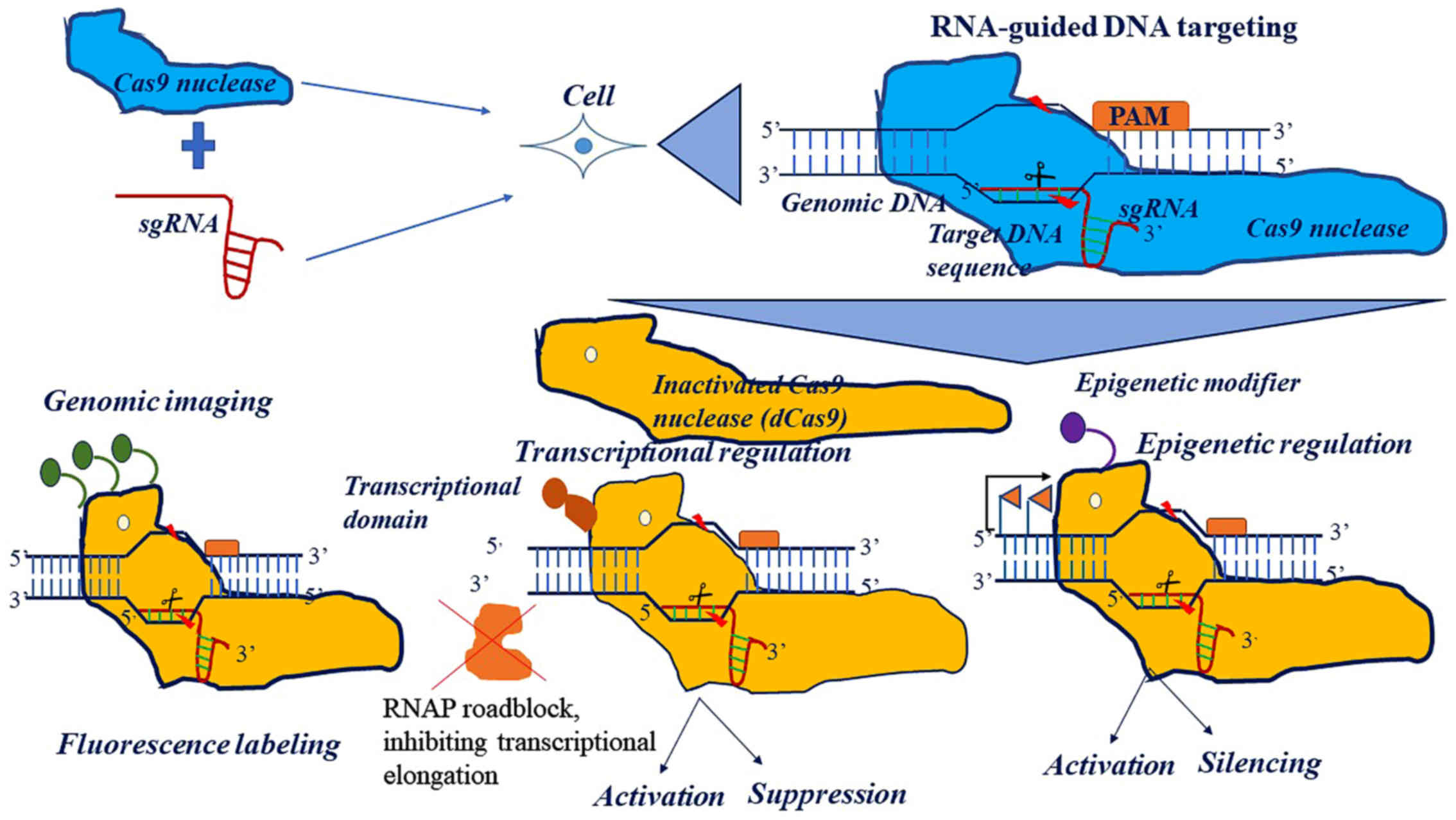
|
1
|
Wright AV, Nuñez JK and Doudna JA: Biology
and applications of CRISPR systems: Harnessing nature's toolbox for
genome engineering. Cell. 164:29–44. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wiedenheft B, Sternberg SH and Doudna JA:
RNA-guided genetic silencing systems in bacteria and archaea.
Nature. 482:331–338. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
van der Oost J, Westra ER, Jackson RN and
Wiedenheft B: Unravelling the structural and mechanistic basis of
CRISPR-cas systems. Nat Rev Microbiol. 12:479–492. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mougiakos I, Bosma EF, de Vos WM, van
Kranenburg R and van der Oost J: Next generation prokaryotic
engineering: The CRISPR-cas toolkit. Trends Biotechnol. 34:575–587.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bolotin A, Quinquis B, Sorokin A and
Ehrlich SD: Clustered regularly interspaced short palindrome
repeats (CRISPRs) have spacers of extrachromosomal origin.
Microbiology. 151:2551–2561. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Westra ER, Semenova E, Datsenko KA,
Jackson RN, Wiedenheft B, Severinov K and Brouns SJ: Type I-E
CRISPR-cas systems discriminate target from non-target DNA through
base pairing-independent PAM recognition. PLoS Genet.
9:e10037422013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mojica FJ, Díez-Villaseñor C,
García-Martínez J and Almendros C: Short motif sequences determine
the targets of the prokaryotic CRISPR defence system. Microbiology.
155:733–740. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jackson AL, Bartz SR, Schelter J,
Kobayashi SV, Burchard J, Mao M, Li B, Cavet G and Linsley PS:
Expression profiling reveals off-target gene regulation by RNAi.
Nat Biotechnol. 21:635–637. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Marine S, Bahl A, Ferrer M and Buehler E:
Common seed analysis to identify off-target effects in siRNA
screens. J Biomol Screen. 17:370–378. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Moore JD: The impact of CRISPR-Cas9 on
target identification and validation. Drug Discov Today.
20:450–457. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ran FA, Hsu PD, Wright J, Agarwala V,
Scott DA and Zhang F: Genome engineering using the CRISPR-Cas9
system. Nat Protoc. 8:2281–2308. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kuscu C, Arslan S, Singh R, Thorpe J and
Adli M: Genome-wide analysis reveals characteristics of off-target
sites bound by the Cas9 endonuclease. Nat Biotechnol. 32:677–683.
2014. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fu Y, Sander JD, Reyon D, Cascio VM and
Joung JK: Improving CRISPR-cas nuclease specificity using truncated
guide RNAs. Nat Biotechnol. 32:279–284. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Qi LS, Larson MH, Gilbert LA, Doudna JA,
Weissman JS, Arkin AP and Lim WA: Repurposing CRISPR as an
RNA-guided platform for sequence-specific control of gene
expression. Cell. 152:1173–1183. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Boettcher M and McManus MT: Choosing the
right tool for the job: RNAi, TALEN, or CRISPR. Mol Cell.
58:575–585. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lawhorn IE, Ferreira JP and Wang CL:
Evaluation of sgRNA target sites for CRISPR-mediated repression of
TP53. PLoS One. 9:e1132322014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gilbert LA, Larson MH, Morsut L, Liu Z,
Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH,
Doudna JA, et al: CRISPR-mediated modular RNA-guided regulation of
transcription in eukaryotes. Cell. 154:442–451. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Konermann S, Brigham MD, Trevino A, Hsu
PD, Heidenreich M, Cong L, Platt RJ, Scott DA, Church GM and Zhang
F: Optical control of mammalian endogenous transcription and
epigenetic states. Nature. 500:472–476. 2013.PubMed/NCBI
|
|
19
|
Mandegar MA, Huebsch N, Frolov EB, Shin E,
Truong A, Olvera MP, Chan AH, Miyaoka Y, Holmes K, Spencer CI, et
al: CRISPR interference efficiently induces specific and reversible
gene silencing in human iPSCs. Cell Stem Cell. 18:541–553. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Larson MH, Gilbert LA, Wang X, Lim WA,
Weissman JS and Qi LS: CRISPR interference (CRISPRi) for
sequence-specific control of gene expression. Nat Protoc.
8:2180–2196. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho
QH and Joung JK: CRISPR RNA-guided activation of endogenous human
genes. Nat Methods. 10:977–979. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mali P, Aach J, Stranges PB, Esvelt KM,
Moosburner M, Kosuri S, Yang L and Church GM: CAS9 transcriptional
activators for target specificity screening and paired nickases for
cooperative genome engineering. Nat Biotechnol. 31:833–838. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Perez-Pinera P, Kocak DD, Vockley CM,
Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout
DG, Leong KW, et al: RNA-guided gene activation by
CRISPR-Cas9-based transcription factors. Nat Methods. 10:973–976.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tanenbaum ME, Gilbert LA, Qi LS, Weissman
JS and Vale RD: A protein-tagging system for signal amplification
in gene expression and fluorescence imaging. Cell. 159:635–646.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
La Russa MF and Qi LS: The new state of
the art: Cas9 for gene activation and repression. Mol Cell Biol.
35:3800–3809. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Konermann S, Brigham MD, Trevino AE, Joung
J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS,
Nishimasu H, et al: Genome-scale transcriptional activation by an
engineered CRISPR-Cas9 complex. Nature. 517:583–588. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chavez A, Scheiman J, Vora S, Pruitt BW,
Tuttle MPR, Iyer E, Lin S, Kiani S, Guzman CD, Wiegand DJ, et al:
Highly efficient Cas9-mediated transcriptional programming. Nat
Methods. 12:326–328. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zalatan JG, Lee ME, Almeida R, Gilbert LA,
Whitehead EH, La Russa M, Tsai JC, Weissman JS, Dueber JE, Qi LS
and Lim WA: Engineering complex synthetic transcriptional programs
with CRISPR RNA scaffolds. Cell. 160:339–350. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Braun CJ, Bruno PM, Horlbeck MA, Gilbert
LA, Weissman JS and Hemann MT: Versatile in vivo regulation of
tumor phenotypes by dCas9-mediated transcriptional perturbation.
Proc Natl Acad Sci USA. 113:E3892–E3900. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mansour MR, Abraham BJ, Anders L,
Berezovskaya A, Gutierrez A, Durbin AD, Etchin J, Lawton L, Sallan
SE, Silverman LB, et al: Oncogene regulation. An oncogenic
super-enhancer formed through somatic mutation of a noncoding
intergenic element. Science. 346:1373–1377. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Inoue F, Kircher M, Martin B, Cooper GM,
Witten DM, McManus MT, Ahituv N and Shendure J: A systematic
comparison reveals substantial differences in chromosomal versus
episomal encoding of enhancer activity. Genome Res. 27:38–52. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xie S, Duan J, Li B, Zhou P and Hon GC:
Multiplexed engineering and analysis of combinatorial enhancer
activity in single cells. Molecular cell. 66:285–99. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chiba K, Johnson JZ, Vogan JM, Wagner T,
Boyle JM and Hockemeyer D: Cancer-associated TERT promoter
mutations abrogate telomerase silencing. eLife. 4:42015. View Article : Google Scholar
|
|
34
|
Polstein LR and Gersbach CA: A
light-inducible CRISPR-Cas9 system for control of endogenous gene
activation. Nat Chem Biol. 11:198–200. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hemphill J, Borchardt EK, Brown K, Asokan
A and Deiters A: Optical Control of CRISPR/Cas9 gene editing. J Am
Chem Soc. 137:5642–5645. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nihongaki Y, Kawano F, Nakajima T and Sato
M: Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat
Biotechnol. 33:755–760. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu KI, Ramli MN, Woo CW, Wang Y, Zhao T,
Zhang X, Yim GR, Chong BY, Gowher A, Chua MZ, et al: A
chemical-inducible CRISPR-Cas9 system for rapid control of genome
editing. Nat Chem Biol. 12:980–987. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zetsche B, Volz SE and Zhang F: A
split-Cas9 architecture for inducible genome editing and
transcription modulation. Nat Biotechnol. 33:139–142. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gilbert LA, Horlbeck MA, Adamson B,
Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh
HL, Bassik MC, et al: Genome-scale CRISPR-mediated control of gene
repression and activation. Cell. 159:647–661. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Haemmerle M and Gutschner T: Long
non-coding RNAs in cancer and development: Where do we go from
here? Int J Mol Sci. 16:1395–1405. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hale CR, Zhao P, Olson S, Duff MO,
Graveley BR, Wells L, Terns RM and Terns MP: RNA-guided RNA
cleavage by a CRISPR RNA-Cas protein complex. Cell. 139:945–956.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gutschner T: Silencing long noncoding RNAs
with genome-editing tools. Methods Mol Biol. 1239:241–250. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sauvageau M, Goff LA, Lodato S, Bonev B,
Groff AF, Gerhardinger C, Sanchez-Gomez DB, Hacisuleyman E, Li E,
Spence M, et al: Multiple knockout mouse models reveal lincRNAs are
required for life and brain development. eLife. 2:e017492013.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yin Y, Yan P, Lu J, Song G, Zhu Y, Li Z,
Zhao Y, Shen B, Huang X, Zhu H, et al: Opposing roles for the
lncRNA haunt and its genomic locus in regulating HOXA gene
activation during embryonic stem cell differentiation. Cell Stem
Cell. 16:504–516. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Xiao A, Wang Z, Hu Y, Wu Y, Luo Z, Yang Z,
Zu Y, Li W, Huang P, Tong X, et al: Chromosomal deletions and
inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic
Acids Res. 41:e1412013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Li X, Chen W, Zeng W, Wan C, Duan S and
Jiang S: microRNA-137 promotes apoptosis in ovarian cancer cells
via the regulation of XIAP. Br J Cancer. 116:66–76. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhen S, Hua L, Liu YH, Sun XM, Jiang MM,
Chen W, Zhao L and Li X: Inhibition of long non-coding RNA UCA1 by
CRISPR/Cas9 attenuated malignant phenotypes of bladder cancer.
Oncotarget. 8:9634–9646. 2017.PubMed/NCBI
|
|
48
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Singh R, Gupta SC, Peng WX, Zhou N,
Pochampally R, Atfi A, Watabe K, Lu Z and Mo YY: Regulation of
alternative splicing of Bcl-x by BC200 contributes to breast cancer
pathogenesis. Cell Death Dis. 7:e22622016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yin Y, Zhong J, Li SW, Li JZ, Zhou M, Chen
Y, Sang Y and Liu L: TRIM11, a direct target of miR-24-3p, promotes
cell proliferation and inhibits apoptosis in colon cancer.
Oncotarget. 7:86755–86765. 2016.PubMed/NCBI
|
|
51
|
Cheng J, Roden CA, Pan W, Zhu S, Baccei A,
Pan X, Jiang T, Kluger Y, Weissman SM, Guo S, et al: A molecular
chipper technology for CRISPR sgRNA library generation and
functional mapping of noncoding regions. Nat Commun. 7:111782016.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Shechner DM, Hacisuleyman E, Younger ST
and Rinn JL: Multiplexable, locus-specific targeting of long RNAs
with CRISPR-Display. Nat Methods. 12:664–670. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Plummer RJ, Guo Y and Peng Y: A CRISPR
reimagining: New twists and turns of CRISPR beyond the
genome-engineering revolution. J Cell Biochem. 2017, https://doi.org/10.1002/jcb.26406 View Article : Google Scholar
|
|
54
|
Vojta A, Dobrinić P, Tadić V, Bočkor L,
Korać P, Julg B, Klasić M and Zoldoš V: Repurposing the CRISPR-Cas9
system for targeted DNA methylation. Nucleic Acids Res.
44:5615–5628. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Amabile A, Migliara A, Capasso P, Biffi M,
Cittaro D, Naldini L and Lombardo A: Inheritable silencing of
endogenous genes by hit-and-run targeted epigenetic editing. Cell.
167:219–232. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Liu XS, Wu H, Ji X, Stelzer Y, Wu X,
Czauderna S, Shu J, Dadon D, Young RA and Jaenisch R: Editing DNA
methylation in the mammalian genome. Cell. 167:233–247. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Hilton IB, D'Ippolito AM, Vockley CM,
Thakore PI, Crawford GE, Reddy TE and Gersbach CA: Epigenome
editing by a CRISPR-Cas9-based acetyltransferase activates genes
from promoters and enhancers. Nat Biotechnol. 33:510–517. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Pineda M, Moghadam F, Ebrahimkhani MR and
Kiani S: Engineered CRISPR systems for next generation gene
therapies. ACS Synth Biol. 6:1614–1626. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Beliveau BJ, Joyce EF, Apostolopoulos N,
Yilmaz F, Fonseka CY, McCole RB, Chang Y, Li JB, Senaratne TN,
Williams BR, et al: Versatile design and synthesis platform for
visualizing genomes with oligopaint FISH probes. Proc Natl Acad Sci
USA. 109:21301–21306. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Heun P, Laroche T, Shimada K, Furrer P and
Gasser SM: Chromosome dynamics in the yeast interphase nucleus.
Science. 294:2181–2186. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Chen B, Gilbert LA, Cimini BA,
Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS,
Qi LS and Huang B: Dynamic imaging of genomic loci in living human
cells by an optimized CRISPR/Cas system. Cell. 155:1479–1491. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Miyanari Y, Ziegler-Birling C and
Torres-Padilla ME: Live visualization of chromatin dynamics with
fluorescent TALEs. Nat Struct Mol Biol. 20:1321–1324. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Roukos V, Voss TC, Schmidt CK, Lee S,
Wangsa D and Misteli T: Spatial dynamics of chromosome
translocations in living cells. Science. 341:660–664. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
van Steensel B and Dekker J: Genomics
tools for unraveling chromosome architecture. Nat Biotechnol.
28:1089–1095. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Nelles DA, Fang MY, O'Connell MR, Xu JL,
Markmiller SJ, Doudna JA and Yeo GW: Programmable RNA tracking in
live cells with CRISPR/Cas9. Cell. 165:488–496. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Wang S, Su JH, Zhang F and Zhuang X: An
RNA-aptamer-based two-color CRISPR labeling system. Sci Rep.
6:268572016. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Jaitin DA, Weiner A, Yofe I, Lara-Astiaso
D, Keren-Shaul H, David E, Salame TM, Tanay A, van Oudenaarden A
and Amit I: Dissecting immune circuits by linking CRISPR-pooled
screens with single-Cell RNA-Seq. Cell. 167:1883–1896. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Guernet A, Mungamuri SK, Cartier D,
Sachidanandam R, Jayaprakash A, Adriouch S, Vezain M, Charbonnier
F, Rohkin G, Coutant S, et al: CRISPR-barcoding for intratumor
genetic heterogeneity modeling and functional analysis of oncogenic
driver mutations. Mol Cell. 63:526–538. 2016. View Article : Google Scholar : PubMed/NCBI
|