Introduction
Gliomas are the most frequent types of primary tumor
observed in the central nervous system (1). Despite progress in treatments,
including surgery, radiation therapy, and chemotherapy (2,3), the
overall survival rate for patients suffering from glioma is among
the lowest of all the main types of cancer and has not improved
during recent decades (4). Thus,
it is vitally important to discover an effective marker for early
detection and as a target molecule for the development of glioma
treatments.
MicroRNAs (miRNAs) are small, endogenous, noncoding
RNAs that regulate gene transcription by complementary base pairing
to specific mRNAs. miRNAs regulate a wide variety of biological
processes including cell migration, invasion, proliferation,
apoptosis, tumorigenesis and tissue morphogenesis (5–8).
Numerous miRNAs have been reported to function in glioma
development, including miR-132 (9), miR-503 (10), miR-661 (11), miR-16 (12), miR-21 (13), and miR-218 (14,15).
Among these miRNA molecules, miR-218 has been demonstrated to be
downregulated in low-grade glioma tissues and glioma cells compared
with normal brain tissues (16).
Previous studies have demonstrated that the overexpression of
miR-218 contributes to not only the inhibition of proliferation,
invasion and migration of glioma cells, however additionally the
induction of apoptosis by downregulating transcription of miR-218
target genes, including inhibitor of nuclear factor κB (NF-κB)
kinase subunit β (IKK-β), lymphoid enhancer binding factor 1, NF-κB
and cyclin-dependent kinase 6 (CDK6) (14,17–19).
However, the role of miR-218 in the regulation of glioma cell
proliferation through targeting a transcription factor called Yin
Yang-1 (YY1) remains to be fully elucidated.
YY1 is a universal and multifunctional zinc-finger
transcription factor that can activate or repress a variety of
genes (20). Increased expression
of YY1 has been reported in prostate cancer, colon cancer, ovarian
cancer and in breast cancer (21).
By contrast, reduced expression of YY1 has been reported in types
of melanoma, urothelial carcinomas and osteosarcomas (21). YY1 is constitutively elevated
during the progression of brain gliomas compared with that of
normal brain tissues, which shows a positive correlation with the
progression of gliomas and meningiomas (22). However, at present there is no
clear explanation about the role and regulated miRNAs of YY1 in
gliomas. The current study aimed to elucidate the role of YY1 and
miR-218 in human glioma cells and determine their regulatory
association. It was demonstrated that YY1 was a promoting factor in
glioma cell proliferation and miR-218 could inhibit glioma cell
proliferation by targeting YY1.
Materials and methods
Antibodies
YY1 antibodies were obtained from Santa Cruz (Santa
Cruz Biotechnology, Inc., Dallas, TX, USA). Antibodies specific for
p53 (cat. no. 2524) and β-actin (cat. no. 3700) were obtained from
Cell Signaling Technology, Inc. (Danvers, MA, USA). Goat anti-mouse
IgG (cat. no. AP124B) and goat anti-rabbit IgG (cat. no. AP307P)
were purchased from EMD Millipore (Billerica, MA, USA).
Cell culture
Human glioma cells U251MG and 293T cells were
obtained from the Cell Bank of Shanghai Institutes of Chinese
Academy of Sciences (Shanghai, China). Cells were grown at 37°C
with 5% CO2 in DMEM medium (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) containing 10% heat-inactivated
fetal bovine serum (Evergreen Biological Engineering, Hangzhou,
China).
miR-218 target prediction
The miR-218 target prediction was performed using
bioinformatics algorithms from TargetScan (http://www.targetscan.org/) and PicTar (http://pictar.mdc-berlin.de/).
Constructs and production of
lentivirus
For overexpression of miR-218, the miR-218 cDNA was
inserted into the pGlV3/H1 plasmid (Shanghai GenePharma Co., Ltd.,
Shanghai, China) using BamHI and MluI sites. For
silencing of miR-218, the short-hairpin RNA (shRNA) oligomer
(target sequence: GAT CCA CAT GGT TAG ATC AAG CAC AAC GAT ACA TGG
TTA GAT CAA GCA CAA ACC GGT ACA TGG TTA GAT CAA GCA CAA TCA CAC ATG
GTT AGA TCA A GC ACA ATT TTT TG) was annealed and then subcloned
into the pGLV3/H1 plasmid by BamHI and MluI cloning
sites. For knockdown of endogenous YY1, a specific shRNA oligomer
(target sequence: CCT CCT GAT TAT TCA GAA TAT) was annealed and
then introduced into the pLV-shRNA plasmid by BamHI and
EcoRI cloning sites (23).
For overexpression of YY1, the YY1 cDNA was inserted into the 3X
Flag plasmid using EcoRI and XbaI sites. 3X Flag-p53
were obtained from Addgene, Inc. (Cambridge, MA, USA). Cells were
transfected with Polyjet (SignaGen Laboratories, Gaithersburg, MD,
USA) according to the manufacturer's protocol. The viruses were
packaged in 293T cells by cotransfecting the corresponding plasmids
with the helper plasmids using Polyjet transfection reagent.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
RNA was extracted from stable lines using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) following manufacturers' instructions and 1 µg total RNA was
subjected for cDNA production using reverse transcription reagents
(Roche Diagnostics GmbH, Basel, Switzerland) according to the
manufacturer's protocol. RT-qPCR reactions were performed with 3 µl
template from each reaction with an ABI 7300 Real-Time PCR
instrument (Applied Biosystems; Thermo Fisher Scientific, Inc.)
using SYBR Green (Roche, Diagnostics GmbH). Primers for the
amplification of YY1 and β-actin were as follows: YY1, forward CCT
GGC ATT GAC CTC TCA GAT CCA and reverse GGG CAA GCT ATT GTT CTT GGA
GCA; β-actin, forward CAT GTA CGT TGC TAT CCA GGC and reverse CGC
TCG GTG AGG ATC TTC ATG. The products for YY1 and β-actin were 101
and 195 bp, respectively. PCR reaction mixture (10 µM forward and
reverse primers, 10 µl of LightCycler 480 SYBR Green I Master
(Roche, Diagnostics GmbH), 3 µl of cDNA template and 6 µl of
RNA-free water) was added into the reaction plate, and then run by
PCR instrument. The following thermocycling conditions were used
for the PCR: Initial denaturation at 95°C for 5 min; followed by 40
cycles of 95°C for 15 sec and 65°C for 35 sec. For each sample, the
Cquantification cycle (Cq) was determined and normalized
to the average of the housekeeping gene (ΔCq=Cq Unknown-Cq
Housekeeping gene). The determination of gene transcript levels in
each sample was performed using the 2−ΔΔCq method
(24).
Luciferase reporter assay
The YY1 3′untranslated region (UTR)-Luc reporter was
created by ligation of the YY1 3′UTR PCR product into the
SacI and XbaI site of the pmirGLO control vector
(Shanghai GenePharma Co., Ltd., Shanghai, China). The mutant
reporter was generated from pmirGLO-wild-type (WT)-YY1 3′UTR-Luc by
deleting the binding site for miR-218 ‘UGAAUGU’. miR-21
promoter-containing (miPPR-21-containing) pmirGLO-basic plasmids
were constructed (Shanghai GenePharma Co., Ltd.). Luciferase
activity was measured using the Dual-Luciferase Reporter Assay
System (Promega Corporation, Madison, WI, USA).
EdU assay
The proliferation of cells was evaluated by
5-ethynyl-2′-deoxyuridine (EdU) incorporation assay using an EdU
assay kit (Guangzhou RiboBio Co., Ltd., China) according to the
manufacturer's protocol. U251MG cell (1×104 cells per
well) were incubated in triplicate in a 96-well plate for 48 h and
then exposed to 50 µM EdU for additional 2 h at 37°C. Cells were
fixed with 4% formaldehyde for 30 min at 25°C and 2 mg/ml glycine
was added to neutralize the formaldehyde. Cells were then incubated
with 0.5% Triton X-100 for 15 min at 25°C for permeabilization.
After washing with PBS three times, the cells were treated with 100
µl 1X Apollo Reaction Cocktail (Guangzhou RiboBio Co., Ltd.) for 30
min. DNA was stained by Hoechst staining for 30 min to determine
total number of cells and calculate percentage proliferation. The
images were photographed under the Olympus IX-71 inverted
microscope (Olympus Corporation, Tokyo, Japan).
Cell Counting Kit-8 (CCK-8) assay
A total of 5,000 cells in 100 µl DMEM medium were
plated in 96-well plates and grown under normal conditions. After
cells had adhered to the plates,
2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-benzene
disulfonate)-2H-tetrazolium monosodium salt (AAT Bioquest, Inc.,
Sunnyvale, CA, USA) was added into the medium and cells were
incubated for 4 h at 37°C. Living cells were counted daily by
reading the absorbance at 450 nm using SynergyMx Multi-Mode
Microplate Reader (Biotek Instruments, Inc., Winooski, VT,
USA).
Colony formation assays
A total of 200 cells were plated in a 6-cm dish and
incubated under normal conditions for 10 days. The cells were fixed
with methanol and dyed with 0.05% crystal violet to assess colony
staining. Following repeated washing with PBS, images were taken
with a camera. Colonies containing more than 50 cells were
counted.
Western blotting
Cells were lysed and equal amounts of cell lysates
were subjected to 10% SDS-PAGE and then transferred onto
polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA,
USA). Subsequent to blocking with 5% skimmed milk, the membrane was
incubated with primary antibodies [YY1 (1:1,000), β-actin
(1:1,000)] at 4°C overnight and then secondary antibodies (1:5,000)
at room temperature for 1 h. Bound antibodies were detected by the
enhanced chemiluminescence Q6 system (Amersham Pharmacia Biotech,
Piscataway, NJ, USA). Band densities were measured using ImageJ
software 1.42q (National Institutes of Health, Bethesda, MD, USA).
All examined gene expression levels were determined by normalizing
the densitometry value of interest to that of the loading
control.
Statistical analysis
SPSS software was used to perform statistical
analyses. Data were presented as the mean ± standard error.
Statistical analyses were performed using SPSS, version 13.0 (SPSS,
Inc., Chicago, IL, USA). Differences in multiple groups were
determined by using one-way analysis of variance followed by
Student-Newman-Keuls test. Comparison between two groups was
performed by Student's t-test. P<0.05 was considered to indicate
a statistically significant difference.
Results
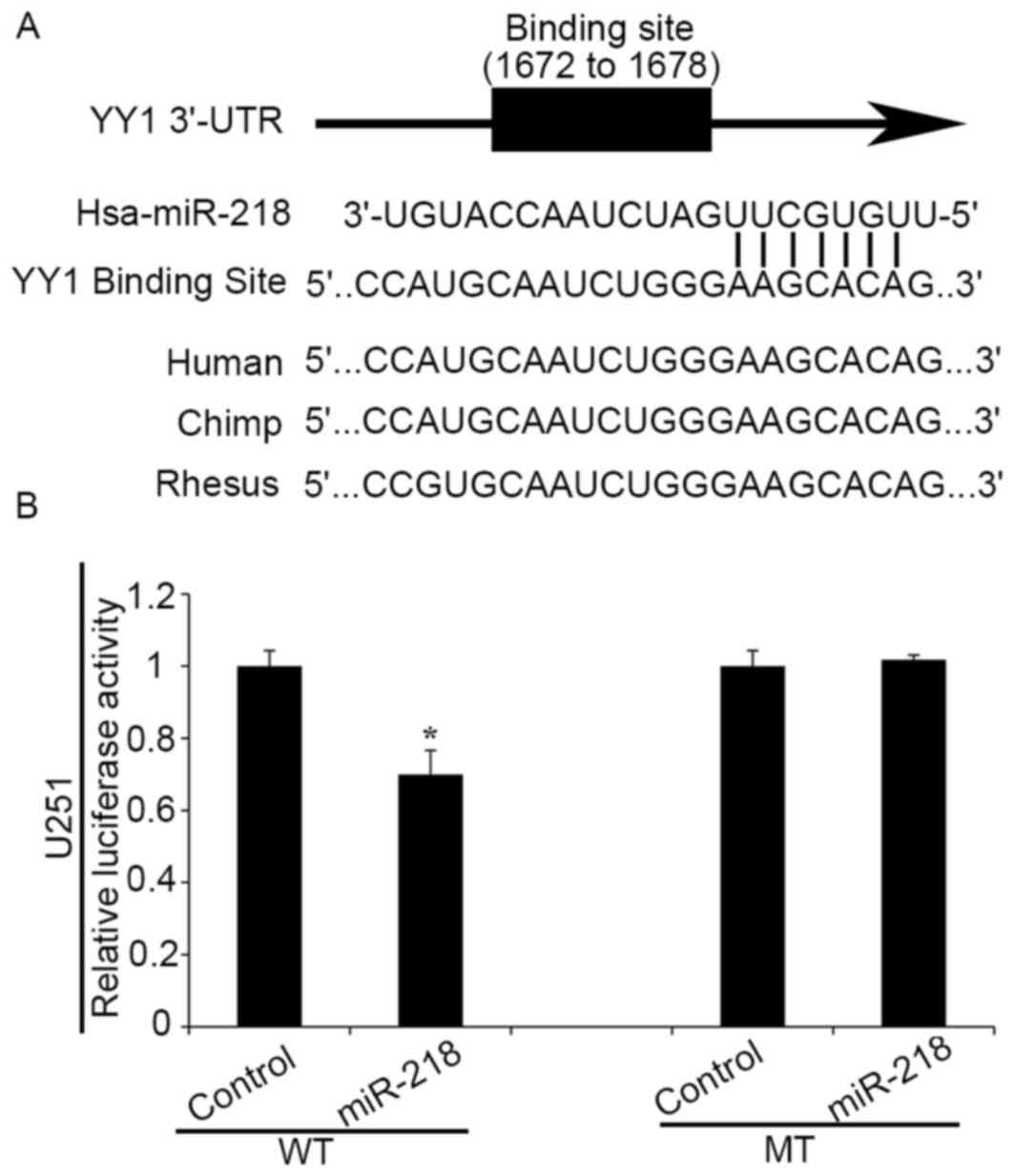
YY1 is a direct target of miR-218
Bioinformatics analysis using TargetScan and PicTar
algorithms indicated that YY1 is a hypothetical target gene of
miR-218 (Fig. 1A). To further
confirm the direct interaction between miR-218 and YY1 3′UTRs,
plasmids containing WT (pmirGLO-WT-YY1 3′UTR) and mutant (MT)-type
(pmirGLO-MT-YY1 3′UTR) YY1 3′UTRs were constructed. Luciferase
reporter assays demonstrated that upregulation of miR-218 led to a
notable decrease of luciferase activity of pmirGLO-WT-YY1 3′UTR in
human glioma cells, without any significant change in luciferase
activity of pmirGLO-MT-YY1 3′UTR (Fig.
1B). These results suggest that YY1 may be a direct target of
miR-218 in human glioma cells.
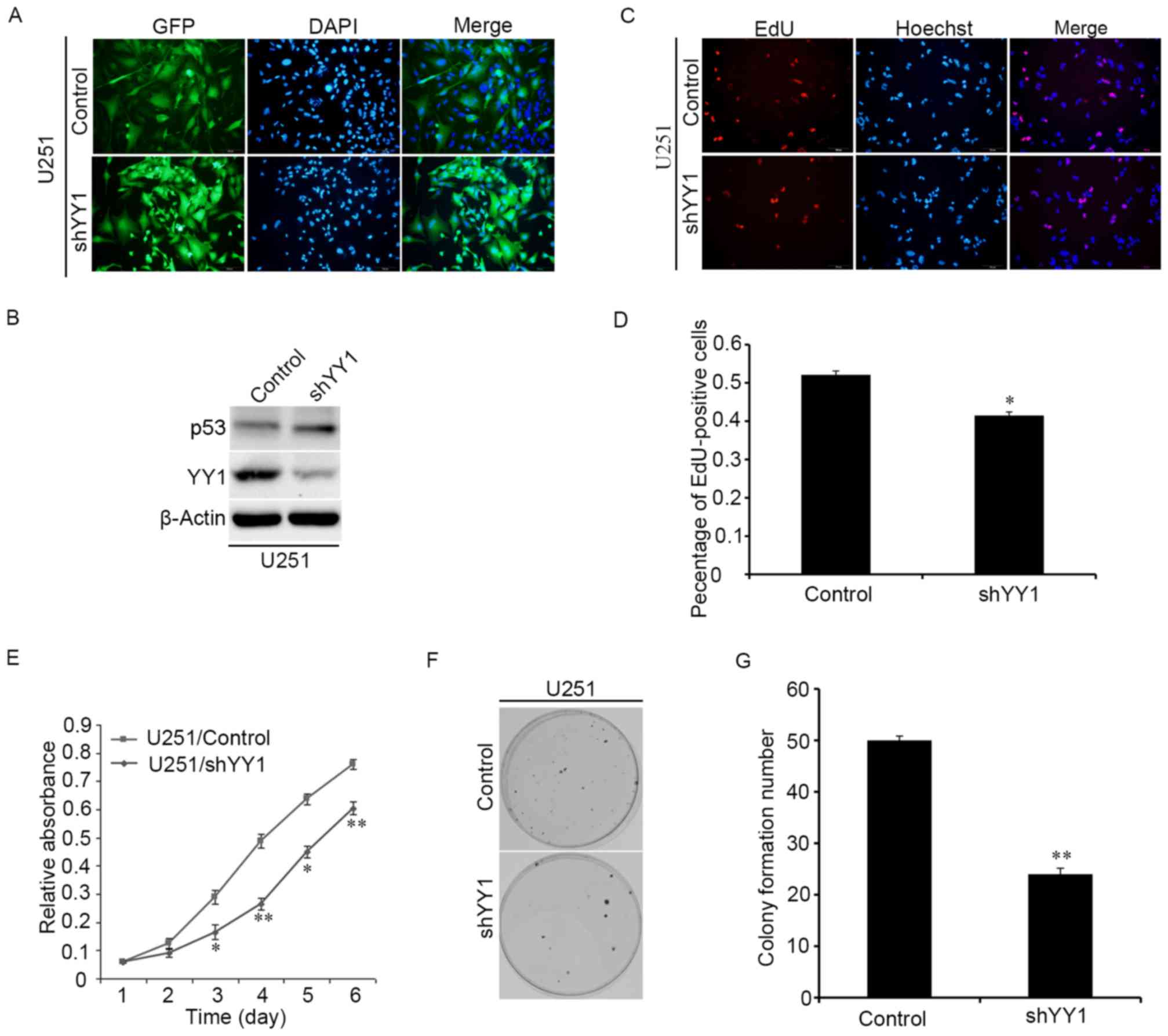
YY1 promotes the proliferation of
glioma cells
Whether YY1 participates in the proliferation of
human glioma cells was investigated in vitro by
overexpressing shRNA against YY1 to downregulate YY1 expression.
The YY1-downregulated stable cell line was estabilshed by
lentiviral infection and the infection efficiency was verified
through the immunofluorescence images (Fig. 2A). The western blotting assay
indicated that knockdown of YY1 expression could increase the
expressions of p53 protein (Fig.
2B). Proliferation of glioma cells was examined by EdU, CCK-8
and colony formation experiments. Knocking down YY1 significantly
decreased the proportion of EdU positive cells (Fig. 2C and D). CCK-8 experiments
demonstrated that knocking down YY1 reduced the proliferation of
glioma cells (Fig. 2E). The number
of colonies was also reduced upon downregulating YY1 (Fig. 2F and G). Therefore, it was
concluded that knocking down YY1 reduces the proliferation of
glioma cells.
Downregulation of miR-218 promotes the
proliferation of human glioma cells
Several studies have demonstrated that miR-218 has
tumor-suppressive functions in human glioma (14,15,18).
However, the role of miR-218 in glioma remains unclear. Thus, the
current study aimed to investigate the effects of miR-218 on
proliferation by regulating YY1 in human glioma cells. miR-218
expression was first knocked down with an miRNA sponge specific for
miR-218 (miR-218 sponge). Stable cell lines expressing the miR-218
sponge were constructed in U251MG cell by lentiviral infection
(Fig. 3A). The infection
efficiency of miR-218 was performed by monitoring the mRNA
expression of YY1 (Fig. 3B). CCK-8
experiments demonstrated that knocking down miR-218 accelerated
cell proliferation (Fig. 3C).
Furthermore, colony formation ability was increased upon
downregulation of miR-218 (Fig. 3D and
E). Taken together, it was concluded that knocking down miR-218
induced the promotion of proliferation in glioma cells.
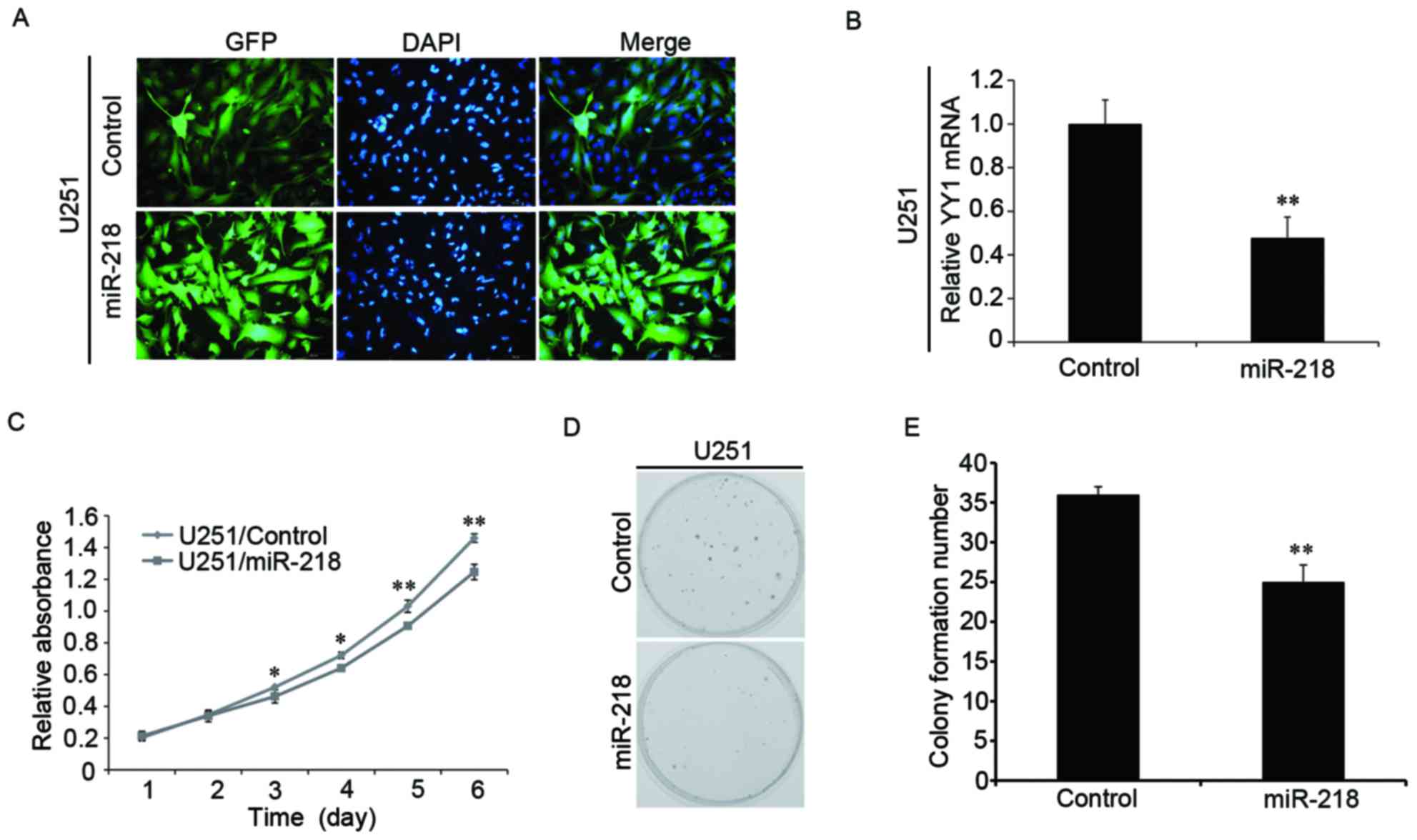
Overexpression of miR-218 inhibits the
proliferation of glioma cells
To further explore the biological function of
miR-218 in glioma cells, miR-218 was upregulated by lentiviral
infection (Fig. 4A). The
expression of YY1 mRNA was verified through RT-qPCR experiments and
was observed to be significantly decreased in response to
overexpressed miR-218 (Fig. 4B).
Overexpression of miR-218 led to inhibition of glioma cell
proliferation, as determined by CCK-8 (Fig. 4C) and colony formation experiments
(Fig. 4D and E). Therefore, it was
concluded that upregulating miR-218 inhibited the proliferation of
human glioma cells.
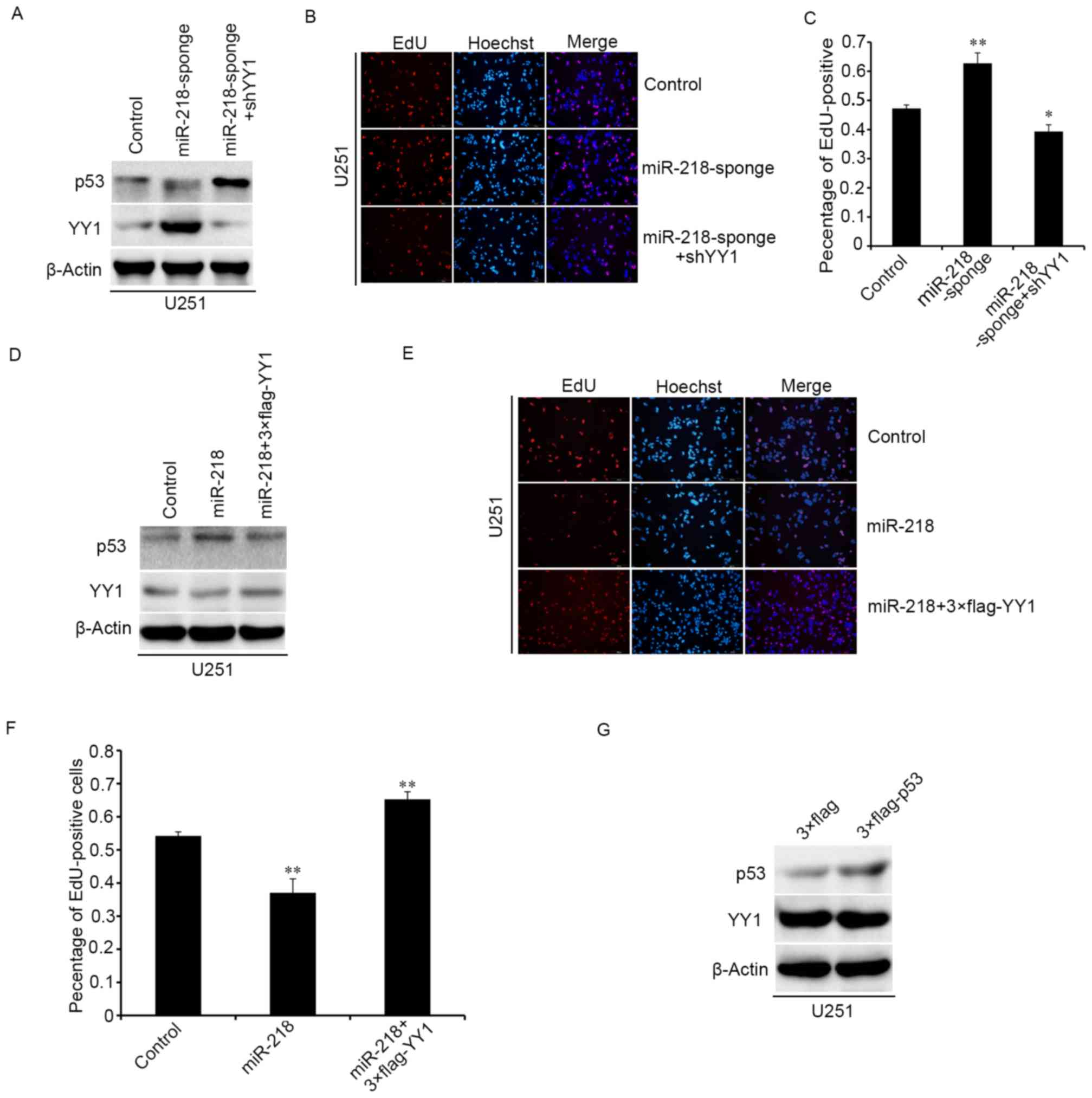
miR-218 inhibits the proliferation of
glioma cells through the YY1/p53 pathway
To clarify the potential mechanism of miR-218 in
glioma cells, the YY1 and p53 expression in sponge cells
overexpressing miR-218 were detected. Western blot analysis
demonstrated that downregulation of miR-218 upregulated the protein
level of YY1, promoted the degradation of p53, and when we
cotransfected the pLV-shYY1 and miR-218-sponge plasmids into
U251MG, the expression levels of p53 were restored compared to
upregulating miR-218-sponge (Fig.
5A). The results of EdU experiments and western blot analysis
were consistent, and pLV-shYY1 could reverse the effect of
miR-218-sponge in cell proliferation (Fig. 5B and C). Upregulating 3X Flag-YY1
and miR-218 led to similar results and the same protein expression
trend with overexpressing pLV-shYY1 and miR-218-sponge, as
determined by western blotting and EdU (Fig. 5D-F). However, when 3X Flag-p53 was
overexpressed in U251MG cell, the expression of YY1 was not
detected to be significantly different (Fig. 5G). Therefore, it was inferred that
inhibition of glioma cell proliferation by overexpressing miR-218
in glioma cells may be mediated by downregulation of YY1 and
upregulation of p53 expression which may be due to the reduction of
p53 degradation by YY1.
Together the data indicated that YY1 is a direct
target of miR-218 and serves a role in promoting the proliferation
of glioma cells. The results suggest that miR-218 suppresses the
proliferation of human glioma cells through the YY1/p53
pathway.
Discussion
The current study demonstrated that YY1 is a direct
target of miR-218. To explore the exact role of miR-218 in glioma
cells, miR-218 was overexpressed and silenced in human glioma cells
by establishing stable cell lines using lentivirus. Silencing of
miR-218 significantly promoted glioma cell proliferation by
activating the YY1/p53 pathway, whereas overexpression of miR-218
inhibited glioma cell proliferation by suppressing the YY1/p53
pathway. It was concluded that post-transcriptional regulation of
YY1 acts via miR-218 and that YY1/p53 signaling is an important
mediator of the effects of miR-218 in glioma.
Previous studies have indicated that miR-218 is
dramatically downregulated in human gliomas compared with normal
brain tissues (14,25,26).
miR-218 has frequently been reported as a direct target of Bmi1
(15), IKK-β (14), RTK (27), and CDK6 (16,19)
to prevent the proliferation, migration, invasion of glioma cells.
To identify more mRNA targets of miR-218 in glioma cells,
bioinformatics analysis was performed and it was identified that
YY1 was a potential target of miR-218. The present study identified
that miR-218 inhibited glioma cell proliferation by indirectly
regulating p53 expression via directly targeting YY1. It is
suggested that the miR-218-mediated inhibition of YY1/p53 signaling
pathway may be a promising method of treating glioma.
The transcription factor YY1 has been identified as
a potential novel prognostic biomarker and therapeutic target. The
role of YY1 in cancer is mediated by regulating several proteins in
cancer development and progression including c-myc, c-fos, Erb-B2
receptor tyrosine kinase 2, and p53 (20,28).
YY1 also interacts with numerous molecules that modulate cell
proliferation and apoptosis, including p53, mouse double minute 2
homolog, enhancer of zeste homolog 2, caspases and histone
deacetylases (20). YY1 is an
important negative regulator of the tumor suppressor gene p53.
Because of this, the level of p53 expression was detected. The
current study exhibited that downregulation of YY1 inhibits the
proliferation of glioma cells by increasing p53 expression. The
data indicated that YY1 functions as a growth promoter of glioma
cells by inhibiting tumor suppressor p53.
Taken together, the results suggested that miR-218
serves an important role in preventing the proliferation of glioma
cells, and the results present a novel mechanism of
miR-218-mediated direct suppression of the YY1/p53 pathway in
glioma.
Acknowledgements
The present study was supported by the Foundation of
Jiangsu Provincial Health Department (grant no. YG201514).
References
|
1
|
Davis FG and McCarthy BJ: Current
epidemiological trends and surveillance issues in brain tumors.
Expert Rev Anticancer Ther. 1:395–401. 2001. View Article : Google Scholar
|
|
2
|
Buonerba C, Di Lorenzo G, Marinelli A,
Federico P, Palmieri G, Imbimbo M, Conti P, Peluso G, De Placido S
and Sampson JH: A comprehensive outlook on intracerebral therapy of
malignant gliomas. Crit Rev Oncol Hematol. 80:54–68. 2011.
View Article : Google Scholar
|
|
3
|
Sherman JH, Hoes K, Marcus J, Komotar RJ,
Brennan CW and Gutin PH: Neurosurgery for brain tumors: Update on
recent technical advances. Curr Neurol Neurosci Rep. 11:313–319.
2011. View Article : Google Scholar
|
|
4
|
Castro MG, Candolfi M, Kroeger K, King GD,
Curtin JF, Yagiz K, Mineharu Y, Assi H, Wibowo M, Muhammad Ghulam
AK, et al: Gene therapy and targeted toxins for glioma. Curr Gene
Ther. 11:155–180. 2011. View Article : Google Scholar :
|
|
5
|
Hwang HW and Mendell JT: MicroRNAs in cell
proliferation, cell death, and tumorigenesis. Br J Cancer.
94:776–780. 2006. View Article : Google Scholar :
|
|
6
|
Gabriely G, Wurdinger T, Kesari S, Esau
CC, Burchard J, Linsley PS and Krichevsky AM: MicroRNA 21 promotes
glioma invasion by targeting matrix metalloproteinase regulators.
Mol Cell Biol. 28:5369–5380. 2008. View Article : Google Scholar :
|
|
7
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar
|
|
8
|
Yang L, Li Q, Wang Q, Jiang Z and Zhang L:
Silencing of miRNA-218 promotes migration and invasion of breast
cancer via Slit2-Robo1 pathway. Biomed Pharmacother. 66:535–540.
2012. View Article : Google Scholar
|
|
9
|
Wang H, Li XT, Wu C, Wu ZW, Li YY, Yang
TQ, Chen GL, Xie XS, Huang YL, Du ZW and Zhou YX: miR-132 can
inhibit glioma cells invasion and migration by target MMP16 in
vitro. Onco Targets Ther. 8:3211–3218. 2015.
|
|
10
|
Liu H, Song Z, Liao D, Zhang T, Liu F,
Zheng W, Luo K and Yang L: miR-503 inhibits cell proliferation and
invasion in glioma by targeting L1CAM. Int J Clin Exp Med.
8:18441–18447. 2015.
|
|
11
|
Li Z, Liu YH, Diao HY, Ma J and Yao YL:
miR-661 inhibits glioma cell proliferation, migration and invasion
by targeting hTERT. Biochem Biophys Res Commun. 468:870–876. 2015.
View Article : Google Scholar
|
|
12
|
Han J and Chen Q: miR-16 modulate
temozolomide resistance by regulating BCL-2 in human glioma cells.
Int J Clin Exp Pathol. 8:12698–12707. 2015.
|
|
13
|
Costa PM, Cardoso AL, Custódia C, Cunha P,
de Almeida Pereira L and de Lima Pedroso MC: miRNA-21 silencing
mediated by tumor-targeted nanoparticles combined with sunitinib: A
new multimodal gene therapy approach for glioblastoma. J Control
Release. 207:31–39. 2015. View Article : Google Scholar
|
|
14
|
Song L, Huang Q, Chen K, Liu L, Lin C, Dai
T, Yu C, Wu Z and Li J: miR-218 inhibits the invasive ability of
glioma cells by direct downregulation of IKK-β. Biochem Biophys Res
Commun. 402:135–140. 2010. View Article : Google Scholar
|
|
15
|
Tu Y, Gao X, Li G, Fu H, Cui D, Liu H, Jin
W and Zhang Y: MicroRNA-218 inhibits glioma invasion, migration,
proliferation, and cancer stem-like cell self-renewal by targeting
the polycomb group gene Bmi1. Cancer Res. 73:6046–6055. 2013.
View Article : Google Scholar
|
|
16
|
Jun GJ, Zhong GG and Ming ZS: miR-218
inhibits the proliferation of glioma U87 cells through the
inactivation of the CDK6/cyclin D1/p21 pathway. Oncol Lett.
9:2743–2749. 2015.
|
|
17
|
Liu Y, Yan W, Zhang W, Chen L, You G, Bao
Z, Wang Y, Wang H, Kang C and Jiang T: miR-218 reverses high
invasiveness of glioblastoma cells by targeting the oncogenic
transcription factor LEF1. Oncol Rep. 28:1013–1021. 2012.
View Article : Google Scholar
|
|
18
|
Xia H, Yan Y, Hu M, Wang Y, Wang Y, Dai Y,
Chen J, Di G, Chen X and Jiang X: miR-218 sensitizes glioma cells
to apoptosis and inhibits tumorigenicity by regulating
ECOP-mediated suppression of NF-κB activity. Neuro Oncol.
15:413–422. 2013. View Article : Google Scholar
|
|
19
|
Zhang JM, Sun CY, Yu SZ, Wang Q, An TL, Li
YY, Kong YL and Wen YJ: Relationship between miR-218 and CDK6
expression and their biological impact on glioma cell proliferation
and apoptosis. Zhonghua Bing Li Xue Za Zhi. 40:454–459. 2011.(In
Chinese).
|
|
20
|
Gordon S, Akopyan G, Garban H and Bonavida
B: Transcription factor YY1: Structure, function, and therapeutic
implications in cancer biology. Oncogene. 25:1125–1142. 2006.
View Article : Google Scholar
|
|
21
|
Kashyap V and Bonavida B: Role of YY1 in
the pathogenesis of prostate cancer and correlation with
bioinformatic data sets of gene expression. Genes Cancer. 5:71–83.
2014.
|
|
22
|
Baritaki S, Chatzinikola AM, Vakis AF,
Soulitzis N, Karabetsos DA, Neonakis I, Bonavida B and Spandidos
DA: YY1 Over-expression in human brain gliomas and meningiomas
correlates with TGF-beta1, IGF-1 and FGF-2 mRNA levels. Cancer
Invest. 27:184–192. 2009. View Article : Google Scholar
|
|
23
|
Liao WR, Hsieh RH, Hsu KW, Wu MZ, Tseng
MJ, Mai RT, Lee Wu YH and Yeh TS: The CBF1-independent Notch1
signal pathway activates human c-myc expression partially via
transcription factor YY1. Carcinogenesis. 28:1867–1876. 2007.
View Article : Google Scholar
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
25
|
Setty M, Helmy K, Khan AA, Silber J, Arvey
A, Neezen F, Agius P, Huse JT, Holland EC and Leslie CS: Inferring
transcriptional and microRNA-mediated regulatory programs in
glioblastoma. Mol Syst Biol. 8:6052012. View Article : Google Scholar :
|
|
26
|
Skalsky RL and Cullen BR: Reduced
expression of brain-enriched microRNAs in glioblastomas permits
targeted regulation of a cell death gene. PLoS One. 6:e242482011.
View Article : Google Scholar :
|
|
27
|
Mathew LK, Skuli N, Mucaj V, Lee SS, Zinn
PO, Sathyan P, Imtiyaz HZ, Zhang Z, Davuluri RV, Rao S, et al:
miR-218 opposes a critical RTK-HIF pathway in mesenchymal
glioblastoma. Proc Natl Acad Sci USA. 111:291–296. 2014. View Article : Google Scholar
|
|
28
|
Sui G, el Affar B and Shi Y, Brignone C,
Wall NR, Yin P, Donohoe M, Luke MP, Calvo D, Grossman SR and Shi Y:
Yin Yang 1 is a negative regulator of p53. Cell. 117:859–872. 2004.
View Article : Google Scholar
|