Introduction
Epilepsies are disorders of neuronal excitability,
characterized by spontaneous and recurrent seizures. Potassium ion
channels serve an important role in the pathogenesis of epilepsy.
They are the core targets for excitability regulation and are
involved in potassium homeostasis. Potassium ion channels adjust
the cell membrane resting potential and determine the amplitude and
frequency of action potentials, and buffer the extracellular
potassium ions. Over 80 subtypes of potassium ion channels have
been identified to date on the surface of neuronal and glial cells
(1). Several epileptic phenotypes
have been associated with dysfunctions of potassium channels.
Changes in the extracellular K+ concentration represent
one of the earliest and most thoroughly described examples of
activity-induced changes to ion concentration gradients (2). Epileptic seizures are associated with
the expression, distribution and density of various subtypes of
potassium channels. A microarray of several potassium channel
genes, Kcnmb4, Kcnd2 and Kcnc1 were downregulated during the acute
phase of epilepsy in the CA3 region of the hippocampus (3). Although the aforementioned studies
revealed that potassium channels can be regulated during epilepsy,
the exact mechanisms underlying these observations remain to be
elucidated.
An increasing amount of evidence suggests that
epigenetic mechanisms are involved in the regulation of potassium
channels in various neural pathogeneses (4,5). The
epigenetic mechanism involved in the potassium homeostasis and the
dynamics of epilepsy remain to be elucidated. Downregulated Kir4.1
protein expression is observed in several human central nervous
system (CNS) pathologies, including CNS ischemic injury, spinal
cord injury, epilepsy, amyotrophic lateral sclerosis and
Alzheimer's disease (reviewed in 6). Studies on Kir4.1 knockout
mice indicated that DNA methylation is a key regulator of Kir4.1
transcription during CNS development (7). DNA hydroxymethylation also regulates
the differential expression of different subtypes of potassium
channels in drug-addiction behaviours (8). Nerve injury increases the
dimethylation of histone H3 at lysine 9 (H3K9me2) at potassium
voltage-gated channel subfamily A member 4, subfamily D member 2
(Kcnd2), subfamily KQT member 2 and calcium-activated potassium
channel subunit alpha-1 promoters, but does not affect the levels
of DNA methylation in the dorsal root ganglion (9). Histone deacetylase inhibitors
modulate ATP-sensitive potassium channel subunit transcription at
the SUR2 promoter in HL-1 cardiomyocytes (10). In a genome-wide DNA methylation
analysis, KCNN4 (KCa 3.1 channel) promoter was identified to be
hypo-methylated in an aggressive non-small-cell lung carcinoma cell
line and in patient samples (11).
One of the major epigenetic modulations is histone
H3 at lysine 9 (H3K9) methylation. It includes mono-, di- and
tri-methylations, which are catalysed by several histone
methyltransferases, such as histone-lysine N-methyltransferase
EHMT2 (G9a) and histone-lysine N-methyltransferase SUV39H1. H3K9me2
is the main repressive mechanism of the transcriptional regulation
of gene expression, and is commonly associated with gene silencing.
G9a mediates H3K9 mono- and di-methylation (H3K9me1/2) and targets
euchromatic loci (12). In the
present study, it was identified that the expression of H3K9me2
decreased markedly during the epileptic process, especially in the
acute phase of epileptic seizure. The downregulation of H3K9me2 was
associated with an upregulated expression of several subtypes of
potassium channels, including Kir4.1. These observations suggest
that H3K9me2 serves a role in the regulation of epileptic seizures
and the transcriptional regulation of potassium channel genes,
which was investigated further in the present study.
Materials and methods
Animals and epilepsy model
All experimental procedures in the present study
were approved by the Animal Ethics and Use Committees of Ningxia
Medical University (Yinchuan, China).
Healthy male Sprague Dawley rats (weight, 280–300 g;
aged 8–12 weeks) obtained from the Animal Centre of Ningxia Medical
University were reared at 6 per cage at 24.0±0.2°C in a humidified
atmosphere (54±5%). Rats were fed with a standard rodent animal
feed, were specific-pathogen-free and kept under a natural
circadian rhythm, with natural lighting and regular ventilation.
Rats were randomly divided into healthy control and pilocarpine
epilepsy groups. Totally 40 rats were used, with 25 in the epilepsy
group and 15 in the control group. More specifically, 15 epileptic
rats and 5 control rats for western blotting; 5 epileptic rats and
5 control rats for RT-qPCR and 5 epileptic rats and 5 control rats
for ChIP assay (Table I). The
epilepsy model was established using lithium chloride
(LiCl)/Pilocarpine (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany)
as previously described (13).
Rats were injected intraperitoneally with LiCl (127 mg/kg) and then
with pilocarpine (30 mg/kg) 18 to 24 h following the first
injection. Rats with Racine IV–V grade epileptic seizures were
administered chloral hydrate (300 mg/kg) injection following 1 h of
seizure, and were subsequently used in further experiments.
Pilocarpine was administered to each animal up to 4 times (Table I). Time intervals between the first
pilocarpine injections to the first IV–V grade epileptic seizure
were recorded as seizure latency and are presented in Table I. Rats that received pilocarpine
more than 6 times were not included in the present study. Rats in
the control group were administered a 0.9% saline injection for 2
treatments with about 1 h interval.
 | Table I.Records of the behaviour of epileptic
rats. |
Table I.
Records of the behaviour of epileptic
rats.
| Experiment |
Western
blotting | RT-qPCR | ChIP |
|---|
|
|
|
|
|---|
| Rat no. | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 |
|---|
|
|---|
| Time point of death
following epilepsy | 1 h | 5 h | 24 h | 5 h | 5 h |
|
|
|
|
|
|
| Number of pilocarpine
injections | 2 | 2 | 1 | 4 | 4 | 1 | 2 | 2 | 3 | 3 | 1 | 1 | 2 | 3 | 4 | 3 | 4 | 2 | 3 | 2 | 3 | 2 | 2 | 3 | 3 |
| Seizure latency
(min) | 50 | 59 | 28 | 104 | 118 | 29 | 44 | 60 | 88 | 67 | 20 | 24 | 48 | 65 | 106 | 76 | 102 | 38 | 96 | 37 | 62 | 51 | 54 | 63 | 62 |
Brain tissue preparation and western
blotting
Rat brains were dissected, and hippocampal and
insular cortex tissues were isolated and processed immediately for
protein or RNA extraction. Each assay included 5 rats. For western
blotting, a Total Protein Extraction kit (Nanjing KeyGen Biotech.
Co., Ltd., Nanjing, China) was used, and the Bicinchoninic Acid
assay was used for protein quantification (Nanjing KeyGen Biotech.
Co., Ltd.) according to the manufacturer's protocol. The following
antibodies were used: Anti-EHMT2/G9A (dilution, 1:1,000; cat. no.
ab40542), anti-Kcnj10 (dilution, 1:1,000; cat. no. ab192404),
anti-histone H3 (di methyl K9; dilution, 1:1,000, cat. no. ab1220;
all from Abcam, Cambridge, MA, USA) and anti-GAPDH (dilution,
1:5,000; cat. no. bsm-0798m; BIOSS, Beijing, China).
Peroxidase-conjugated goat anti-rabbit IgG (cat. no. bs-0295G) and
goat anti-mouse IgG (cat. no. bs-0296G) were used as the secondary
antibodies (both dilutions, 1:10,000; BIOSS). All antibodies were
diluted with 5% skimmed milk (BD Biosciences, Franklin Lakes, NJ,
CA, USA).
RT-qPCR
Total RNA was purified from rat brain tissues with
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) according to manufacturer's protocol. The RNA was
reverse-transcribed to cDNA using a Reverse Transcription system
(Pomega Corporation, Madison, WI, USA) according to the
manufacturer's protocol: 70°C for 5 min, 25°C for 5 min, 42°C for
60 min, and 70°C for 15 min. A total of 16 primer pairs specific to
different potassium channel subtypes were designed and tested by
the melting curve analysis (Table
II). The following thermo cycling conditions were used qPCR:
50°C for 2 min, 95°C for 10 min and 40 cycles of 95°C for 15 sec,
followed by 60°C for 1 min. GAPDH served as a housekeeping gene.
All RT-qPCR experiments were run in triplicate. The
2−∆∆Cq method (14) was
used to determine the relative fold change in mRNA expression.
Samples were analysed using a Smart Spec Plus spectrophotometer
(Bio-Rad Laboratories, Inc., Hercules, CA, USA).
 | Table II.Primer sequences for RT-qPCR and
ChIP. |
Table II.
Primer sequences for RT-qPCR and
ChIP.
|
| RT-qPCR primer
sequence (5′→3′) |
|---|
|
|
|
|---|
| Gene | Forward | Reverse |
|---|
| KCNT2 |
ACATCCCCAAAGCCCACA |
TGGGAGCAGATTTTACGAGTTC |
| KCNA5 |
GAGAATGACCAGGACCGACAC |
TCATCAAGGAAGAGGAGAAGCC |
| KCNQ2 |
GGATTCCGCCGTTTCTCA |
ACCCTCATTGGTGTCTCATTCTT |
| KCNJ10 |
GGGGACGCCACTTTCACAA |
GAGATCCTCTGGGGCTACGA |
| KCNS2 |
TGCCCAAGTTAGCCAAGGT |
AATCGTGGAGCACTTTGGC |
| KCNJ16 |
GGGTCGGAAGTCACCTATGC |
CATTGGGGCAGCCTTGG |
| KCNH2 |
GCCCCTCGGAATGGTTTG |
TTCCATCAGATTCCCAACCC |
| KCNJ4 |
CGGAGATGACAGCGTGGTG |
GCGGTCATCGCAGTGGTT |
| KCNA3 |
TTACTGGGGCAAGCAAAGAAT |
TCTTCTGCTTGGAGACACTACCC |
| KCNC3 |
CAACGCCCAACTGCTACG |
GACTCCCGTTCCCTCTTCG |
| KPNB1 |
TGGATCATTGGCCTAGCTTCTA |
AGCCCAAGTGGACAAGTCAGA |
| KPNA3 |
TGGGACAGGACATCGCAGTT |
AGCCACCAGGAAGTCAAAGTT |
| KCNK10 |
CCCTGCCACAAAATCACCA |
ATCCCGCTCTTCGGTTTCT |
| KCNAB2 |
GCTCCAGCGTGATGTGCC |
GAAGAAGGGGTGGAGACGG |
| KCTD2 |
CGTTGTCCCGTATCCTTTCC |
ACTCGGACAAGGATGAGACTGG |
| Kcnip1 |
ATGATGTTGTCGTCCTCCTGAC |
CATCAACAAGGACGGATACATAAAC |
| GAPDH |
GGTCCACCACTGACACGTTG |
ACAACAGCCTCAAGATCATCAG |
| Gene | ChIP primer
sequence (5′→3′) |
| 1-KCNJ10 |
AAGAAGGAGGGAAGAACA |
ACATAGTGGATGGGAAGAT |
| 2-KCNJ10 |
TGAAGGAACCAGGACGAG |
ACAGGGAACAACGAAAAC |
| 3-KCNJ10 |
GGTAGTGGGACAGATGAAGA |
GACAAACCCTTATCTGATTC |
| 4-KCNJ10 |
TGCCAACCGCCAACTACA |
TCCCTCGTGCTTTCATCG |
| 5-KCNJ10 |
CTCTGCCCTCTACACTCAT |
GATAATCTCCCATTCCTAAC |
ChIP assay
A total of 5 epileptic and 5 control rats were used
for ChIP assay. Anti-histone H3 (di methyl K9) antibody (dilution,
1:1,000; cat. no. ab1220; Abcam) and anti-EHMT2/G9A (dilution,
1:1,000; cat. no. ab40542; Abcam) antibodies, and an EZ-ChIP kit
(Haoran Biological Technology Co., Ltd., Shanghai, China) were
used. DNA fragments were sonicated to 100–500 bp. RT-qPCR was
performed as described in the previous section. A total of 5 primer
pairs specific to the Kcnj10 promoter region were designed, and the
sequences are presented in Table
II. Total genome DNA was used as a template. All samples were
tested in triplicate and the results from each sample were
normalized relative to the input genomic DNA.
Cell culture and treatments
The C6 glioma cell line (Shanghai Jing Kang
Biological Engineering. Co., Ltd., Shanghai, China; www.shjingk.com/) were cultured in high glucose
Dulbecco's modified Eagle's Medium with 1% penicillin/streptomycin
and 5% fetal bovine serum in a 5% CO2 incubator at 37°C
for 24 h. A total of 1 mM/ml2-
(Hexahydro-4-methyl-1H-1,4-diazepin-1-yl)
−6,7-di-methoxy-N-(1-(phenyl-methyl)-4-piperidinyl)-4-quinazolinamine
tri-hydrochloride hydrate (Bix01294; Sigma-Aldrich; Merck KGaA,)
was added to the medium at the final concentration of 1, 2, 4 and 8
µM/ml for 5 h. Proteins were extracted using the Cell Lysis &
Protein Extraction kit, containing inhibitors of phosphatases and
protease inhibitors, from brain tissues or C6 cells (Cell Lysis
& Protein Extraction kit; Nanjing KeyGen Biotech. Co., Ltd.,
Nanjing, China). Briefly, tissues were pipetted in the lysis buffer
on ice and rotated at 4°C for 30 min and then centrifuged for 40
min in 10,800 × g at 4°C.
Statistical analysis
Statistical analyses were performed using SPSS
(version 17.0; SPSS, Inc., Chicago, IL, USA). All data are
presented as the mean ± standard error. Differences between groups
were analysed using Student's t-test or one-way analysis of
variance with Dunnett's post hoc test. P<0.05 was considered to
indicate a statistically significant difference.
Results
Screening of the potassium channel
subtypes by RT-qPCR
A total of 16 potassium channel subtypes including
voltage-gated, inwardly rectifying, ATP sensitive, M type and
Ca2+−stimulated, were screened by RT-qPCR. In the
insular cortex, and in hippocampus, 5 h after seizure, Kcnj10 was
upregulated (Fig. 1A and B). The
other upregulated gene was Kcnk10. Many subtypes such as Kcna5,
Kcnj16, Kcnh2, Kcnj4 andKctd2 in the insular (Fig. 1A) and Kcna5, Kcns2, Kcnj16, Kcna3
andKctd2 in the hippocampus (Fig.
1B), were downregulated. In the present study, the focus was on
Kcnj10.
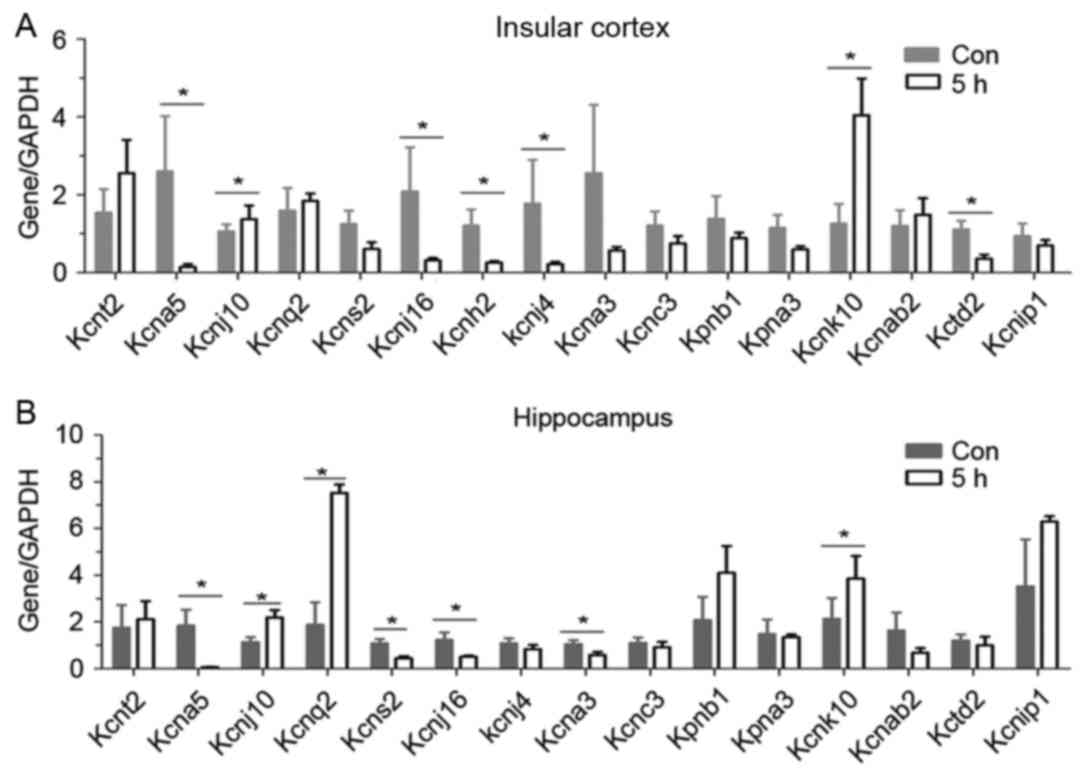 | Figure 1.Screening of potassium channel
subtypes by reverse transcription-quantitative polymerase chain
reaction. Sixteen potassium subtypes were screened in (A and B) two
brain regions of rats 5 h following epilepsy. Five rats were used
in each group (n=5). Data are expressed as the mean ± standard
error. *P<0.05 vs. the control group. Kcnt2, potassium channel
subfamily T member 2; Kcna5, potassium voltage-gated channel
subfamily A member 5; Kcnq2, potassium voltage-gated channel;
Kcnq2, subfamily KQT member 2; Kcns2, subfamily S member 2; Kcnh2,
subfamily H member 2; Kcna3, subfamily A member 3; Kcnj10,
ATP-sensitive inward rectifier potassium channel 10; Kcnj10,
ATP-sensitive inward rectifier potassium channel 10; Kcnj16, inward
rectifier potassium channel 16; Kcnj4, inward rectifier potassium
channel; Kpnb1, importin subunit beta-1; Kpna3, inward rectifier
potassium channel alpha-4; Kcnk10 importin subunit potassium
channel subfamily K member 10; Kcnab2, voltage-gated potassium
channel subunit beta-2; Kctd2, BTB/POZ domain-containing protein
KCTD2; Kcnip1, Kv channel-interacting protein 1; Con, control. |
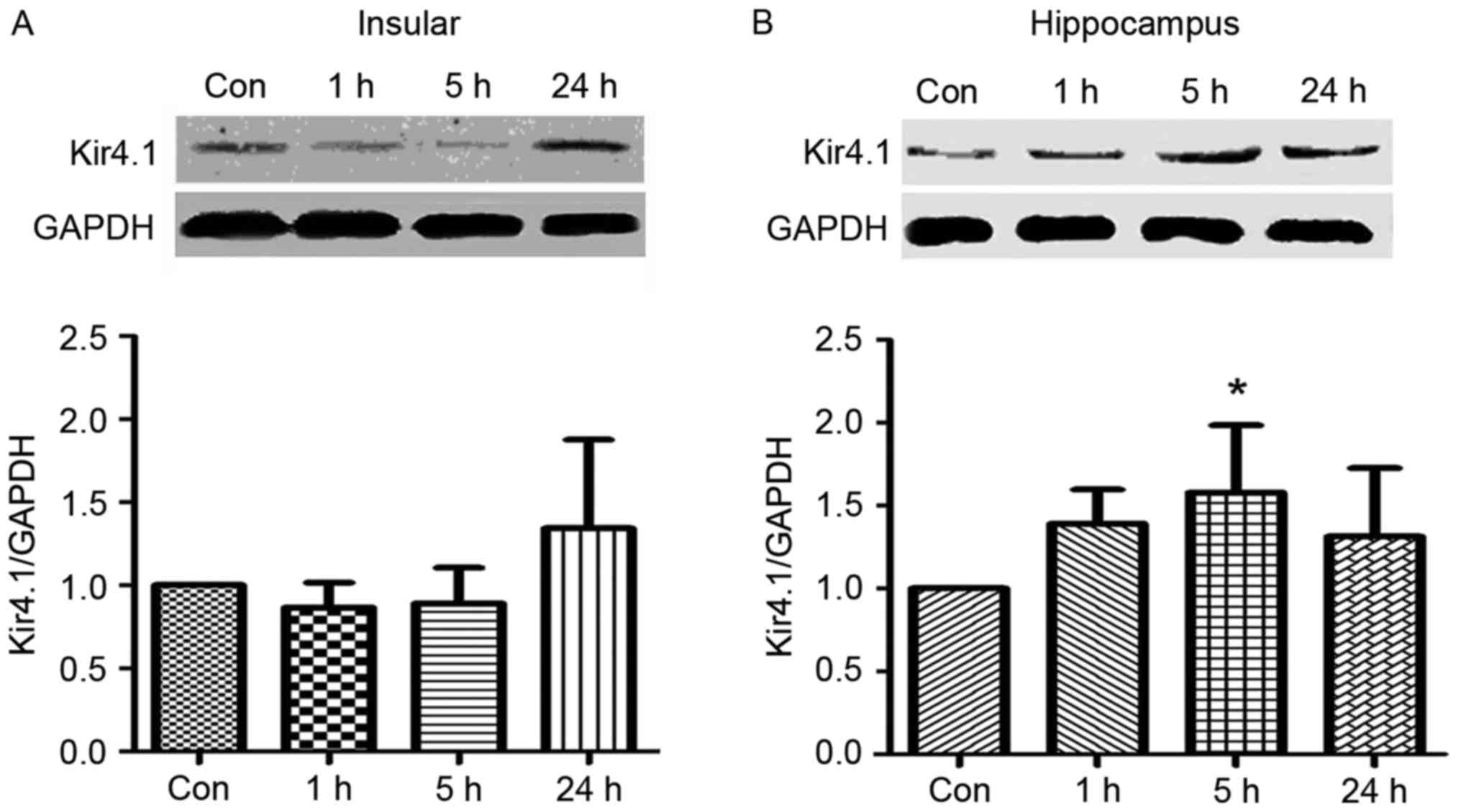
Upregulation of Kcnj10-encoded Kir4.1
protein in hippocampus
The expression of Kir4.1 was tested by western
blotting with five epileptic and five control rats. Tissues were
analysed 1, 5 and 24 h following an epileptic seizure. The protein
expression levels of the Kir4.1 were upregulated at 24 h following
an epileptic seizure in the insular cortex (Fig. 2A) and at 5 h following an epileptic
seizure in the hippocampus (Fig.
2B). Compared with the control group, the expression of Kir4.1
proteins at 5 h following a seizure was increased in the
hippocampus (P<0.05).
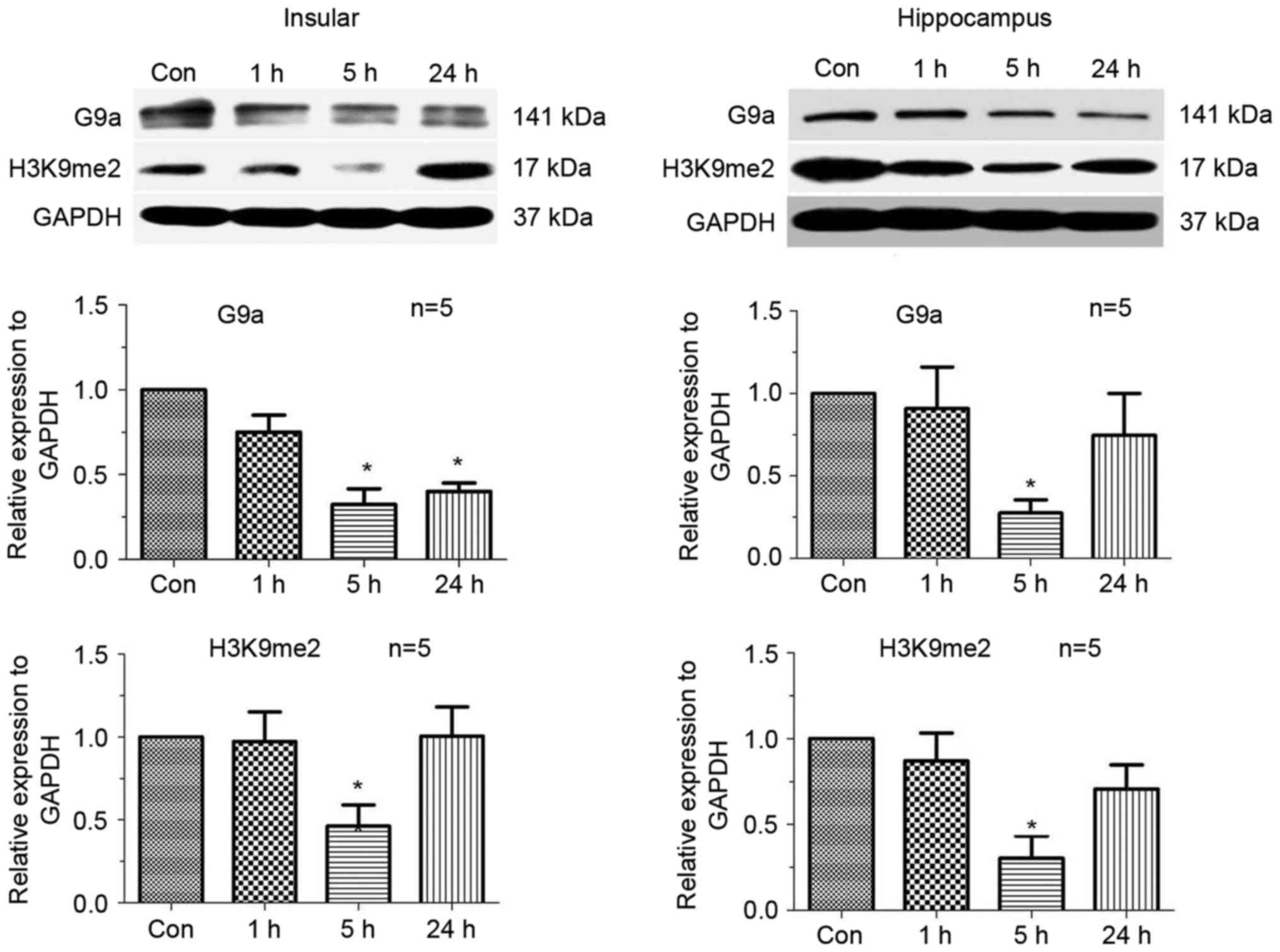
Downregulation of H3K9me2 5 h
following an epileptic seizure
In order to analyse the mechanism of epigenetic
regulation of the potassium channel genes, the expression of
H3K9me2 and its enzyme G9a was tested by a western blotting at
three different time points: 1, 5 and 24 h. As presented in
Fig. 3, H3K9me2 and G9a were
significantly downregulated 5 h following a seizure, both in the
hippocampus and insular cortex (P<0.05). The initial expression
level was restored at 24 h (Fig.
3).
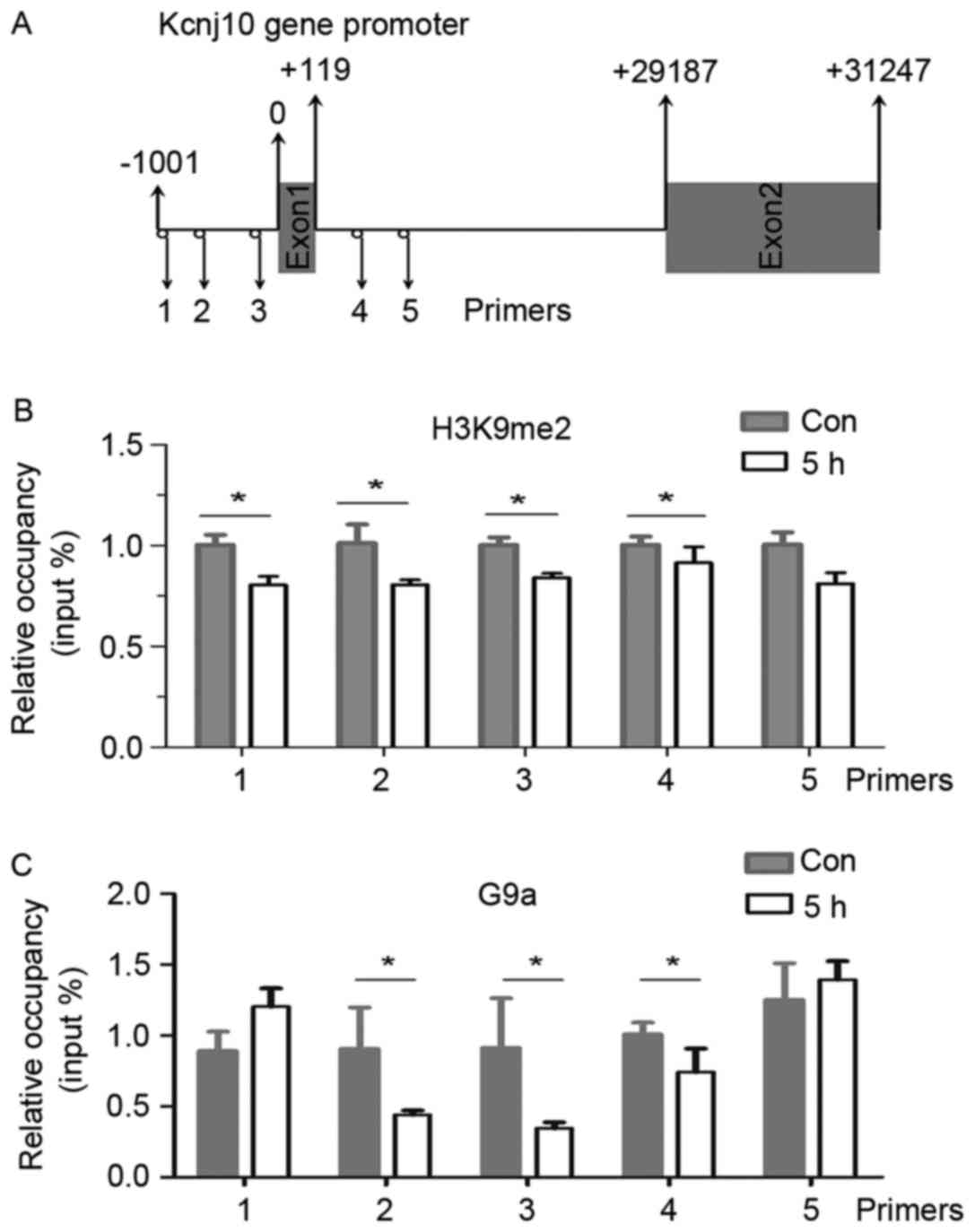
Decrease of H3K9me2 and G9a occupancy
in the Kcnj10 promoter
The aforementioned results suggested an association
between the H3K9me2 modification and the transcriptional regulation
of Kcnj10 gene. ChIP experiments were designed to analyse changes
in levels of H3K9me2 in the promoter region of the Kcnj10 gene. A
total of 5 primer pairs specific to the Kcnj10 promoter region were
designed to anneal across exon1 (Fig.
4A).
Relative levels of H3K9me2 at primer annealing sites
1, 2, 3, and 4 decreased significantly 5 h following an epileptic
seizure in the hippocampus (Fig.
4B).
In order to confirm whether the observed changes in
methylation were catalysed by G9a, ChIP experiments using the
anti-G9a antibody were performed to identify on-going methylation
events. The abundance of G9a at primer annealing sites 2, 3 and 4
also decreased significantly 5 h following an epileptic seizure in
the hippocampus (Fig. 4C).
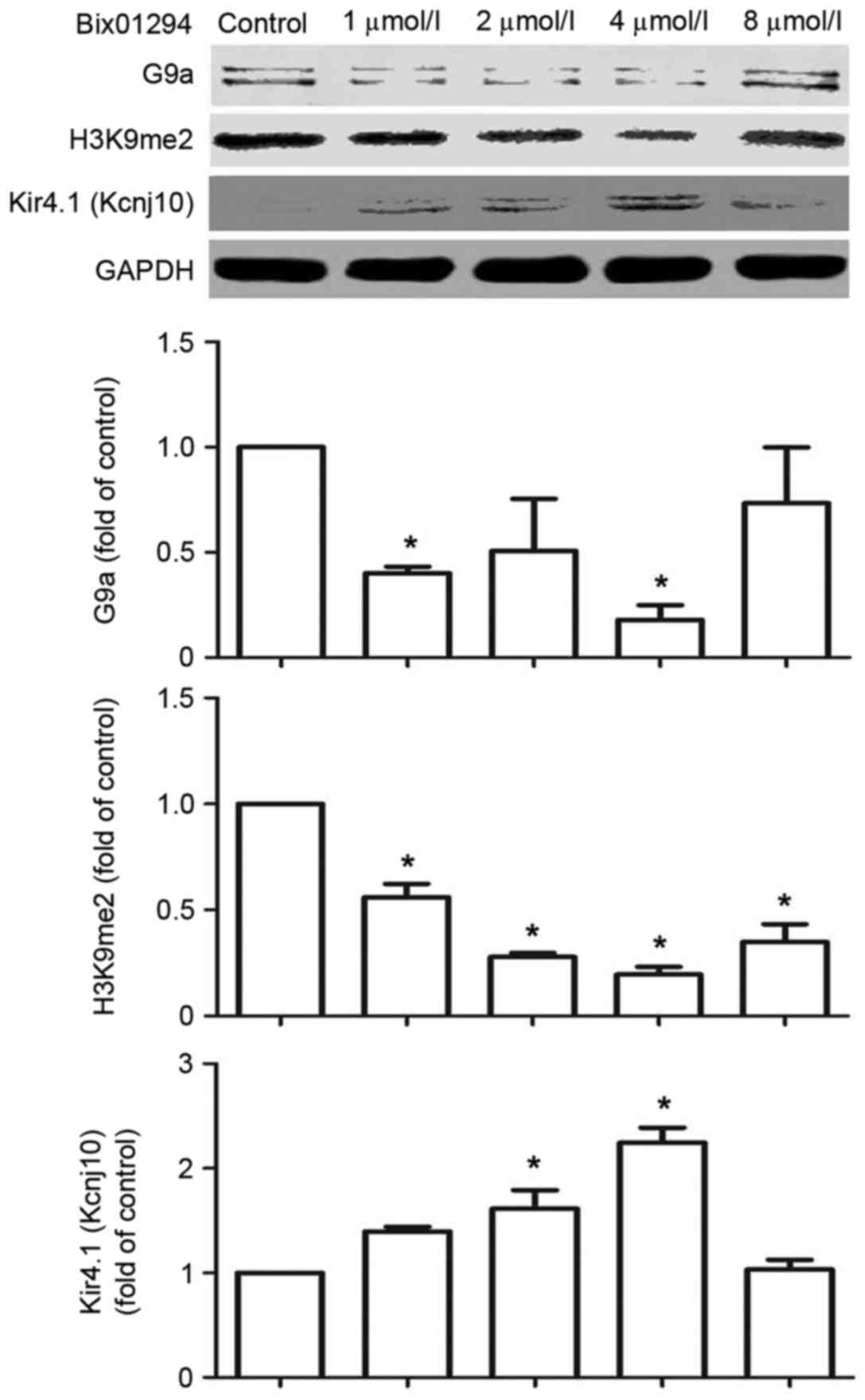
Bix01294 inhibits G9a and H3K9me2 and
increases the expression of Kir4.1 (Kcnj10)
Bix01294, a specific G9a inhibitor, was used to
verify Kcnj10 regulation by G9a. Following a 1 and 4 µmol/l
treatments with bix01294 for 5 h, the expression of G9a and H3K9me2
decreased in C6 cells, while the expression of Kir4.1 (Kcnj10)
demonstrated an increase in a dose-dependent manner between 1 and 4
µmol/l doses (Fig. 5).
Discussion
Seizures can cause long-lasting changes in gene
expression patterns. Epigenetic mechanisms may alter gene
expression in response to seizure. Without causing mutations in
gene sequences, epigenetic modifications, including histone
methylation, may respond to changes inside the cell caused by
seizure and alter gene transcription through chromatin adjustments.
The present study investigated the association between epigenetic
modification of the potassium channel expression and epileptic
activities.
The present study demonstrated that H3K9
dimethylation and its enzyme G9a were sensitive to the seizure
activity. Following a 1 h-long seizure interrupted with
LiCl/pilocarpine, H3K9me2 and G9a were downregulated. The
downregulation lasted for <24 h, was lowest at 5 h following the
seizure and then returned to its original level after 1 day. A
likely explanation for these observations is that H3K9me2 and G9a
were sensitive to the stress caused by seizure, and responded to
modulate the acute phase of epilepsy. Similar mechanisms have been
previously reported in a study investigating seizures in cocaine
addicts (15). In mouse nucleus
accumbens, repeated cocaine administration reduces overall levels
of H3K9 dimethylation catalysed by G9a, and the acute response may
be mediated by the immediate FosB proto-oncogene (15).
An alternative mechanism of G9a and H3K9me2 response
to epileptic activity is through RE1 silencing transcription factor
(REST). REST can respond to epileptic activity within 5 h, and
recruit co-repressors including HDACs, KDMs, Brg1 and G9a to become
more abundant in dentate granule neurons (16). G9a alone cannot directly bind to
DNA and needs to be recruited by a transcription factor, such as
REST (17). In the present study,
the expression of G9a was decreased, suggesting that there may be a
REST-independent mechanism of G9a binding.
Potassium channels may be regulated by G9a and
H3K9me2. In a neuropathic pain model, persistent pain stimulus can
lead to a continuous upregulation of G9a and H3K9me2, and
downregulation of some subtypes of potassium channels, such as
Kcnq4, Kcnd2, Kcnq2 and Kcnq1 in dorsal root ganglion neurons
(9). The screening performed in
the present study demonstrated that Kcnj10 was upregulated while
G9a and H3K9me2 were downregulated. ChIP assay demonstrated that,
following a seizure, H3K9me2 in the promoter region of Kcnj10 gene
decreased causing its de-repression.
Kir4.1 encoded by Kcnj10, is a potassium channel
expressed on the membrane of the astrocytes. Kir4.1 is an inwardly
rectifying ATP-activated potassium channel that is mainly expressed
in astrocytes, and serves a crucial role in the regulation of
potassium ion homeostasis during neuronal activity (6). The phenotype of the Kir4.1-knockout
mouse is characterized by severe epilepsy (18). Mutations in the human Kcnj10 gene
have been reported in patients suffering from mesial temporal lobe
epilepsy (19). As part of a
feedback mechanism, Kir4.1 could respond to epileptic stress and
become upregulated. These results suggest that astrocytes can
respond to seizure-induced K+ fluctuation.
The association between G9a and H3K9me2
downregulation, and the upregulation of the Kir4.1 potassium
channel suggest that G9a and H3K9me2 mediate epileptic
seizure-induced changes in the Kcnj10 expression. ChIP results
obtained in the present study confirmed that G9a and H3K9me2
accumulation in the Kcnj10 promoter region decreased 5 h following
epileptic activity. Inhibition of G9a and H3K9me2 with bix01294 in
astrocyte tumour cell line promoted the expression of Kcnj10 in a
dose dependent manner, which further confirmed the inhibitory
regulation of G9a and H3K9me2 on Kcnj10 transcription.
The other upregulated gene was Kcnk10, which encodes
for arachidonic acid sensitive TREK-2 K(+) potassium channel. The
TREK channels are expressed highly in the human central nervous
system, and can be activated by temperature, membrane stretch and
internal acidosis (20). Due to
its differences from Kir4.1, and unknown functions that need
further investigation Trek2/Kcnk10 was not investigated further in
the present study.
In conclusion, H3K9me2 and G9a are sensitive to
epileptic seizure activity during the acute phase of epilepsy and
can affect the transcriptional regulation of the Kcnj10 (Kir4.1)
channel. Our findings may provide a potential epigenetic link
between the regulation of potassium channel subtypes and epileptic
development which may be helpful for understanding epilepsy.
Acknowledgements
The present study was funded by the National Natural
Science Foundation of China (grant nos. 31460300 and 31260246); the
Ningxia Medical University Foundation of China (grant nos. XY201406
and XY2016055); the National Basic Research Program of China (973
Program; grant no. 2012CB722408) and the 13.5 Major Scientific and
Technological Projects of Ningxia Hui Autonomous Region (grant no.
2016BZ07).
References
|
1
|
Villa C and Combi R: Potassium channels
and human epileptic phenotypes: An updated overview. Front Cell
Neurosci. 10:812016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Raimondo JV, Burman RJ, Katz AA and
Akerman CJ: Ion dynamics during seizures. Front Cell Neurosci.
9:4192015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gorter JA, van Vliet EA, Aronica E, Breit
T, Rauwerda H, da Silva Lopes FH and Wadman WJ: Potential new
antiepileptogenic targets indicated by microarray analysis in a rat
model for temporal lobe epilepsy. J Neurosci. 26:11083–11110. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang KC and Nerbonne JM: Mechanisms
contributing to myocardial potassium channel diversity, regulation
and remodeling. Trends Cardiovasc Med. 26:209–218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ryland KE, Hawkins AG, Weisenberger DJ,
Punj V, Borinstein SC, Laird PW, Martens JR and Lawlor ER: Promoter
methylation analysis reveals that KCNA5 ion channel silencing
supports ewing sarcoma cell proliferation. Mol Cancer Res.
14:26–34. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Olsen ML and Sontheimer H: Functional
implications for Kir4.1 channels in glial biology: From
K+ buffering to cell differentiation. J Neurochem.
107:589–601. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nwaobi SE, Lin E, Peramsetty SR and Olsen
ML: DNA methylation functions as a critical regulator of Kir4.1
expression during CNS development. Glia. 62:411–427. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cadet JL, Brannock C, Krasnova IN,
Jayanthi S, Ladenheim B, McCoy MT, Walther D, Godino A, Piroozina M
and Lee RS: Genome-wide DNA hydroxymethylation identifies potassium
channels in the nucleus accumbens as discriminators of
methamphetamine addiction and abstinence. Mol Psychiatry.
22:1196–1204. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Laumet G, Garriga J, Chen SR, Zhang Y, Li
DP, Smith TM, Dong Y, Jelinek J, Cesaroni M, Issa JP and Pan HL:
G9a is essential for epigenetic silencing of K(+) channel genes in
acute-to-chronic pain transition. Nat Neurosci. 18:1746–1755. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fatima N, Cohen DC, Sukumar G, Sissung TM,
Schooley JF Jr, Haigney MC, Claycomb WC, Cox RT, Dalgard CL, Bates
SE and Flagg TP: Histone deacetylase inhibitors modulate KATP
subunit transcription in HL-1 cardiomyocytes through effects on
cholesterol homeostasis. Front Pharmacol. 6:1682015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bulk E, Ay AS, Hammadi M, Ouadid-Ahidouch
H, Schelhaas S, Hascher A, Rohde C, Thoennissen NH, Wiewrodt R,
Schmidt E, et al: Epigenetic dysregulation of KCa 3.1 channels
induces poor prognosis in lung cancer. Int J Cancer. 137:1306–1317.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shankar SR, Bahirvani AG, Rao VK, Bharathy
N, Ow JR and Taneja R: G9a, a multipotent regulator of gene
expression. Epigenetics. 8:16–22. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang L, Feng D, Tao H, DE X, Chang Q and
Hu Q: Increased stathmin expression strengthens fear conditioning
in epileptic rats. Biomed Rep. 3:28–32. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Maze I, Covington HE III, Dietz DM,
LaPlant Q, Renthal W, Russo SJ, Mechanic M, Mouzon E, Neve RL,
Haggarty SJ, et al: Essential role of the histone methyltransferase
G9a in cocaine-induced plasticity. Science. 327:213–216. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Roopra A, Dingledine R and Hsieh J:
Epigenetics and epilepsy. Epilepsia. 53 Suppl 9:S2–S10. 2012.
View Article : Google Scholar
|
|
17
|
Hwang JY, Aromolaran KA and Zukin RS:
Epigenetic mechanisms in stroke and epilepsy.
Neuropsychopharmacology. 38:167–182. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Haj-Yasein NN, Jensen V, Vindedal GF,
Gundersen GA, Klungland A, Ottersen OP, Hvalby O and Nagelhus EA:
Evidence that compromised K+ spatial buffering
contributes to the epileptogenic effect of mutations in the human
Kir4.1 gene (KCNJ10). Glia. 59:1635–1642. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Guo Y, Yan KP, Qu Q, Qu J, Chen ZG, Song
T, Luo XY, Sun ZY, Bi CL and Liu JF: Common variants of KCNJ10 are
associated with susceptibility and anti-epileptic drug resistance
in Chinese genetic generalized epilepsies. PLoS One.
10:e01248962015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Franks NP and Honoré E: The TREK K2P
channels and their role in general anaesthesia and neuroprotection.
Trends Pharmacol Sci. 25:601–608. 2004. View Article : Google Scholar : PubMed/NCBI
|