, the mouse spleen cells were
stained at room temperature for 30 min with Annexin V-fluorescein
isothiocyanate (FITC)/propidium iodide apoptosis detection kit
(cat. no. KGA107; Nanjing KeyGen Biotech Co., Ltd., Nanjing, China)
to analyze apoptosis, or with FITC-labeled anti-cluster of
differentiation (CD)69 (cat. no. 11–0691-81; eBioscience; Thermo
Fisher Scientific, Inc.) to detect activation of spleen cells. The
cells were then stored at 4°C for 30 min, washed with 1X PBS and
analyzed by flow cytometry (FACSCalibur; BD Biosciences, San Jose,
CA, USA) using FlowJo version is 7.6 (FlowJo LLC, Ashland, OR,
USA). For flow cytometric detection, lymphocytes and monocytes were
gated, and ≤1×106 splenocytes were acquired per
test.
Western blot analysis
TCL was prepared from Lewis cells (1×106)
and was separated by 10% SDS-PAGE at 20 µg per lane. The proteins
were then transferred onto a polyvinylidene fluoride membrane. The
membrane was blocked with 5% evaporated milk for 1 h at room
temperature and was then incubated overnight at 4°C with anti-RACK1
(cat. no. 5432; 1:1,000; Cell Signaling Technology, Inc., Danvers,
MA, USA), anti-CTNNBL1 (cat. no. ab95170; 1:1,000; Abcam,
Cambridge, MA, USA), anti-Cullin 3 (cat. no. ab108407; 1:1,000;
Abcam) or anti-GAPDH monoclonal antibodies (cat. no. 5174; 1:1,000;
Cell Signaling Technology, Inc., Danvers, MA, USA). The membrane
was then incubated for 1.5 h with IRDye 800CW goat anti-mouse
secondary antibody (cat. no. P/N 925-32211; 1:10,000; LI-COR
Biosciences, Lincoln, NE, USA) at room temperature. To visualize
the protein on the membrane, the membrane was scanned and analyzed
using the Odyssey fluorescent scanning system (LI-COR
Biosciences).
Statistical analysis
Data are presented as the mean ± standard deviation
of 3 independent experiments. Statistical significance was
determined using χ2 test. P<0.05 was considered to
indicate a statistically significant difference. Statistical
analysis was performed using SPSS software version 22.0 (IBM Corp.,
Armonk, NY, USA).
Results
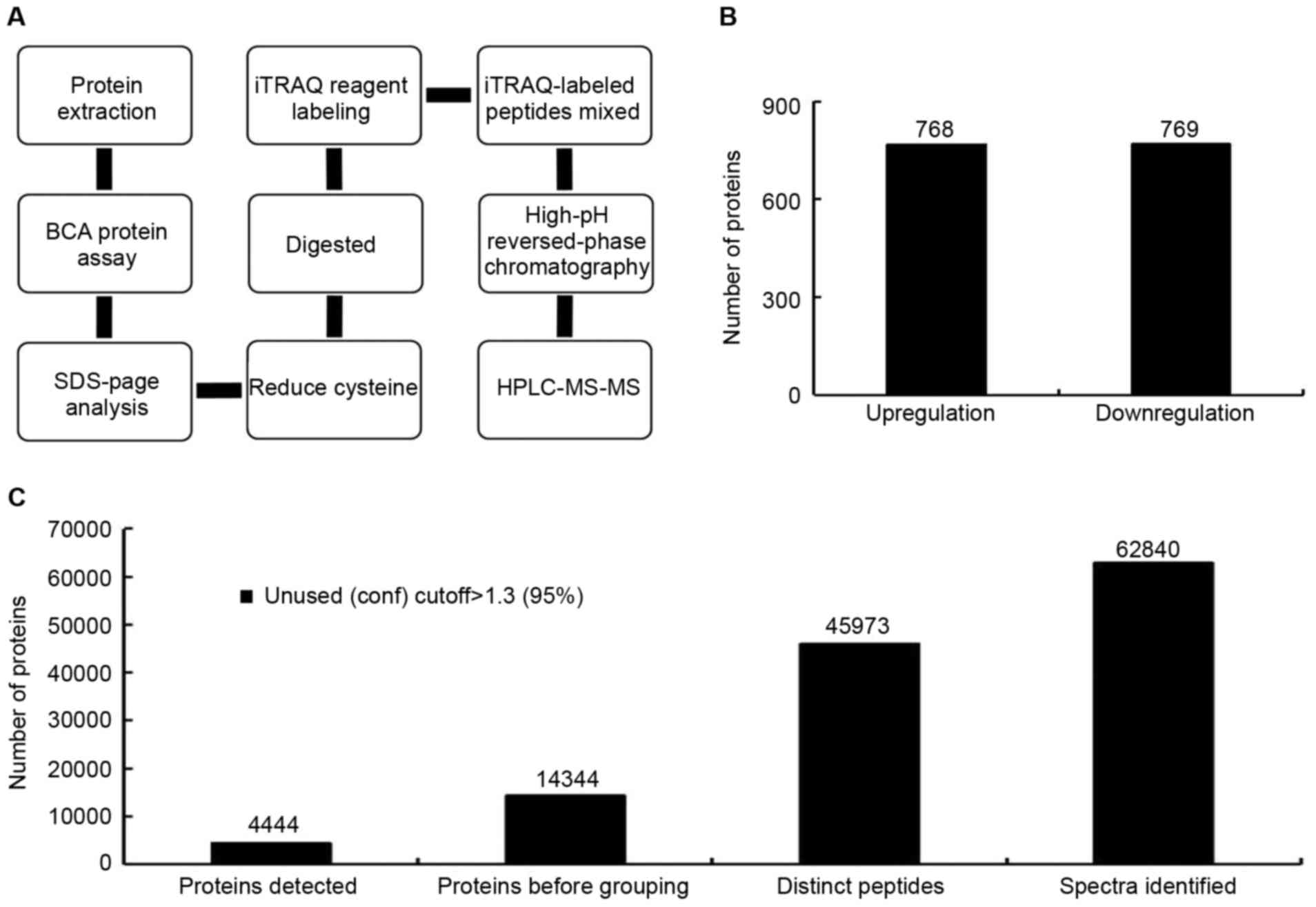
iTRAQ detection of proteins in Lewis
lung cancer TCL
Theoretically, all the proteins expressed in the
tumor cells should exist in the TCL prepared with from these cells.
To determine protein variety and quantitation in TCL, a TCL was
prepared from Lewis lung cancer cells and was analyzed using iTRAQ
(Fig. 1). Using the iTRAQ
detection method, 4,444 confidence proteins were detected in the
TCL (Fig. 1C). A confidence
protein is one that can be searched in the currently available
protein information databases, with high quantitation and
structural integrity. Among these proteins, 768 proteins were
upregulated in Lewis lung cancer TCL compared with mouse type II
alveolar epithelial cell lysate. The Lewis lung cancer TCL/type II
alveolar epithelial cell lysate ratio was >0.67 (Fig. 1B). A total of 769 proteins were
downregulated in the Lewis lung cancer TCL compared with the mouse
type II alveolar epithelial cell lysate with a ratio of >1.5
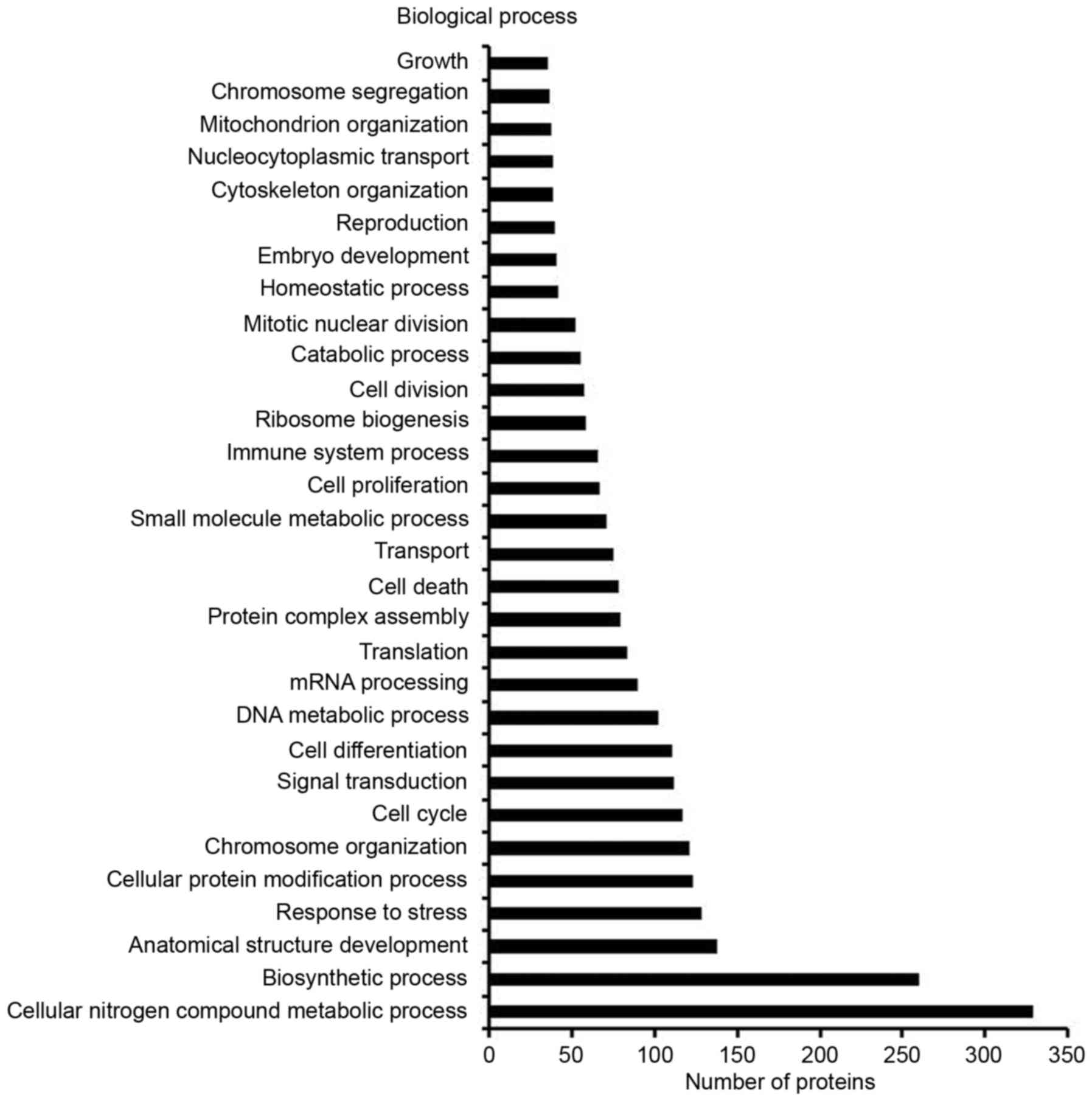
(Fig. 1B). These proteins could be
classified based on molecular function, cellular component or
biological process (Fig. 2),
including cell growth, cell division, cell death, cell
differentiation, cell proliferation and cell cycle. Among the
detected proteins that were differentially expressed between the
TCL and the alveolar epithelial cell lysate, 4.56% were associated
with cell growth, 7.42% were associated with cell division, 10.16%
were associated with cell death, 14.32% were associated with cell
differentiation, 8.72% were associated with cell proliferation and
15.23% were associated with cell cycle (Fig. 2). These proteins were selected as
the target proteins in the present study, since they serve an
important role in cell proliferation and apoptosis.
Target protein screen in Lewis lung
cancer TCL
Using the iTRAQ method, a large number of proteins
were detected in the TCL that are correlated with cell
proliferation. To determine the proteins that have the most
influence on cell proliferation and apoptosis, the functions of
these proteins were determined using protein databases and
previously published literature, and the ratio of protein
expression in Lewis lung cancer cell TCL/type II alveolar
epithelial cell lysate was analyzed. Subsequently, the following
proteins were selected as targets in the present study: RACK1,
CTNNBL1, Cullin 3, BCLAF1, RPS6, SOD1, HMGB1, ING1 and ERCC3. These
proteins have been revealed to promote cell proliferation and
inhibit apoptosis in numerous protein databases (Panther,
http://pantherdb.org/; UniProt, http://www.uniprot.org/; and NCBI Protein, https://www.ncbi.nlm.nih.gov/protein)
and in the literature (8,9,15–21).
In addition, these proteins exist in ≥2 signaling pathways that are
associated with cell proliferation or apoptosis, and the value of
type II alveolar epithelial cell lysate/Lewis lung cancer cell TCL
calculated by iTRAQ was <0.3 for these proteins. Western blot
analysis and RT-qPCR were used to detect the expression levels of
these proteins in TCL. The results demonstrated that RACK1, CTNNBL1
and Cullin 3 can be detected by WB, and the expression levels of
RACK1 and CTNNBL1 were higher than Cullin 3 in TCL (Fig. 3A). However, BCLAF1, RPS6, SOD1,
HMGB1, ING1 and ERCC3 can only be detected by RT-qPCR, which is
known to be more sensitive (Fig.
3B). In addition, protein expression was increased in whole TCL
compared with the TCL supernatant as determined by western blotting
(Fig. 3A). This may be caused by
the accumulation of proteins in the lysis cells debris; therefore,
the whole TCL was used to investigate its function in subsequent
experiments.
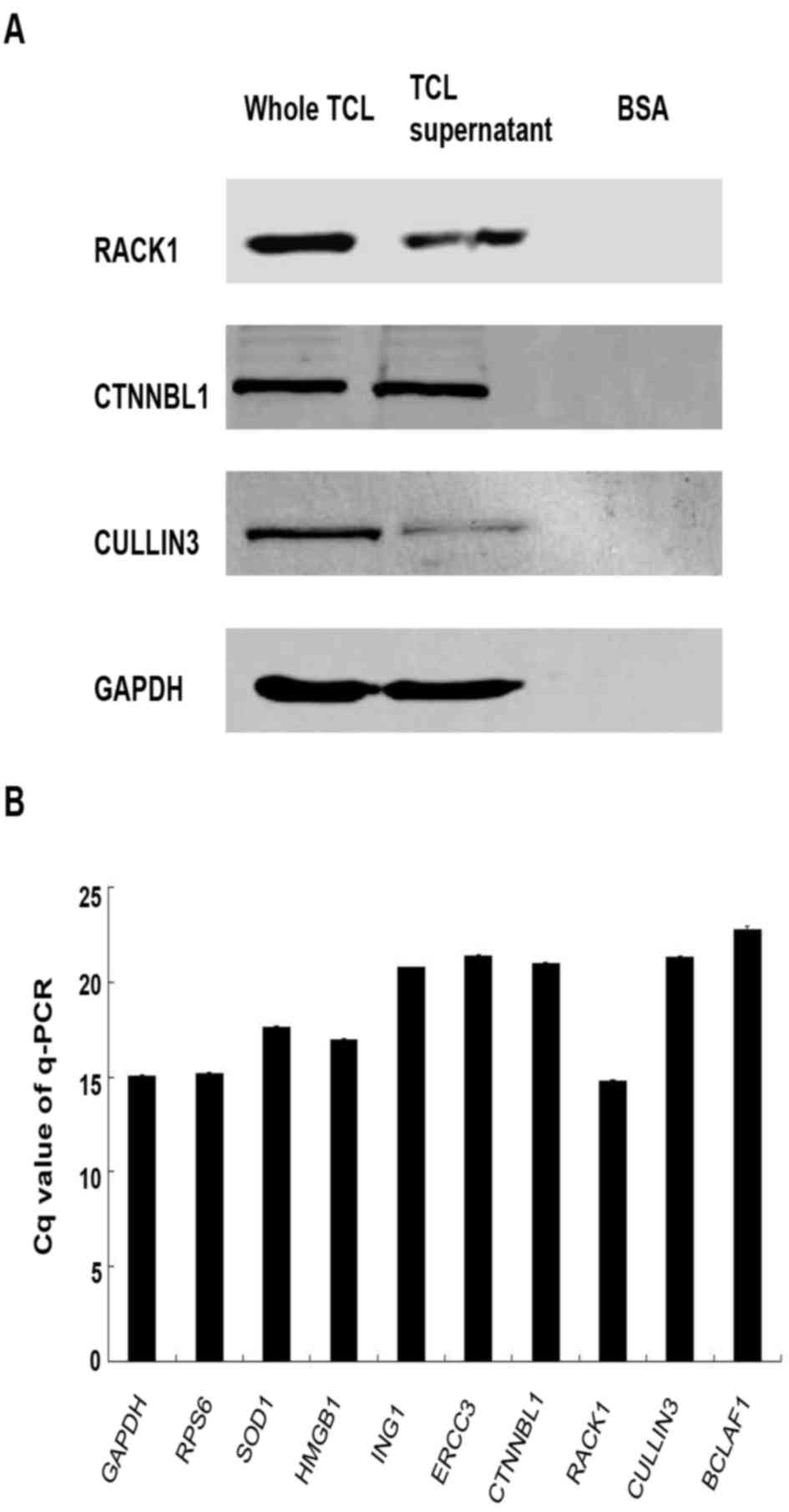 | Figure 3.RACK1, CTNNBL1, Cullin 3, BCLAF1,
RPS6, SOD1, HMGB1, ING1 and ERCC3 expression in TCL. (A) TCL was
prepared from 1×106 Lewis lung cancer cells. Immunoblot
analysis was performed to detect the protein expression levels of
RACK1, CTNNBL1 and Cullin 3 in TCL with specific antibodies. Based
on band intensity, RACK1 and CTNNBL1 appeared to be upregulated
compared with Cullin 3. (B) Reverse transcription-qPCR was used to
detect RACK1, CTNNBL1, Cullin 3, BCLAF1, RPS6, SOD1, HMGB1, ING1
and ERCC3 mRNA expression in 1×106 Lewis lung cancer
cells. Each histogram represents the mean Cq value of the PCR
products. All genes exhibited increased mRNA expression in Lewis
lung cancer cells compared with GAPDH. BSA, bovine serum albumin;
BCLAF1, B-cell lymphoma 2-associated transcription factor 1; Cq,
quantification cycle; CTNNBL1, catenin β-like 1; ERCC3, ERCC
excision repair 3, TFIIH, core complex helicase subunit; HMGB1,
high mobility group box 1; ING1, inhibitor of growth family member
1; qPCR, quantitative polymerase chain reaction; RACK1, receptor
for activated C kinase 1; RPS6, ribosomal protein S6; SOD1,
superoxide dismutase 1; TCL, tumor cell lysate. |
Removal of RACK1 and CTNNBL1 from
Lewis lung cancer TCL
As determined by western blot analysis and RT-qPCR,
RACK1 and CTNNBL1 were selected as targets to investigate the
function of TCL on mouse splenocytes. The present study used a
novel protein extraction method based on IP. To remove both
proteins, the method was repeated two times. In the first round, a
RACK1 antibody was used to remove RACK1 from the TCL. Following IP,
western blotting demonstrated that RACK1 was removed from TCL;
however, CTNNBL1 was still detected in the flow-through (Fig. 4A). Similarly, in the second round,
CTNNBL1 was removed from the processed TCL after the first round
using an anti-CTNNBL1 antibody (Fig.
4B). This resulted in the removal of CTNNBL1 from TCL but not
GAPDH; the flow-through of the second round of IP did not contain
RACK1 or CTNNBL1; however, other proteins, including GAPDH, were
still present. Using two rounds of IP, RACK1 and CTNNBL1 were
successfully removed from TCL. The subsequent experiments were
performed using TCL that was devoid of RACK1 and CTNNBL1.
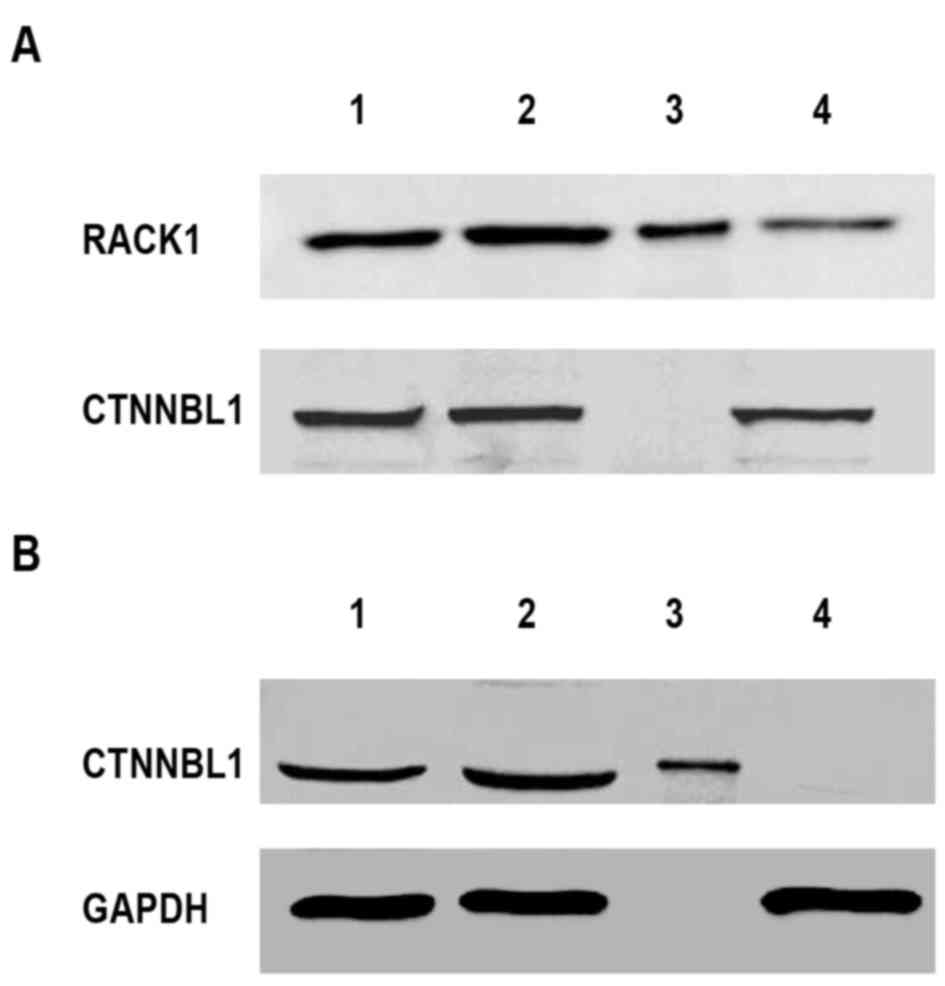 | Figure 4.Isolation of RACK1 and CTNNBL1 from
TCL. The TCL was prepared from 1×106 Lewis lung cancer
cells. Subsequently, RACK1 and CTNNBL1 were removed from TCL using
IP. Immunoblot analysis was used to detect RACK1, CTNNBL1 and GAPDH
expression in whole TCL, TCL supernatant, IP sample or
flow-through. (A) First round IP was used to remove RACK1 with an
anti-RACK1 antibody. (B) Second round IP was used to remove CTNNBL1
with an anti-CTNNBL1 antibody. Lane 1, TCL supernatant; lane 2,
whole TCL; lane 3, IP sample; lane 4, flow-through. CTNNBL1,
catenin β-like 1; IP, immunoprecipitation; RACK1, receptor for
activated C kinase 1; TCL, tumor cell lysate. |
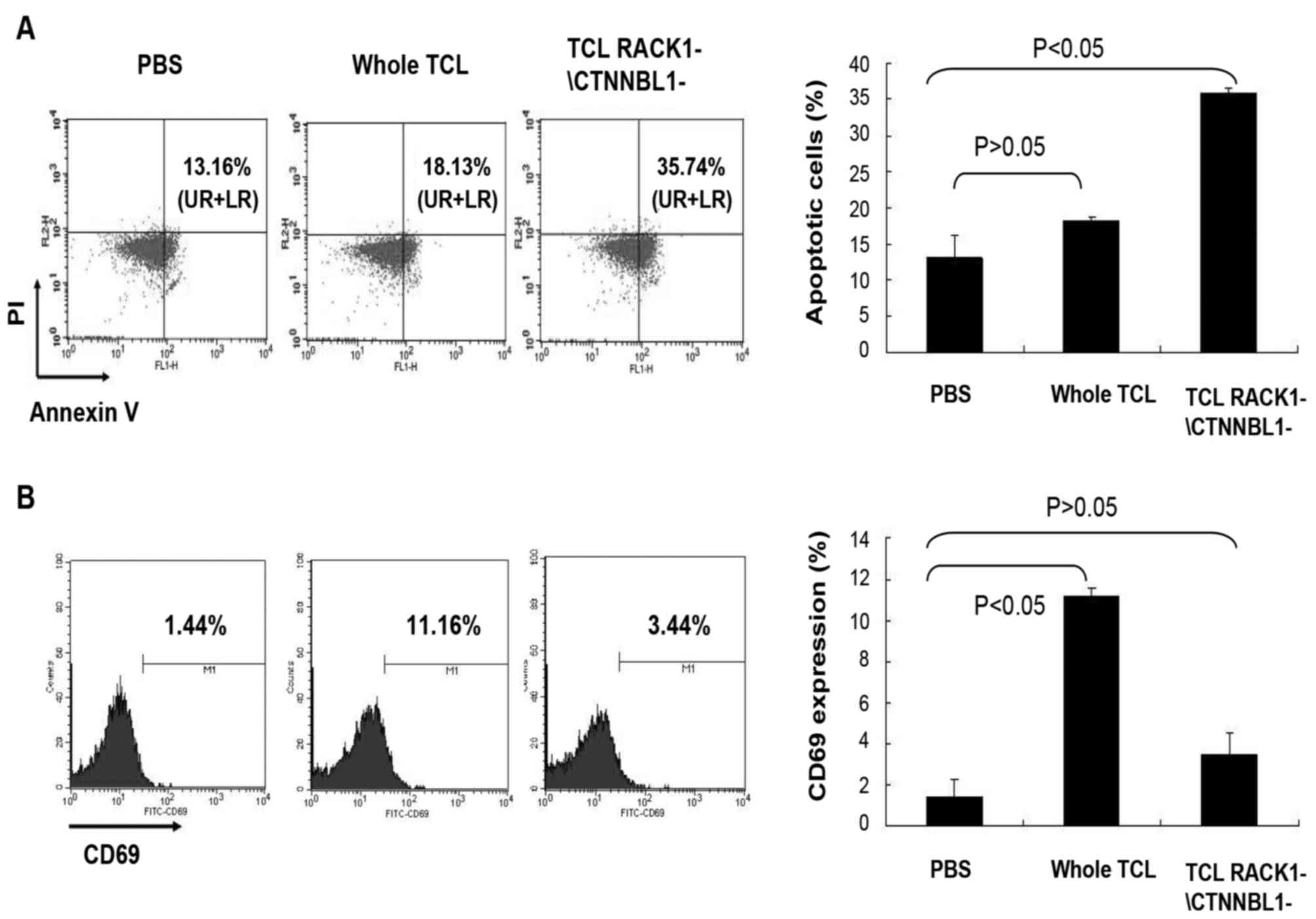
Function of RACK1 and CTNNBL1 in mouse
splenocytes
To investigate the function of RACK1 and CTNNBL1 in
splenocyte activation and apoptosis, mouse splenocytes were
incubated with TCL lacking RACK1 and CTNNBL1, and were then
analyzed by flow cytometry. The results demonstrated that the
expression of the early apoptosis marker Annexin V was higher in
mouse splenocytes stimulated with whole TCL compared with in the
control splenocytes stimulated with PBS; however, the result was
not statistically significant (P>0.05; Fig. 5A). In addition, the early
activation marker CD69 was significantly upregulated in splenocytes
stimulated with whole TCL (P<0.05; Fig. 5B). Conversely, CD69 expression in
mouse splenocytes stimulated with TCL lacking RACK1 and CTNNBL1 was
downregulated (P<0.05; Fig.
5B), whereas Annexin V expression was upregulated in these
cells (P>0.05; Fig. 5A). These
results indicated that RACK1 and CTNNBL1 may be the main elements
in TCL that affect activation and apoptosis of mouse
splenocytes.
Discussion
Activation of immunocytes has a key role in
antitumor immunotherapy. Numerous studies have reported that TCL
can activate immunocytes (22–24);
however, it remains unclear as to what components present in TCL
possess this ability. TCL is a protein mixture produced by lysed
tumor cells; the majority of these proteins are involved in
signaling pathways associated with cell division and activation.
Therefore, it is hypothesized that TCL depends on these proteins to
activate immunocytes. The present study identified the proteins in
Lewis lung cancer cell TCL using iTRAQ detection. Among the
proteins, RACK1, CTNNBL1, Cullin 3, BCLAF1, RPS6, SOD1, HMGB1, ING1
and ERCC3 have been reported to activate cells and inhibit
apoptosis, according to protein databases and previous studies
(8,9,15–21).
Among these proteins, only RACK1, CTNNBL1 and Cullin 3 could be
detected by western blotting, and the protein expression levels of
RACK1 and CTNNBL1 in TCL were higher than the levels of Cullin 3.
Therefore, RACK1 and CTNNBL1 may serve a more important role in
immunocyte activation.
Pathways involved in cell activation and apoptosis
in almost all animal cells require RACK1, a Sonic hedgehog signal
pathway protein (8), and CTNNBL1,
a Wnt signal pathway protein (9).
Therefore, RACK1 and CTNNBL1 may be involved in the activation of
non-immunocytes and immunocytes. RACK1 knockdown in monocytes has
been reported to lead to impaired proliferation and decreased
interleukin (IL)-6 and tumor necrosis factor (TNF)-α expression,
thus indicating that RACK1 can activate monocytes (25). Conversely, to the best of our
knowledge, there is no direct evidence to suggest that CTNNBL1 can
activate immunocytes. However, B lymphocyte differentiation and
antibody production require CTNNBL1 to stabilize activation-induced
cytidine deaminase, which is a critical enzyme associated with B
cell differentiation and antibody production (12). Therefore, the role of CTNNBL1 may
be similar to RACK1, but with the ability to activate B
lymphocytes, depending on its cell activation signaling
pathway.
To investigate the role of these proteins in the
TCL-induced activation of immunocytes, the proteins were removed
from the TCL and the difference in TCL functionality was
determined. RACK1 and CTNNBL1 were isolated from Lewis lung cancer
TCL using an IP-based method. IP is used to separate individual
proteins from cells to study the relationship between various
proteins. In this process, the cell lysate should be prepared
first, and then the protein of interest can be isolated using a
specific antibody and collected using agarose beads to further
analyze the protein function. Alternatively, to remove RACK1 and
CTNNBL1, the unbound flow-through was collected after IP, rather
than the RACK1 and CTNNBL1 isolate. In theory, three or more
proteins can be removed from TCL by repeating this procedure, and
this method may be of value in understanding TCL-based cancer
immunotherapy.
Incubation with the RACK1 and CTNNBL1-deficient TCL
induced apoptosis of mouse splenocytes; however, their activation
was reduced compared with those stimulated by whole TCL. This
finding indicated that RACK1 and CTNNBL1 in TCL serve a key role in
mouse splenocyte activation. Mouse splenocytes consist of
lymphocytes and monocytes. Among these cells, macrophages and DCs
are able to ingest exogenous protein (26,27).
It has been reported that purified glutathione S-transferase can be
ingested and transferred the cell membrane in RAW264.7 mouse
macrophages, where it participates in signaling pathways associated
with cell proliferation, apoptosis and cell division (28). Similarly, ovalbumin protein can be
ingested by DCs as an exogenous protein, which may be used to
estimate DC immunoactivity (29).
B lymphocytes are also able to present antigens during the immune
response (30). Based on these
findings, it is hypothesized that RACK1 and CTNNBL1 may activate
mouse splenocytes as follows: When TCL is cocultured with mouse
splenocytes, RACK1 and CTNNBL1 may be obtained by monocytes or B
lymphocytes, which are consequently activated. The activated
monocytes or B lymphocytes possess a stronger antigen-presenting
ability and secrete more IL-6 and TNF-α, which promotes activation
of T and B lymphocytes, and natural killer cells. In addition,
expression of the early immunocyte marker, CD69, is increased in
these mouse splenocytes. The present study suggested that RACK1 and
CTNNBL1-induced activation of monocytes or B lymphocytes is a key
stage in mouse splenocyte activation. In these cells, RACK1 and
CTNNBL1, via cell signaling pathways, inhibit apoptosis and promote
activation. With regards to the proliferation of non-immunocytes,
RACK1 and CTNNBL1 function via different cell signaling pathways.
RACK1 promotes tumor cell proliferation and inhibits apoptosis via
the Sonic hedgehog signaling pathway (8), whereas CTNNBL1 enhances proliferation
through the Wnt signaling pathway (9). Therefore, it may be hypothesized that
RACK1 and CTNNBL1 activate the same signaling pathways to induce
activation of mouse monocytes or B lymphocytes, and inhibit their
apoptosis. However, this hypothesis requires further experimental
verification, which we aim to pursue in future experiments.
Furthermore, although low quantities were detected in the TCL in
the present study, seven other proteins (CULLIN 3, BCLAF1, RPS6,
SOD1, HMGB1, ING1 and ERCC3) may influence immunocyte activation or
apoptosis; further studies are required to clarify their roles.
In conclusion, the present study identified a novel
IP-based method for the removal of proteins from TCL. Using this
method, it was demonstrated that RACK1 and CTNNBL1 are the key
elements in Lewis lung cancer TCL, which are involved in the
activation of mouse splenocytes. This approach may be used to
generate TCL-based tumor vaccines for human use.
Acknowledgements
The present study was supported by the National
Natural Science Foundation (grant no. 81402351), the 2015 Anhui
Province Funded Project of Study Abroad (grant no. 5), and the
Anhui Laboratory of Biological Macro-molecules Research Foundation
(grant no. 1306C083008).
References
|
1
|
Ogino S, Galon J, Fuchs CS and Dranoff G:
Cancer immunology-analysis of host and tumor factors for
personalized medicine. Nat Rev Clin Oncol. 8:711–719. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Borghaei H, Smith MR and Campbell KS:
Immunotherapy of cancer. Eur J Pharmacol. 625:41–54. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chiang CL, Coukos G and Kandalaft LE:
Whole tumor antigen vaccines: Where are we? Vaccines (Basel).
3:344–372. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
González FE, Gleisner A, Falcón-Beas F,
Osorio F, López MN and Salazar-Onfray F: Tumor cell lysates as
immunogenic sources for cancer vaccine design. Hum Vaccin
Immunother. 10:3261–3269. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Son CH, Bae JH, Shin DY, Lee HR, Yang K
and Park YS: Antitumor effect of dendritic cell loaded ex vivo and
in vivo with tumor-associated antigens in lung cancermodel. Immunol
Invest. 43:447–462. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dong B, Sun L, Wu X, Zhang P and Wang L,
Wei H, Zhou L, Hu X, Yu Y, Hua S and Wang L: Vaccination with TCL
plus MHSP65 induces anti-lung cancer immunity in mice. Cancer
Immunol Immunother. 59:899–908. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li JJ and Xie D: RACK1, a versatile hub in
cancer. Oncogene. 34:1890–1898. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shi S, Deng YZ, Zhao JS, Ji XD, Shi J,
Feng YX, Li G, Li JJ, Zhu D, Koeffler HP, et al: RACK1 promotes
non-small-cell lung cancer tumorigenicity through activating sonic
hedgehog signaling pathway. J Biol Chem. 287:7845–7858. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huhn S, Ingelfinger D, Bermejo JL, Bevier
M, Pardini B, Naccarati A, Steinke V, Rahner N, Holinski-Feder E,
Morak M, et al: Polymorphisms in CTNNBL1 in relation to colorectal
cancer with evolutionary implications. Int J Mol Epidemiol Genet.
2:36–50. 2011.PubMed/NCBI
|
|
10
|
van Maldegem F, Maslen S, Johnson CM,
Chandra A, Ganesh K, Skehel M and Rada C: CTNNBL1 facilitates the
association of CWC15 with CDC5L and is required to maintain the
abundance of the Prp19 spliceosomal complex. Nucleic Acids Res.
43:7058–7069. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ganesh K, Adam S, Taylor B, Simpson P,
Rada C and Neuberger M: CTNNBL1 is a novel nuclear localization
sequence-binding protein that recognizes RNA-splicing factors CDC5L
and Prp31. J Biol Chem. 286:17091–17102. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Conticello SG, Ganesh K, Xue K, Lu M, Rada
C and Neuberger MS: Interaction between antibody-diversification
enzyme AID and spliceosome-associated factor CTNNBL1. Mol Cell.
31:474–484. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Svec D, Tichopad A, Novosadova V, Pfaffl
MW and Kubista M: How good is a PCR efficiency estimate:
Recommendations for precise and robust qPCR efficiency assessments.
Biomol Detect Quantif. 3:9–16. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dorr C, Janik C, Weg M, Been RA, Bader J,
Kang R, Ng B, Foran L, Landman SR, O'Sullivan MG, et al: Transposon
mutagenesis screen identifies potential lung cancer drivers and
CUL3 as a tumor suppressor. Mol Cancer Res. 13:1238–1247. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhou X, Li X, Cheng Y, Wu W, Xie Z, Xi Q,
Han J, Wu G, Fang J and Feng Y: BCLAF1 and its splicing regulator
SRSF10 regulate the tumorigenic potential of colon cancer cells.
Nat Commun. 5:45812014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Knoll M, Macher-Goeppinger S, Kopitz J,
Duensing S, Pahernik S, Hohenfellner M, Schirmacher P and Roth W:
The ribosomal protein S6 in renal cell carcinoma: Functional
relevance and potential as biomarker. Oncotarget. 7:418–432. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Skrzycki M, Czeczot H, Chrzanowska A and
Otto-Ślusarczyk D: The level of superoxide dismutase expression in
primary and metastatic colorectal cancer cells in hypoxia and
tissue normoxia. Pol Merkur Lekarski. 39:281–286. 2015.(In Polish).
PubMed/NCBI
|
|
19
|
Sharma S, Evans A and Hemers E:
Mesenchymal-epithelial signalling in tumour microenvironment: Role
of high-mobility group Box 1. Cell Tissue Res. 365:357–366. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Han X, Chen Y, Yao N, Liu H and Wang Z:
MicroRNA let-7b suppresses human gastric cancer malignancy by
targeting ING1. Cancer Gene Ther. 22:122–129. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Terashita Y, Ishiguro H, Haruki N, Sugiura
H, Tanaka T, Kimura M, Shinoda N, Kuwabara Y and Fujii Y: Excision
repair cross complementing 3 expression is involved in patient
prognosis and tumor progression in esophageal cancer. Oncol Rep.
12:827–831. 2004.PubMed/NCBI
|
|
22
|
Kawahara M and Takaku H: A tumor lysate is
an effective vaccine antigen for the stimulation of CD4(+) T-cell
function and subsequent induction of antitumor immunity mediated by
CD8(+) T cells. Cancer Biol Ther. 16:1616–1625. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gershan JA, Barr KM, Weber JJ, Jing W and
Johnson BD: Immune modulating effects of cyclophosphamide and
treatment with tumor lysate/CpG synergize to eliminate murine
neuroblastoma. J Immunother Cancer. 3:242015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yuan X, Li W, Cui Y, Zhan Q, Zhang C, Yang
Z, Li X, Li S, Guan Q and Sun X: Specific cellular immune response
elicited by the necrotic tumor cell-stimulated macrophages. Int
Immunopharmacol. 27:171–176. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang DL: The role of scaffold RACK1 in
the production of proinflammatory cytokines [Dissertation]. Guang
Zhou: Zhongnan University; 2013
|
|
26
|
O'Neill LA and Pearce EJ: Immunometabolism
governs dendritic cell and macrophage function. J Exp Med.
213:15–23. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cybulsky MI, Cheong C and Robbins CS:
Macrophages and dendritic cells: Partners in atherogenesis. Circ
Res. 118:637–652. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang Y, Shen JY, Luo L and Yin ZM: The
Location of FITC-GSTP1 in the Murine Macrophages. J Nanjing Norm
Univ (Nat Sci Ed). 1–110. 2008.
|
|
29
|
Wirsdörfer F, Bangen JM, Pastille E,
Schmitz D, Flohé S, Schumak B and Flohé SB: Dendritic cell-like
cells accumulate in regenerating murine skeletal muscle after
injury and boost adaptive immune responses only upon a microbial
challenge. PLoS One. 11:e01558702016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wennhold K, Weber TM, Klein-Gonzalez N,
Thelen M, Garcia-Marquez M, Chakupurakal G, Fiedler A, Schlösser
HA, Fischer R, Theurich S, et al: CD40-activated B cells induce
anti-tumor immunity in vivo. Oncotarget. 8:27740–27753.
2017.PubMed/NCBI
|